
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:FRMD3-BRD4 (FusionGDB2 ID:31316) |
Fusion Gene Summary for FRMD3-BRD4 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: FRMD3-BRD4 | Fusion gene ID: 31316 | Hgene | Tgene | Gene symbol | FRMD3 | BRD4 | Gene ID | 257019 | 23476 |
| Gene name | FERM domain containing 3 | bromodomain containing 4 | |
| Synonyms | 4.1O|EPB41L4O|EPB41LO|P410 | CAP|HUNK1|HUNKI|MCAP | |
| Cytomap | 9q21.32 | 19p13.12 | |
| Type of gene | protein-coding | protein-coding | |
| Description | FERM domain-containing protein 3band 4.1-like protein 4band 4.1-like protein 4Oovary type protein 4.1protein 4.1O | bromodomain-containing protein 4chromosome-associated proteinmitotic chromosome-associated protein | |
| Modification date | 20200313 | 20200329 | |
| UniProtAcc | A2A2Y4 | O60885 | |
| Ensembl transtripts involved in fusion gene | ENST00000304195, ENST00000376438, ENST00000328788, ENST00000376434, ENST00000465485, | ENST00000602230, ENST00000263377, ENST00000360016, ENST00000371835, | |
| Fusion gene scores | * DoF score | 5 X 5 X 5=125 | 15 X 19 X 13=3705 |
| # samples | 5 | 28 | |
| ** MAII score | log2(5/125*10)=-1.32192809488736 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(28/3705*10)=-3.72597481024823 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: FRMD3 [Title/Abstract] AND BRD4 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | FRMD3(85987828)-BRD4(15365073), # samples:2 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Tgene | BRD4 | GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter | 19103749|23086925 |
| Tgene | BRD4 | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling | 19103749 |
| Tgene | BRD4 | GO:0045944 | positive regulation of transcription by RNA polymerase II | 23086925|23317504|24360279 |
| Tgene | BRD4 | GO:0050727 | regulation of inflammatory response | 19103749 |
| Tgene | BRD4 | GO:1901407 | regulation of phosphorylation of RNA polymerase II C-terminal domain | 23086925 |
 Fusion gene breakpoints across FRMD3 (5'-gene) Fusion gene breakpoints across FRMD3 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
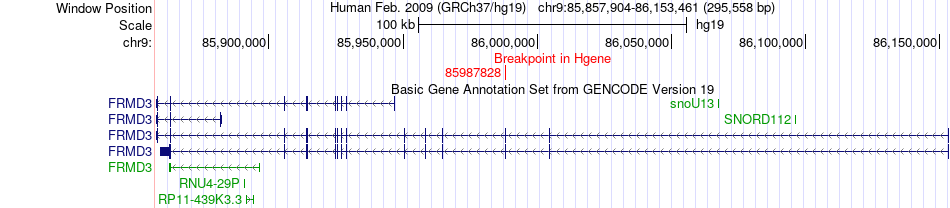 |
 Fusion gene breakpoints across BRD4 (3'-gene) Fusion gene breakpoints across BRD4 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
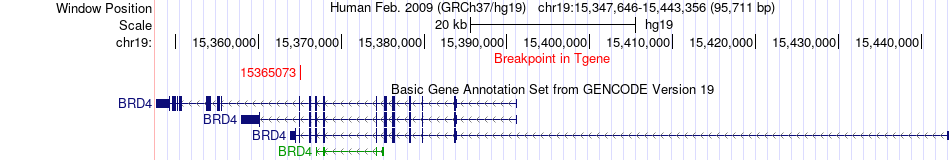 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | BRCA | TCGA-E2-A14N-01A | FRMD3 | chr9 | 85987828 | - | BRD4 | chr19 | 15365073 | - |
Top |
Fusion Gene ORF analysis for FRMD3-BRD4 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-intron | ENST00000304195 | ENST00000602230 | FRMD3 | chr9 | 85987828 | - | BRD4 | chr19 | 15365073 | - |
| 5CDS-intron | ENST00000376438 | ENST00000602230 | FRMD3 | chr9 | 85987828 | - | BRD4 | chr19 | 15365073 | - |
| In-frame | ENST00000304195 | ENST00000263377 | FRMD3 | chr9 | 85987828 | - | BRD4 | chr19 | 15365073 | - |
| In-frame | ENST00000304195 | ENST00000360016 | FRMD3 | chr9 | 85987828 | - | BRD4 | chr19 | 15365073 | - |
| In-frame | ENST00000304195 | ENST00000371835 | FRMD3 | chr9 | 85987828 | - | BRD4 | chr19 | 15365073 | - |
| In-frame | ENST00000376438 | ENST00000263377 | FRMD3 | chr9 | 85987828 | - | BRD4 | chr19 | 15365073 | - |
| In-frame | ENST00000376438 | ENST00000360016 | FRMD3 | chr9 | 85987828 | - | BRD4 | chr19 | 15365073 | - |
| In-frame | ENST00000376438 | ENST00000371835 | FRMD3 | chr9 | 85987828 | - | BRD4 | chr19 | 15365073 | - |
| intron-3CDS | ENST00000328788 | ENST00000263377 | FRMD3 | chr9 | 85987828 | - | BRD4 | chr19 | 15365073 | - |
| intron-3CDS | ENST00000328788 | ENST00000360016 | FRMD3 | chr9 | 85987828 | - | BRD4 | chr19 | 15365073 | - |
| intron-3CDS | ENST00000328788 | ENST00000371835 | FRMD3 | chr9 | 85987828 | - | BRD4 | chr19 | 15365073 | - |
| intron-3CDS | ENST00000376434 | ENST00000263377 | FRMD3 | chr9 | 85987828 | - | BRD4 | chr19 | 15365073 | - |
| intron-3CDS | ENST00000376434 | ENST00000360016 | FRMD3 | chr9 | 85987828 | - | BRD4 | chr19 | 15365073 | - |
| intron-3CDS | ENST00000376434 | ENST00000371835 | FRMD3 | chr9 | 85987828 | - | BRD4 | chr19 | 15365073 | - |
| intron-3CDS | ENST00000465485 | ENST00000263377 | FRMD3 | chr9 | 85987828 | - | BRD4 | chr19 | 15365073 | - |
| intron-3CDS | ENST00000465485 | ENST00000360016 | FRMD3 | chr9 | 85987828 | - | BRD4 | chr19 | 15365073 | - |
| intron-3CDS | ENST00000465485 | ENST00000371835 | FRMD3 | chr9 | 85987828 | - | BRD4 | chr19 | 15365073 | - |
| intron-intron | ENST00000328788 | ENST00000602230 | FRMD3 | chr9 | 85987828 | - | BRD4 | chr19 | 15365073 | - |
| intron-intron | ENST00000376434 | ENST00000602230 | FRMD3 | chr9 | 85987828 | - | BRD4 | chr19 | 15365073 | - |
| intron-intron | ENST00000465485 | ENST00000602230 | FRMD3 | chr9 | 85987828 | - | BRD4 | chr19 | 15365073 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000376438 | FRMD3 | chr9 | 85987828 | - | ENST00000263377 | BRD4 | chr19 | 15365073 | - | 4193 | 610 | 207 | 2651 | 814 |
| ENST00000376438 | FRMD3 | chr9 | 85987828 | - | ENST00000371835 | BRD4 | chr19 | 15365073 | - | 2955 | 610 | 602 | 0 | 201 |
| ENST00000376438 | FRMD3 | chr9 | 85987828 | - | ENST00000360016 | BRD4 | chr19 | 15365073 | - | 1513 | 610 | 207 | 947 | 246 |
| ENST00000304195 | FRMD3 | chr9 | 85987828 | - | ENST00000263377 | BRD4 | chr19 | 15365073 | - | 4085 | 502 | 99 | 2543 | 814 |
| ENST00000304195 | FRMD3 | chr9 | 85987828 | - | ENST00000371835 | BRD4 | chr19 | 15365073 | - | 2847 | 502 | 99 | 623 | 174 |
| ENST00000304195 | FRMD3 | chr9 | 85987828 | - | ENST00000360016 | BRD4 | chr19 | 15365073 | - | 1405 | 502 | 99 | 839 | 246 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000376438 | ENST00000263377 | FRMD3 | chr9 | 85987828 | - | BRD4 | chr19 | 15365073 | - | 0.02579781 | 0.9742022 |
| ENST00000376438 | ENST00000371835 | FRMD3 | chr9 | 85987828 | - | BRD4 | chr19 | 15365073 | - | 0.08950321 | 0.9104968 |
| ENST00000376438 | ENST00000360016 | FRMD3 | chr9 | 85987828 | - | BRD4 | chr19 | 15365073 | - | 0.062464703 | 0.9375353 |
| ENST00000304195 | ENST00000263377 | FRMD3 | chr9 | 85987828 | - | BRD4 | chr19 | 15365073 | - | 0.026292447 | 0.9737075 |
| ENST00000304195 | ENST00000371835 | FRMD3 | chr9 | 85987828 | - | BRD4 | chr19 | 15365073 | - | 0.1378494 | 0.8621506 |
| ENST00000304195 | ENST00000360016 | FRMD3 | chr9 | 85987828 | - | BRD4 | chr19 | 15365073 | - | 0.08509716 | 0.91490287 |
Top |
Fusion Genomic Features for FRMD3-BRD4 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
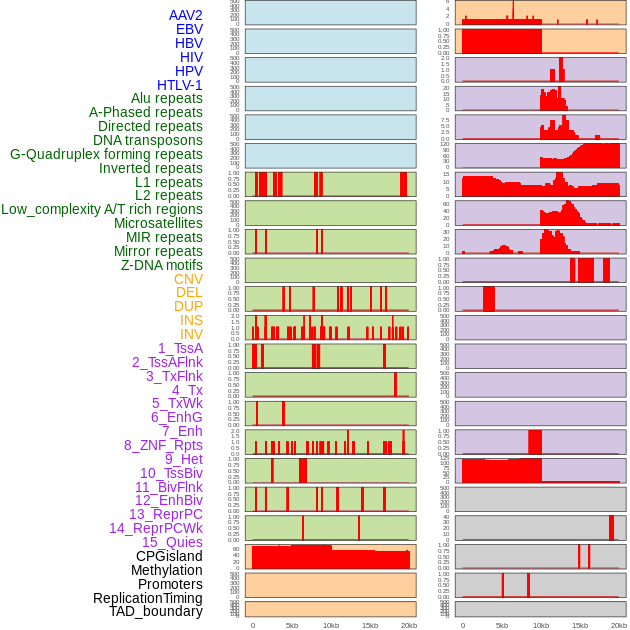 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for FRMD3-BRD4 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr9:85987828/chr19:15365073) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| FRMD3 | BRD4 |
| FUNCTION: Putative tumor suppressor gene that may be implicated in the origin and progression of lung cancer. {ECO:0000269|PubMed:17260017}. | FUNCTION: Chromatin reader protein that recognizes and binds acetylated histones and plays a key role in transmission of epigenetic memory across cell divisions and transcription regulation. Remains associated with acetylated chromatin throughout the entire cell cycle and provides epigenetic memory for postmitotic G1 gene transcription by preserving acetylated chromatin status and maintaining high-order chromatin structure (PubMed:23589332, PubMed:23317504, PubMed:22334664). During interphase, plays a key role in regulating the transcription of signal-inducible genes by associating with the P-TEFb complex and recruiting it to promoters. Also recruits P-TEFb complex to distal enhancers, so called anti-pause enhancers in collaboration with JMJD6. BRD4 and JMJD6 are required to form the transcriptionally active P-TEFb complex by displacing negative regulators such as HEXIM1 and 7SKsnRNA complex from P-TEFb, thereby transforming it into an active form that can then phosphorylate the C-terminal domain (CTD) of RNA polymerase II (PubMed:23589332, PubMed:19596240, PubMed:16109377, PubMed:16109376, PubMed:24360279). Promotes phosphorylation of 'Ser-2' of the C-terminal domain (CTD) of RNA polymerase II (PubMed:23086925). According to a report, directly acts as an atypical protein kinase and mediates phosphorylation of 'Ser-2' of the C-terminal domain (CTD) of RNA polymerase II; these data however need additional evidences in vivo (PubMed:22509028). In addition to acetylated histones, also recognizes and binds acetylated RELA, leading to further recruitment of the P-TEFb complex and subsequent activation of NF-kappa-B (PubMed:19103749). Also acts as a regulator of p53/TP53-mediated transcription: following phosphorylation by CK2, recruited to p53/TP53 specific target promoters (PubMed:23317504). {ECO:0000269|PubMed:16109376, ECO:0000269|PubMed:16109377, ECO:0000269|PubMed:19103749, ECO:0000269|PubMed:19596240, ECO:0000269|PubMed:22334664, ECO:0000269|PubMed:22509028, ECO:0000269|PubMed:23086925, ECO:0000269|PubMed:23317504, ECO:0000269|PubMed:23589332, ECO:0000269|PubMed:24360279}.; FUNCTION: [Isoform B]: Acts as a chromatin insulator in the DNA damage response pathway. Inhibits DNA damage response signaling by recruiting the condensin-2 complex to acetylated histones, leading to chromatin structure remodeling, insulating the region from DNA damage response by limiting spreading of histone H2AX/H2A.x phosphorylation. {ECO:0000269|PubMed:23728299}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | BRD4 | chr9:85987828 | chr19:15365073 | ENST00000263377 | 9 | 20 | 1011_1014 | 682 | 1363.0 | Compositional bias | Note=Poly-Pro | |
| Tgene | BRD4 | chr9:85987828 | chr19:15365073 | ENST00000263377 | 9 | 20 | 1028_1033 | 682 | 1363.0 | Compositional bias | Note=Poly-Pro | |
| Tgene | BRD4 | chr9:85987828 | chr19:15365073 | ENST00000263377 | 9 | 20 | 1283_1300 | 682 | 1363.0 | Compositional bias | Note=Poly-Gln | |
| Tgene | BRD4 | chr9:85987828 | chr19:15365073 | ENST00000263377 | 9 | 20 | 1301_1308 | 682 | 1363.0 | Compositional bias | Note=Poly-Ala | |
| Tgene | BRD4 | chr9:85987828 | chr19:15365073 | ENST00000263377 | 9 | 20 | 1335_1338 | 682 | 1363.0 | Compositional bias | Note=Poly-Arg | |
| Tgene | BRD4 | chr9:85987828 | chr19:15365073 | ENST00000263377 | 9 | 20 | 692_717 | 682 | 1363.0 | Compositional bias | Note=Ser-rich | |
| Tgene | BRD4 | chr9:85987828 | chr19:15365073 | ENST00000263377 | 9 | 20 | 703_714 | 682 | 1363.0 | Compositional bias | Note=Poly-Ser | |
| Tgene | BRD4 | chr9:85987828 | chr19:15365073 | ENST00000263377 | 9 | 20 | 738_743 | 682 | 1363.0 | Compositional bias | Note=Poly-His | |
| Tgene | BRD4 | chr9:85987828 | chr19:15365073 | ENST00000263377 | 9 | 20 | 757_761 | 682 | 1363.0 | Compositional bias | Note=Poly-Pro | |
| Tgene | BRD4 | chr9:85987828 | chr19:15365073 | ENST00000263377 | 9 | 20 | 764_770 | 682 | 1363.0 | Compositional bias | Note=Poly-Pro | |
| Tgene | BRD4 | chr9:85987828 | chr19:15365073 | ENST00000263377 | 9 | 20 | 771_775 | 682 | 1363.0 | Compositional bias | Note=Poly-Gln | |
| Tgene | BRD4 | chr9:85987828 | chr19:15365073 | ENST00000263377 | 9 | 20 | 776_783 | 682 | 1363.0 | Compositional bias | Note=Poly-Pro | |
| Tgene | BRD4 | chr9:85987828 | chr19:15365073 | ENST00000263377 | 9 | 20 | 954_964 | 682 | 1363.0 | Compositional bias | Note=Poly-Pro | |
| Tgene | BRD4 | chr9:85987828 | chr19:15365073 | ENST00000263377 | 9 | 20 | 974_986 | 682 | 1363.0 | Compositional bias | Note=Poly-Pro | |
| Tgene | BRD4 | chr9:85987828 | chr19:15365073 | ENST00000360016 | 9 | 12 | 1011_1014 | 682 | 795.0 | Compositional bias | Note=Poly-Pro | |
| Tgene | BRD4 | chr9:85987828 | chr19:15365073 | ENST00000360016 | 9 | 12 | 1028_1033 | 682 | 795.0 | Compositional bias | Note=Poly-Pro | |
| Tgene | BRD4 | chr9:85987828 | chr19:15365073 | ENST00000360016 | 9 | 12 | 1283_1300 | 682 | 795.0 | Compositional bias | Note=Poly-Gln | |
| Tgene | BRD4 | chr9:85987828 | chr19:15365073 | ENST00000360016 | 9 | 12 | 1301_1308 | 682 | 795.0 | Compositional bias | Note=Poly-Ala | |
| Tgene | BRD4 | chr9:85987828 | chr19:15365073 | ENST00000360016 | 9 | 12 | 1335_1338 | 682 | 795.0 | Compositional bias | Note=Poly-Arg | |
| Tgene | BRD4 | chr9:85987828 | chr19:15365073 | ENST00000360016 | 9 | 12 | 692_717 | 682 | 795.0 | Compositional bias | Note=Ser-rich | |
| Tgene | BRD4 | chr9:85987828 | chr19:15365073 | ENST00000360016 | 9 | 12 | 703_714 | 682 | 795.0 | Compositional bias | Note=Poly-Ser | |
| Tgene | BRD4 | chr9:85987828 | chr19:15365073 | ENST00000360016 | 9 | 12 | 738_743 | 682 | 795.0 | Compositional bias | Note=Poly-His | |
| Tgene | BRD4 | chr9:85987828 | chr19:15365073 | ENST00000360016 | 9 | 12 | 757_761 | 682 | 795.0 | Compositional bias | Note=Poly-Pro | |
| Tgene | BRD4 | chr9:85987828 | chr19:15365073 | ENST00000360016 | 9 | 12 | 764_770 | 682 | 795.0 | Compositional bias | Note=Poly-Pro | |
| Tgene | BRD4 | chr9:85987828 | chr19:15365073 | ENST00000360016 | 9 | 12 | 771_775 | 682 | 795.0 | Compositional bias | Note=Poly-Gln | |
| Tgene | BRD4 | chr9:85987828 | chr19:15365073 | ENST00000360016 | 9 | 12 | 776_783 | 682 | 795.0 | Compositional bias | Note=Poly-Pro | |
| Tgene | BRD4 | chr9:85987828 | chr19:15365073 | ENST00000360016 | 9 | 12 | 954_964 | 682 | 795.0 | Compositional bias | Note=Poly-Pro | |
| Tgene | BRD4 | chr9:85987828 | chr19:15365073 | ENST00000360016 | 9 | 12 | 974_986 | 682 | 795.0 | Compositional bias | Note=Poly-Pro | |
| Tgene | BRD4 | chr9:85987828 | chr19:15365073 | ENST00000371835 | 9 | 12 | 1011_1014 | 682 | 723.0 | Compositional bias | Note=Poly-Pro | |
| Tgene | BRD4 | chr9:85987828 | chr19:15365073 | ENST00000371835 | 9 | 12 | 1028_1033 | 682 | 723.0 | Compositional bias | Note=Poly-Pro | |
| Tgene | BRD4 | chr9:85987828 | chr19:15365073 | ENST00000371835 | 9 | 12 | 1283_1300 | 682 | 723.0 | Compositional bias | Note=Poly-Gln | |
| Tgene | BRD4 | chr9:85987828 | chr19:15365073 | ENST00000371835 | 9 | 12 | 1301_1308 | 682 | 723.0 | Compositional bias | Note=Poly-Ala | |
| Tgene | BRD4 | chr9:85987828 | chr19:15365073 | ENST00000371835 | 9 | 12 | 1335_1338 | 682 | 723.0 | Compositional bias | Note=Poly-Arg | |
| Tgene | BRD4 | chr9:85987828 | chr19:15365073 | ENST00000371835 | 9 | 12 | 692_717 | 682 | 723.0 | Compositional bias | Note=Ser-rich | |
| Tgene | BRD4 | chr9:85987828 | chr19:15365073 | ENST00000371835 | 9 | 12 | 703_714 | 682 | 723.0 | Compositional bias | Note=Poly-Ser | |
| Tgene | BRD4 | chr9:85987828 | chr19:15365073 | ENST00000371835 | 9 | 12 | 738_743 | 682 | 723.0 | Compositional bias | Note=Poly-His | |
| Tgene | BRD4 | chr9:85987828 | chr19:15365073 | ENST00000371835 | 9 | 12 | 757_761 | 682 | 723.0 | Compositional bias | Note=Poly-Pro | |
| Tgene | BRD4 | chr9:85987828 | chr19:15365073 | ENST00000371835 | 9 | 12 | 764_770 | 682 | 723.0 | Compositional bias | Note=Poly-Pro | |
| Tgene | BRD4 | chr9:85987828 | chr19:15365073 | ENST00000371835 | 9 | 12 | 771_775 | 682 | 723.0 | Compositional bias | Note=Poly-Gln | |
| Tgene | BRD4 | chr9:85987828 | chr19:15365073 | ENST00000371835 | 9 | 12 | 776_783 | 682 | 723.0 | Compositional bias | Note=Poly-Pro | |
| Tgene | BRD4 | chr9:85987828 | chr19:15365073 | ENST00000371835 | 9 | 12 | 954_964 | 682 | 723.0 | Compositional bias | Note=Poly-Pro | |
| Tgene | BRD4 | chr9:85987828 | chr19:15365073 | ENST00000371835 | 9 | 12 | 974_986 | 682 | 723.0 | Compositional bias | Note=Poly-Pro | |
| Tgene | BRD4 | chr9:85987828 | chr19:15365073 | ENST00000263377 | 9 | 20 | 1047_1362 | 682 | 1363.0 | Region | Note=C-terminal (CTD) region | |
| Tgene | BRD4 | chr9:85987828 | chr19:15365073 | ENST00000360016 | 9 | 12 | 1047_1362 | 682 | 795.0 | Region | Note=C-terminal (CTD) region | |
| Tgene | BRD4 | chr9:85987828 | chr19:15365073 | ENST00000371835 | 9 | 12 | 1047_1362 | 682 | 723.0 | Region | Note=C-terminal (CTD) region |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | FRMD3 | chr9:85987828 | chr19:15365073 | ENST00000304195 | - | 3 | 14 | 32_312 | 98 | 598.0 | Domain | FERM |
| Hgene | FRMD3 | chr9:85987828 | chr19:15365073 | ENST00000328788 | - | 1 | 3 | 32_312 | 0 | 214.0 | Domain | FERM |
| Hgene | FRMD3 | chr9:85987828 | chr19:15365073 | ENST00000376434 | - | 1 | 10 | 32_312 | 0 | 363.0 | Domain | FERM |
| Hgene | FRMD3 | chr9:85987828 | chr19:15365073 | ENST00000376438 | - | 3 | 15 | 32_312 | 98 | 557.0 | Domain | FERM |
| Hgene | FRMD3 | chr9:85987828 | chr19:15365073 | ENST00000304195 | - | 3 | 14 | 531_551 | 98 | 598.0 | Transmembrane | Helical |
| Hgene | FRMD3 | chr9:85987828 | chr19:15365073 | ENST00000328788 | - | 1 | 3 | 531_551 | 0 | 214.0 | Transmembrane | Helical |
| Hgene | FRMD3 | chr9:85987828 | chr19:15365073 | ENST00000376434 | - | 1 | 10 | 531_551 | 0 | 363.0 | Transmembrane | Helical |
| Hgene | FRMD3 | chr9:85987828 | chr19:15365073 | ENST00000376438 | - | 3 | 15 | 531_551 | 98 | 557.0 | Transmembrane | Helical |
| Tgene | BRD4 | chr9:85987828 | chr19:15365073 | ENST00000263377 | 9 | 20 | 535_594 | 682 | 1363.0 | Compositional bias | Note=Lys-rich | |
| Tgene | BRD4 | chr9:85987828 | chr19:15365073 | ENST00000360016 | 9 | 12 | 535_594 | 682 | 795.0 | Compositional bias | Note=Lys-rich | |
| Tgene | BRD4 | chr9:85987828 | chr19:15365073 | ENST00000371835 | 9 | 12 | 535_594 | 682 | 723.0 | Compositional bias | Note=Lys-rich | |
| Tgene | BRD4 | chr9:85987828 | chr19:15365073 | ENST00000263377 | 9 | 20 | 368_440 | 682 | 1363.0 | Domain | Bromo 2 | |
| Tgene | BRD4 | chr9:85987828 | chr19:15365073 | ENST00000263377 | 9 | 20 | 600_682 | 682 | 1363.0 | Domain | NET | |
| Tgene | BRD4 | chr9:85987828 | chr19:15365073 | ENST00000263377 | 9 | 20 | 75_147 | 682 | 1363.0 | Domain | Bromo 1 | |
| Tgene | BRD4 | chr9:85987828 | chr19:15365073 | ENST00000360016 | 9 | 12 | 368_440 | 682 | 795.0 | Domain | Bromo 2 | |
| Tgene | BRD4 | chr9:85987828 | chr19:15365073 | ENST00000360016 | 9 | 12 | 600_682 | 682 | 795.0 | Domain | NET | |
| Tgene | BRD4 | chr9:85987828 | chr19:15365073 | ENST00000360016 | 9 | 12 | 75_147 | 682 | 795.0 | Domain | Bromo 1 | |
| Tgene | BRD4 | chr9:85987828 | chr19:15365073 | ENST00000371835 | 9 | 12 | 368_440 | 682 | 723.0 | Domain | Bromo 2 | |
| Tgene | BRD4 | chr9:85987828 | chr19:15365073 | ENST00000371835 | 9 | 12 | 600_682 | 682 | 723.0 | Domain | NET | |
| Tgene | BRD4 | chr9:85987828 | chr19:15365073 | ENST00000371835 | 9 | 12 | 75_147 | 682 | 723.0 | Domain | Bromo 1 | |
| Tgene | BRD4 | chr9:85987828 | chr19:15365073 | ENST00000263377 | 9 | 20 | 484_503 | 682 | 1363.0 | Region | Note=NPS region | |
| Tgene | BRD4 | chr9:85987828 | chr19:15365073 | ENST00000263377 | 9 | 20 | 524_579 | 682 | 1363.0 | Region | Note=BID region | |
| Tgene | BRD4 | chr9:85987828 | chr19:15365073 | ENST00000360016 | 9 | 12 | 484_503 | 682 | 795.0 | Region | Note=NPS region | |
| Tgene | BRD4 | chr9:85987828 | chr19:15365073 | ENST00000360016 | 9 | 12 | 524_579 | 682 | 795.0 | Region | Note=BID region | |
| Tgene | BRD4 | chr9:85987828 | chr19:15365073 | ENST00000371835 | 9 | 12 | 484_503 | 682 | 723.0 | Region | Note=NPS region | |
| Tgene | BRD4 | chr9:85987828 | chr19:15365073 | ENST00000371835 | 9 | 12 | 524_579 | 682 | 723.0 | Region | Note=BID region |
Top |
Fusion Gene Sequence for FRMD3-BRD4 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >31316_31316_1_FRMD3-BRD4_FRMD3_chr9_85987828_ENST00000304195_BRD4_chr19_15365073_ENST00000263377_length(transcript)=4085nt_BP=502nt GAGGCGGAGGCGAGCGAACAGAGGGAGGGACCCGCCCGCCGCGCCCCGGCCGCTGGGCATGTGTGTCCGCAGGCGCCCGACGCTGCCGAT GTCCCGGGGCTGAGCCGCGCCCAGGTGTCCCGGACAGTGCGTGCGAGCGTGTGTGTCCGCGCAGGCGAGCACCGCGCCGGCCCTGAGCCT CCCGCTCGCTCCCCACGGCCGCGGTGCATGTTCGCCTCCTGCCACTGTGTGCCGAGAGGCAGGAGGACCATGAAAATGATCCACTTTCGG AGCTCCAGCGTCAAATCGCTCAGCCAGGAGATGAGATGCACCATCCGGCTGCTGGACGACTCGGAGATCTCCTGCCACATCCAGAGGGAA ACCAAAGGGCAGTTTCTCATTGACCACATCTGCAACTACTACAGCCTGCTGGAGAAGGACTACTTTGGCATTCGCTATGTGGACCCAGAG AAGCAAAGGCACTGGCTTGAACCTAACAAGTCCATCTTCAAGCAAATGAAAACTGAGAAAGTTGATGTGATTGCCGGCTCCTCCAAGATG AAGGGCTTCTCGTCCTCAGAGTCGGAGAGCTCCAGTGAGTCCAGCTCCTCTGACAGCGAAGACTCCGAAACAGAGATGGCTCCGAAGTCA AAAAAGAAGGGGCACCCCGGGAGGGAGCAGAAGAAGCACCATCATCACCACCATCAGCAGATGCAGCAGGCCCCGGCTCCTGTGCCCCAG CAGCCGCCCCCGCCTCCCCAGCAGCCCCCACCGCCTCCACCTCCGCAGCAGCAACAGCAGCCGCCACCCCCGCCTCCCCCACCCTCCATG CCGCAGCAGGCAGCCCCGGCGATGAAGTCCTCGCCCCCACCCTTCATTGCCACCCAGGTGCCCGTCCTGGAGCCCCAGCTCCCAGGCAGC GTCTTTGACCCCATCGGCCACTTCACCCAGCCCATCCTGCACCTGCCGCAGCCTGAGCTGCCCCCTCACCTGCCCCAGCCGCCTGAGCAC AGCACTCCACCCCATCTCAACCAGCACGCAGTGGTCTCTCCTCCAGCTTTGCACAACGCACTACCCCAGCAGCCATCACGGCCCAGCAAC CGAGCCGCTGCCCTGCCTCCCAAGCCCGCCCGGCCCCCAGCCGTGTCACCAGCCTTGACCCAAACACCCCTGCTCCCACAGCCCCCCATG GCCCAACCCCCCCAAGTGCTGCTGGAGGATGAAGAGCCACCTGCCCCACCCCTCACCTCCATGCAGATGCAGCTGTACCTGCAGCAGCTG CAGAAGGTGCAGCCCCCTACGCCGCTACTCCCTTCCGTGAAGGTGCAGTCCCAGCCCCCACCCCCCCTGCCGCCCCCACCCCACCCCTCT GTGCAGCAGCAGCTGCAGCAGCAGCCGCCACCACCCCCACCACCCCAGCCCCAGCCTCCACCCCAGCAGCAGCATCAGCCCCCTCCACGG CCCGTGCACTTGCAGCCCATGCAGTTTTCCACCCACATCCAACAGCCCCCGCCACCCCAGGGCCAGCAGCCCCCCCATCCGCCCCCAGGC CAGCAGCCACCCCCGCCGCAGCCTGCCAAGCCTCAGCAAGTCATCCAGCACCACCATTCACCCCGGCACCACAAGTCGGACCCCTACTCA ACCGGTCACCTCCGCGAAGCCCCCTCCCCGCTTATGATACATTCCCCCCAGATGTCACAGTTCCAGAGCCTGACCCACCAGTCTCCACCC CAGCAAAACGTCCAGCCTAAGAAACAGGAGCTGCGTGCTGCCTCCGTGGTCCAGCCCCAGCCCCTCGTGGTGGTGAAGGAGGAGAAGATC CACTCACCCATCATCCGCAGCGAGCCCTTCAGCCCCTCGCTGCGGCCGGAGCCCCCCAAGCACCCGGAGAGCATCAAGGCCCCCGTCCAC CTGCCCCAGCGGCCGGAAATGAAGCCTGTGGATGTCGGGAGGCCTGTGATCCGGCCCCCAGAGCAGAACGCACCGCCACCAGGGGCCCCT GACAAGGACAAACAGAAACAGGAGCCGAAGACTCCAGTTGCGCCCAAAAAGGACCTGAAAATCAAGAACATGGGCTCCTGGGCCAGCCTA GTGCAGAAGCATCCGACCACCCCCTCCTCCACAGCCAAGTCATCCAGCGACAGCTTCGAGCAGTTCCGCCGCGCCGCTCGGGAGAAAGAG GAGCGTGAGAAGGCCCTGAAGGCTCAGGCCGAGCACGCTGAGAAGGAGAAGGAGCGGCTGCGGCAGGAGCGCATGAGGAGCCGAGAGGAC GAGGATGCGCTGGAGCAGGCCCGGCGGGCCCATGAGGAGGCACGTCGGCGCCAGGAGCAGCAGCAGCAGCAGCGCCAGGAGCAACAGCAG CAGCAGCAACAGCAAGCAGCTGCGGTGGCTGCCGCCGCCACCCCACAGGCCCAGAGCTCCCAGCCCCAGTCCATGCTGGACCAGCAGAGG GAGTTGGCCCGGAAGCGGGAGCAGGAGCGAAGACGCCGGGAAGCCATGGCAGCTACCATTGACATGAATTTCCAGAGTGATCTATTGTCA ATATTTGAAGAAAATCTTTTCTGAGCGCACCTAGGTGGCTTCTGACTTTGATTTTCTGGCAAAACATTGACTTTCCATAGTGTTAGGGGC GGTGGTGGAGGTGGGATCAGCGGCCAGGGGATGCCTCAGGGCCTGGCCCTCCTGCATGCTATGCCCGGGGCAGGCCTGACGGGCAGCTGA GGATTGCAGAGCCTGTCTGCCTTACGGCCAGTCGGACAGACGTCCCGCCACCCACCACCCCTCACAGGACGTCCGCTCAGCACACGCCTT GTTACGAGCAAGTGCCGGCTGGACCCAAGCCCTGCATCCCCACATGCGGGGCAGAGGCCCTTCTCTCCGCCAAATGTCTACACAGTATAC ACAGGACATCGTTGCTGCCGCCGTGACTGGTTTTCTGTCCCCAAGAACGTGACGTTCGTGATGTCCTGCCCGCCGGGAGTCTTTCCCCAC ACCCCAGCCATCGCCGCCCGCTCCCAGGAGGCCAGGGCAGGCCTGCGTGGGCTGGAGGCGGGCGAGGCCGGCCCACCCCCTCGCTGGCAC TGACTTTGCCTTGAACAGACCCCCCGACCCTCCCCCACAAGCCTTTAATTGAGAGCCGCTCTCTGTAAGTGTTTGCTTGTGCAAAAGGGA ATAGTGCCGTGGAGGTGTGTGTGTCCATGGCATCCGGAGCGAGGCGACTGTCCTGCGTGGGTAGCCCTCGGCCGGGGAGTGAGGCCACCA ACCAAAGTCAGTTCCTTCCCACCTGTGTTTCTGTTTCGTTTTTTTTTTTCTTTTTTTTCTATATATATTTTTTGTTGAATTCTATTTTAT TTTTAATTCTCTCTTCTCCTCCAGACACAATGGCACTGCTTATCTCCGAAATGGTGTGATCGTCTCCTCATTGAGCAGCGGCTGCCACCG CGCTGTGGGTAGTGTGTGACCGTGGCTGTACTGTATAGTGAACATAGTTGGCATATCTTTGTTTGAAGTTTGTTGGTGACTCCACCAAAC TGGTGTGAAAAAAGAAAAAAGCTCAAAAAAATCCACAAAAAGACAAAACACACAAAAAAAATCCTGCCTATATTTTACTCAGTTTCAAAC TTTATTAGTCTATTTTTAATTATAAAACCAGAAAGCTACAATTTCTTTTCTTTCCCCTCCACCCCCCCCCCCCCCACCCATTTGTTGGCT TTTTTGTTTTTTAATGTCAGATCTGTTGAGTTGGTTTTTTTGGTTTTGGTTTTTGTTTTTGTTTTTGTTTTTTACTGAGAAAGGAAGGGC CAAGGGATGAGGTGGGAACCGGGCCCTGGGGGCGCCACAGACTAAGGCAGAGACTCCCCTACCTGGCGCCCAGCCCCAACCAGCTGGCCG CTCCTGCCCATGCTTTTTTTTTTTTTTTTTTTTAATTTTTATAATTGGAGCCCCTGGTGAGGTTACGCGTGCCATGAGAACCCACTCTAC ACCACGACGCTGGTGCCTCAGTGTTGGCCAAACTCTGGAGTCACTGACTGGTTTGACTTTCATACGGTGAATATGCATTTGGTCTGTACT >31316_31316_1_FRMD3-BRD4_FRMD3_chr9_85987828_ENST00000304195_BRD4_chr19_15365073_ENST00000263377_length(amino acids)=814AA_BP=424 MSRAQVSRTVRASVCVRAGEHRAGPEPPARSPRPRCMFASCHCVPRGRRTMKMIHFRSSSVKSLSQEMRCTIRLLDDSEISCHIQRETKG QFLIDHICNYYSLLEKDYFGIRYVDPEKQRHWLEPNKSIFKQMKTEKVDVIAGSSKMKGFSSSESESSSESSSSDSEDSETEMAPKSKKK GHPGREQKKHHHHHHQQMQQAPAPVPQQPPPPPQQPPPPPPPQQQQQPPPPPPPPSMPQQAAPAMKSSPPPFIATQVPVLEPQLPGSVFD PIGHFTQPILHLPQPELPPHLPQPPEHSTPPHLNQHAVVSPPALHNALPQQPSRPSNRAAALPPKPARPPAVSPALTQTPLLPQPPMAQP PQVLLEDEEPPAPPLTSMQMQLYLQQLQKVQPPTPLLPSVKVQSQPPPPLPPPPHPSVQQQLQQQPPPPPPPQPQPPPQQQHQPPPRPVH LQPMQFSTHIQQPPPPQGQQPPHPPPGQQPPPPQPAKPQQVIQHHHSPRHHKSDPYSTGHLREAPSPLMIHSPQMSQFQSLTHQSPPQQN VQPKKQELRAASVVQPQPLVVVKEEKIHSPIIRSEPFSPSLRPEPPKHPESIKAPVHLPQRPEMKPVDVGRPVIRPPEQNAPPPGAPDKD KQKQEPKTPVAPKKDLKIKNMGSWASLVQKHPTTPSSTAKSSSDSFEQFRRAAREKEEREKALKAQAEHAEKEKERLRQERMRSREDEDA LEQARRAHEEARRRQEQQQQQRQEQQQQQQQQAAAVAAAATPQAQSSQPQSMLDQQRELARKREQERRRREAMAATIDMNFQSDLLSIFE -------------------------------------------------------------- >31316_31316_2_FRMD3-BRD4_FRMD3_chr9_85987828_ENST00000304195_BRD4_chr19_15365073_ENST00000360016_length(transcript)=1405nt_BP=502nt GAGGCGGAGGCGAGCGAACAGAGGGAGGGACCCGCCCGCCGCGCCCCGGCCGCTGGGCATGTGTGTCCGCAGGCGCCCGACGCTGCCGAT GTCCCGGGGCTGAGCCGCGCCCAGGTGTCCCGGACAGTGCGTGCGAGCGTGTGTGTCCGCGCAGGCGAGCACCGCGCCGGCCCTGAGCCT CCCGCTCGCTCCCCACGGCCGCGGTGCATGTTCGCCTCCTGCCACTGTGTGCCGAGAGGCAGGAGGACCATGAAAATGATCCACTTTCGG AGCTCCAGCGTCAAATCGCTCAGCCAGGAGATGAGATGCACCATCCGGCTGCTGGACGACTCGGAGATCTCCTGCCACATCCAGAGGGAA ACCAAAGGGCAGTTTCTCATTGACCACATCTGCAACTACTACAGCCTGCTGGAGAAGGACTACTTTGGCATTCGCTATGTGGACCCAGAG AAGCAAAGGCACTGGCTTGAACCTAACAAGTCCATCTTCAAGCAAATGAAAACTGAGAAAGTTGATGTGATTGCCGGCTCCTCCAAGATG AAGGGCTTCTCGTCCTCAGAGTCGGAGAGCTCCAGTGAGTCCAGCTCCTCTGACAGCGAAGACTCCGAAACAGCTTTCTGCACCAGTGGA GACTTCGTGTCCCCAGGGCCTTCCCCGTATCACAGTCACGTGCAGTGCGGCCGCTTCAGGGAGATGCTTCGCTGGTTTCTGGTGGATGTG GAGCAGACTGCAGCTGGCCAGCCGCATCGCCAGTCTGCTGCTGGCCCTGCCATCACCTGGGCCCCAGCCATTGCCTACCCCTCCCCAGAG TGTGCTCGTTGCTGTGTTGGCTGCTCCTGAATCTGCCCTAACTCCACACGCACCTGGACTTGCGTGTCCCTCCTGCAGTTCTAACTAACA GTCCCTTCTTTTCAAGCCCCGTGGGTCTGCACGTTGGACCCTGGGTTCCCCATTAGAGCCCACCTTCTGAGCAGCAGCCTCAGTGGGAGG TGGAGGCAGGTAGTGATGCTGGGTGCCAGGTGGGAGTGGAGAGGGGACTGCTCTCCTCCAAGTGTGCACTTTCCTCATTATTCTCAGGGG CGCAGATGCTCAGGCTGGCGCAGGGGAGAGGCTAGGGAGGGAGCCATGGTGCCCAGAAGGCCTGGCGACCAGCCCCTGCTGAGAGATGGA GCTAACATCCTGTGTTTACGGCCAACGGGGTTGCCGCTAGGCTGGTGCAGCTGTCAGTGAGCTGGCGTGCTGCAGACCACCTGAGAGCTG GCCCTAGGGTCTCAGGCAGACTGGGGAGTGGGGATCCACAGTGGGAAACCTGTGTTTTGGCAGTAGACTCCTGCATGTTCTCCCACGGGC >31316_31316_2_FRMD3-BRD4_FRMD3_chr9_85987828_ENST00000304195_BRD4_chr19_15365073_ENST00000360016_length(amino acids)=246AA_BP=135 MSRAQVSRTVRASVCVRAGEHRAGPEPPARSPRPRCMFASCHCVPRGRRTMKMIHFRSSSVKSLSQEMRCTIRLLDDSEISCHIQRETKG QFLIDHICNYYSLLEKDYFGIRYVDPEKQRHWLEPNKSIFKQMKTEKVDVIAGSSKMKGFSSSESESSSESSSSDSEDSETAFCTSGDFV -------------------------------------------------------------- >31316_31316_3_FRMD3-BRD4_FRMD3_chr9_85987828_ENST00000304195_BRD4_chr19_15365073_ENST00000371835_length(transcript)=2847nt_BP=502nt GAGGCGGAGGCGAGCGAACAGAGGGAGGGACCCGCCCGCCGCGCCCCGGCCGCTGGGCATGTGTGTCCGCAGGCGCCCGACGCTGCCGAT GTCCCGGGGCTGAGCCGCGCCCAGGTGTCCCGGACAGTGCGTGCGAGCGTGTGTGTCCGCGCAGGCGAGCACCGCGCCGGCCCTGAGCCT CCCGCTCGCTCCCCACGGCCGCGGTGCATGTTCGCCTCCTGCCACTGTGTGCCGAGAGGCAGGAGGACCATGAAAATGATCCACTTTCGG AGCTCCAGCGTCAAATCGCTCAGCCAGGAGATGAGATGCACCATCCGGCTGCTGGACGACTCGGAGATCTCCTGCCACATCCAGAGGGAA ACCAAAGGGCAGTTTCTCATTGACCACATCTGCAACTACTACAGCCTGCTGGAGAAGGACTACTTTGGCATTCGCTATGTGGACCCAGAG AAGCAAAGGCACTGGCTTGAACCTAACAAGTCCATCTTCAAGCAAATGAAAACTGAGAAAGTTGATGTGATTGCCGGCTCCTCCAAGATG AAGGGCTTCTCGTCCTCAGAGTCGGAGAGCTCCAGTGAGTCCAGCTCCTCTGACAGCGAAGACTCCGAAACAGGTCCTGCCTAATCATTG GACACGGACTCTTAATAAAACGGTCTTCAGTTCCAGATTCCTTCCCAGCAAGCTATAGCTTAAGTCCATTTTCTTCCGTGAAAGGGACAG GACTCCATCAAGTTATGGAATTCCTCAGAGCCCTGGGCCTGTCCCCCGGGGTGGATTAGTCATGTCCAGCAGCACACGCCTAGTCCCGCC TTCGGGAAGGCTGCCTGCCTGGCCAGCCGCCCAGGCCTCTCTGTGTAAAGACTGCCTGGCTGTCCTGCCCAGCCTTCCTGGTTCTCTGGG GTCCTCTGGGTGGGTGGCATCTCCTGGAGGGTGATGACAATCCCCAACACATGCATTCATGTGGTGCTACTCTGTGTGCAAAGCCAGACC CCAAGTATGTTTTCTCTCTTTGTCCCATCCCTCTTTTTCTGGGACTTTGGACCCTAACTACTTCCCTCCTGAACCTTGCAGTGACATCAG TCCAGGAGAGCTCTCGTTCAGTGTGCGGAAGAACACTCTGACCTCTAGAGCTGTCCTAGATAAGGAGTGGGAGCTTTAGAGGCAAGGCCT CTAGACCCTGGAAGGCTCAGTGAGGCTCTTCCCACAGCATGCTTCTCACTGGTGCCCTGTAAGGCTCGAGCCACCGCTGACTCTGAGCCT TTTGGAGTCTTTCCTCCTTCGTCTCCATTGTTCCCGTGCATTTCCAAAAGCTTAAGTTGCCTGGTGGGCATTTCCCCAGTTTCTTTGGCC TCCGTCTTCTCAAGTCACATAGGGAAAGTACCTCCTGGAACCAGGCTGCAGTATGCAGGACCTGCCAGGCAGGCACTGGTGAAGGGCCTT GGGCCTATCATCCCCCCAACCCCACCTCACCCCACCCGCCTCCTCTAGTGGGGTGAGTCTGGGCTGGTGGACCAGAGAGGGTGTCACAGA CCCTCAGGGACTGCCCCATGGACACCTCTGACTGGTGTTAACAGTGTGAACATTTTCCCCGTCTTCAGTCCCTTAGAATGACGACAGCCC CTGGGGTTGGGGCAGGCGAGTGTGGCCACATCATCCAAGCCCTCCCAGAGACACAAATAGGCTTTTTTGCTCTAAAAATAAATACCAGCC CTTTTTTGGTCACAAATCCAGCATCTCAGCAGAAAACTGCCTGACATGAAAAGTCCCCTGAGGAACTGCATCTGCGTTTCAGGGGCTTTT CATTTTTTCTCCTTTTTTAAAGTGTAGATTGTGGGTGCTTCCTAGAGGCCTGCCTTCTTCTGGAACTGGAAGTGGGCTATCACCATGGGC AAGCCCTTGGGTGCAGGCTCCCCACCTGCCTGGGAACTCTGGCAGCTCTCCTCAGCTCCTTGGGCTTGAGCAGCTGCAACTGCCCCAGAT TTGCTGTGGAAGCAGGGGCTAGCCCTGGCCTCACCAGGGCCTCCCGGGGCCCTGCATTGATGCTCAGGAGTTCCTGGGCTGCTCTTGATC CTTTCTGGGCATCCAGCTTCCAGTTAAGCTCTGTTTGCCAAACAAACTATTCTCAGCTGCCCTTTGGCCTGCGCCTGATGTGTTCCTGTT GCAGTCCCGCCTGCCTGAGACAGGAGCAGGCAGGAGAGCCTTCATGCCCAGATTCCCACAGGACAATTGGGGAGCTGCTGGCATTGTCTT TCTGGGAAGATTCTGCTTTCTTGGACCAAATGGCAGCCTGATTACCAGTGTCGGGCCTGCATGCTGCCCCCGACACACGCACGCACGCGC ACACACGTGTGCACATGGGCCATAGCCACAAGCCAGCTCTCCTCCAGGGTCCTTTCAACCTCGCTGTCCAGGGACCCTGTCCTTCTTGCC CGTGGGGCTTCCATCTGGCAGAGAACGTTCAGGGCTTGTTGAACTTGAAAGCTCATTAGACTTAAGCTGTCACCTGTGCTTGGTGCCCCA GGAACAGCCAGAGAGGACAGTGCCCACTCACTTCTTGTTGGCAGCCTCCTGTGCAGGAAGTGCCAGCCGGGCCTCGACGCACCAGCTGGC TGTGGGTCCTGAGGAGGGGCGGGAGGCGGCCGCTCAGTGCAGATGGGGACTCCTCTCCTCTGCCCTGACCTTACCCTCCATTACCTCCTT CACTGGAGTGGGGCTGGGGGGTGGGTGGAATCAGTGTTTTAATCGGATTTTTAAAAAACATTTTATTTCTTTGTACAATTACCATCCTAT >31316_31316_3_FRMD3-BRD4_FRMD3_chr9_85987828_ENST00000304195_BRD4_chr19_15365073_ENST00000371835_length(amino acids)=174AA_BP=135 MSRAQVSRTVRASVCVRAGEHRAGPEPPARSPRPRCMFASCHCVPRGRRTMKMIHFRSSSVKSLSQEMRCTIRLLDDSEISCHIQRETKG -------------------------------------------------------------- >31316_31316_4_FRMD3-BRD4_FRMD3_chr9_85987828_ENST00000376438_BRD4_chr19_15365073_ENST00000263377_length(transcript)=4193nt_BP=610nt CGCGAGCCCGCCGGGGGTCTCAGGCTCTGGGCGTCCTCGGCGAGCGAGCCCGGGCAGAGGGAGGCGCACGAGCGCGCGCGACAGAAGGAG GCGGGGAAAGGAGGGGGCGAGGCGGAGGCGAGCGAACAGAGGGAGGGACCCGCCCGCCGCGCCCCGGCCGCTGGGCATGTGTGTCCGCAG GCGCCCGACGCTGCCGATGTCCCGGGGCTGAGCCGCGCCCAGGTGTCCCGGACAGTGCGTGCGAGCGTGTGTGTCCGCGCAGGCGAGCAC CGCGCCGGCCCTGAGCCTCCCGCTCGCTCCCCACGGCCGCGGTGCATGTTCGCCTCCTGCCACTGTGTGCCGAGAGGCAGGAGGACCATG AAAATGATCCACTTTCGGAGCTCCAGCGTCAAATCGCTCAGCCAGGAGATGAGATGCACCATCCGGCTGCTGGACGACTCGGAGATCTCC TGCCACATCCAGAGGGAAACCAAAGGGCAGTTTCTCATTGACCACATCTGCAACTACTACAGCCTGCTGGAGAAGGACTACTTTGGCATT CGCTATGTGGACCCAGAGAAGCAAAGGCACTGGCTTGAACCTAACAAGTCCATCTTCAAGCAAATGAAAACTGAGAAAGTTGATGTGATT GCCGGCTCCTCCAAGATGAAGGGCTTCTCGTCCTCAGAGTCGGAGAGCTCCAGTGAGTCCAGCTCCTCTGACAGCGAAGACTCCGAAACA GAGATGGCTCCGAAGTCAAAAAAGAAGGGGCACCCCGGGAGGGAGCAGAAGAAGCACCATCATCACCACCATCAGCAGATGCAGCAGGCC CCGGCTCCTGTGCCCCAGCAGCCGCCCCCGCCTCCCCAGCAGCCCCCACCGCCTCCACCTCCGCAGCAGCAACAGCAGCCGCCACCCCCG CCTCCCCCACCCTCCATGCCGCAGCAGGCAGCCCCGGCGATGAAGTCCTCGCCCCCACCCTTCATTGCCACCCAGGTGCCCGTCCTGGAG CCCCAGCTCCCAGGCAGCGTCTTTGACCCCATCGGCCACTTCACCCAGCCCATCCTGCACCTGCCGCAGCCTGAGCTGCCCCCTCACCTG CCCCAGCCGCCTGAGCACAGCACTCCACCCCATCTCAACCAGCACGCAGTGGTCTCTCCTCCAGCTTTGCACAACGCACTACCCCAGCAG CCATCACGGCCCAGCAACCGAGCCGCTGCCCTGCCTCCCAAGCCCGCCCGGCCCCCAGCCGTGTCACCAGCCTTGACCCAAACACCCCTG CTCCCACAGCCCCCCATGGCCCAACCCCCCCAAGTGCTGCTGGAGGATGAAGAGCCACCTGCCCCACCCCTCACCTCCATGCAGATGCAG CTGTACCTGCAGCAGCTGCAGAAGGTGCAGCCCCCTACGCCGCTACTCCCTTCCGTGAAGGTGCAGTCCCAGCCCCCACCCCCCCTGCCG CCCCCACCCCACCCCTCTGTGCAGCAGCAGCTGCAGCAGCAGCCGCCACCACCCCCACCACCCCAGCCCCAGCCTCCACCCCAGCAGCAG CATCAGCCCCCTCCACGGCCCGTGCACTTGCAGCCCATGCAGTTTTCCACCCACATCCAACAGCCCCCGCCACCCCAGGGCCAGCAGCCC CCCCATCCGCCCCCAGGCCAGCAGCCACCCCCGCCGCAGCCTGCCAAGCCTCAGCAAGTCATCCAGCACCACCATTCACCCCGGCACCAC AAGTCGGACCCCTACTCAACCGGTCACCTCCGCGAAGCCCCCTCCCCGCTTATGATACATTCCCCCCAGATGTCACAGTTCCAGAGCCTG ACCCACCAGTCTCCACCCCAGCAAAACGTCCAGCCTAAGAAACAGGAGCTGCGTGCTGCCTCCGTGGTCCAGCCCCAGCCCCTCGTGGTG GTGAAGGAGGAGAAGATCCACTCACCCATCATCCGCAGCGAGCCCTTCAGCCCCTCGCTGCGGCCGGAGCCCCCCAAGCACCCGGAGAGC ATCAAGGCCCCCGTCCACCTGCCCCAGCGGCCGGAAATGAAGCCTGTGGATGTCGGGAGGCCTGTGATCCGGCCCCCAGAGCAGAACGCA CCGCCACCAGGGGCCCCTGACAAGGACAAACAGAAACAGGAGCCGAAGACTCCAGTTGCGCCCAAAAAGGACCTGAAAATCAAGAACATG GGCTCCTGGGCCAGCCTAGTGCAGAAGCATCCGACCACCCCCTCCTCCACAGCCAAGTCATCCAGCGACAGCTTCGAGCAGTTCCGCCGC GCCGCTCGGGAGAAAGAGGAGCGTGAGAAGGCCCTGAAGGCTCAGGCCGAGCACGCTGAGAAGGAGAAGGAGCGGCTGCGGCAGGAGCGC ATGAGGAGCCGAGAGGACGAGGATGCGCTGGAGCAGGCCCGGCGGGCCCATGAGGAGGCACGTCGGCGCCAGGAGCAGCAGCAGCAGCAG CGCCAGGAGCAACAGCAGCAGCAGCAACAGCAAGCAGCTGCGGTGGCTGCCGCCGCCACCCCACAGGCCCAGAGCTCCCAGCCCCAGTCC ATGCTGGACCAGCAGAGGGAGTTGGCCCGGAAGCGGGAGCAGGAGCGAAGACGCCGGGAAGCCATGGCAGCTACCATTGACATGAATTTC CAGAGTGATCTATTGTCAATATTTGAAGAAAATCTTTTCTGAGCGCACCTAGGTGGCTTCTGACTTTGATTTTCTGGCAAAACATTGACT TTCCATAGTGTTAGGGGCGGTGGTGGAGGTGGGATCAGCGGCCAGGGGATGCCTCAGGGCCTGGCCCTCCTGCATGCTATGCCCGGGGCA GGCCTGACGGGCAGCTGAGGATTGCAGAGCCTGTCTGCCTTACGGCCAGTCGGACAGACGTCCCGCCACCCACCACCCCTCACAGGACGT CCGCTCAGCACACGCCTTGTTACGAGCAAGTGCCGGCTGGACCCAAGCCCTGCATCCCCACATGCGGGGCAGAGGCCCTTCTCTCCGCCA AATGTCTACACAGTATACACAGGACATCGTTGCTGCCGCCGTGACTGGTTTTCTGTCCCCAAGAACGTGACGTTCGTGATGTCCTGCCCG CCGGGAGTCTTTCCCCACACCCCAGCCATCGCCGCCCGCTCCCAGGAGGCCAGGGCAGGCCTGCGTGGGCTGGAGGCGGGCGAGGCCGGC CCACCCCCTCGCTGGCACTGACTTTGCCTTGAACAGACCCCCCGACCCTCCCCCACAAGCCTTTAATTGAGAGCCGCTCTCTGTAAGTGT TTGCTTGTGCAAAAGGGAATAGTGCCGTGGAGGTGTGTGTGTCCATGGCATCCGGAGCGAGGCGACTGTCCTGCGTGGGTAGCCCTCGGC CGGGGAGTGAGGCCACCAACCAAAGTCAGTTCCTTCCCACCTGTGTTTCTGTTTCGTTTTTTTTTTTCTTTTTTTTCTATATATATTTTT TGTTGAATTCTATTTTATTTTTAATTCTCTCTTCTCCTCCAGACACAATGGCACTGCTTATCTCCGAAATGGTGTGATCGTCTCCTCATT GAGCAGCGGCTGCCACCGCGCTGTGGGTAGTGTGTGACCGTGGCTGTACTGTATAGTGAACATAGTTGGCATATCTTTGTTTGAAGTTTG TTGGTGACTCCACCAAACTGGTGTGAAAAAAGAAAAAAGCTCAAAAAAATCCACAAAAAGACAAAACACACAAAAAAAATCCTGCCTATA TTTTACTCAGTTTCAAACTTTATTAGTCTATTTTTAATTATAAAACCAGAAAGCTACAATTTCTTTTCTTTCCCCTCCACCCCCCCCCCC CCCACCCATTTGTTGGCTTTTTTGTTTTTTAATGTCAGATCTGTTGAGTTGGTTTTTTTGGTTTTGGTTTTTGTTTTTGTTTTTGTTTTT TACTGAGAAAGGAAGGGCCAAGGGATGAGGTGGGAACCGGGCCCTGGGGGCGCCACAGACTAAGGCAGAGACTCCCCTACCTGGCGCCCA GCCCCAACCAGCTGGCCGCTCCTGCCCATGCTTTTTTTTTTTTTTTTTTTTAATTTTTATAATTGGAGCCCCTGGTGAGGTTACGCGTGC CATGAGAACCCACTCTACACCACGACGCTGGTGCCTCAGTGTTGGCCAAACTCTGGAGTCACTGACTGGTTTGACTTTCATACGGTGAAT >31316_31316_4_FRMD3-BRD4_FRMD3_chr9_85987828_ENST00000376438_BRD4_chr19_15365073_ENST00000263377_length(amino acids)=814AA_BP=424 MSRAQVSRTVRASVCVRAGEHRAGPEPPARSPRPRCMFASCHCVPRGRRTMKMIHFRSSSVKSLSQEMRCTIRLLDDSEISCHIQRETKG QFLIDHICNYYSLLEKDYFGIRYVDPEKQRHWLEPNKSIFKQMKTEKVDVIAGSSKMKGFSSSESESSSESSSSDSEDSETEMAPKSKKK GHPGREQKKHHHHHHQQMQQAPAPVPQQPPPPPQQPPPPPPPQQQQQPPPPPPPPSMPQQAAPAMKSSPPPFIATQVPVLEPQLPGSVFD PIGHFTQPILHLPQPELPPHLPQPPEHSTPPHLNQHAVVSPPALHNALPQQPSRPSNRAAALPPKPARPPAVSPALTQTPLLPQPPMAQP PQVLLEDEEPPAPPLTSMQMQLYLQQLQKVQPPTPLLPSVKVQSQPPPPLPPPPHPSVQQQLQQQPPPPPPPQPQPPPQQQHQPPPRPVH LQPMQFSTHIQQPPPPQGQQPPHPPPGQQPPPPQPAKPQQVIQHHHSPRHHKSDPYSTGHLREAPSPLMIHSPQMSQFQSLTHQSPPQQN VQPKKQELRAASVVQPQPLVVVKEEKIHSPIIRSEPFSPSLRPEPPKHPESIKAPVHLPQRPEMKPVDVGRPVIRPPEQNAPPPGAPDKD KQKQEPKTPVAPKKDLKIKNMGSWASLVQKHPTTPSSTAKSSSDSFEQFRRAAREKEEREKALKAQAEHAEKEKERLRQERMRSREDEDA LEQARRAHEEARRRQEQQQQQRQEQQQQQQQQAAAVAAAATPQAQSSQPQSMLDQQRELARKREQERRRREAMAATIDMNFQSDLLSIFE -------------------------------------------------------------- >31316_31316_5_FRMD3-BRD4_FRMD3_chr9_85987828_ENST00000376438_BRD4_chr19_15365073_ENST00000360016_length(transcript)=1513nt_BP=610nt CGCGAGCCCGCCGGGGGTCTCAGGCTCTGGGCGTCCTCGGCGAGCGAGCCCGGGCAGAGGGAGGCGCACGAGCGCGCGCGACAGAAGGAG GCGGGGAAAGGAGGGGGCGAGGCGGAGGCGAGCGAACAGAGGGAGGGACCCGCCCGCCGCGCCCCGGCCGCTGGGCATGTGTGTCCGCAG GCGCCCGACGCTGCCGATGTCCCGGGGCTGAGCCGCGCCCAGGTGTCCCGGACAGTGCGTGCGAGCGTGTGTGTCCGCGCAGGCGAGCAC CGCGCCGGCCCTGAGCCTCCCGCTCGCTCCCCACGGCCGCGGTGCATGTTCGCCTCCTGCCACTGTGTGCCGAGAGGCAGGAGGACCATG AAAATGATCCACTTTCGGAGCTCCAGCGTCAAATCGCTCAGCCAGGAGATGAGATGCACCATCCGGCTGCTGGACGACTCGGAGATCTCC TGCCACATCCAGAGGGAAACCAAAGGGCAGTTTCTCATTGACCACATCTGCAACTACTACAGCCTGCTGGAGAAGGACTACTTTGGCATT CGCTATGTGGACCCAGAGAAGCAAAGGCACTGGCTTGAACCTAACAAGTCCATCTTCAAGCAAATGAAAACTGAGAAAGTTGATGTGATT GCCGGCTCCTCCAAGATGAAGGGCTTCTCGTCCTCAGAGTCGGAGAGCTCCAGTGAGTCCAGCTCCTCTGACAGCGAAGACTCCGAAACA GCTTTCTGCACCAGTGGAGACTTCGTGTCCCCAGGGCCTTCCCCGTATCACAGTCACGTGCAGTGCGGCCGCTTCAGGGAGATGCTTCGC TGGTTTCTGGTGGATGTGGAGCAGACTGCAGCTGGCCAGCCGCATCGCCAGTCTGCTGCTGGCCCTGCCATCACCTGGGCCCCAGCCATT GCCTACCCCTCCCCAGAGTGTGCTCGTTGCTGTGTTGGCTGCTCCTGAATCTGCCCTAACTCCACACGCACCTGGACTTGCGTGTCCCTC CTGCAGTTCTAACTAACAGTCCCTTCTTTTCAAGCCCCGTGGGTCTGCACGTTGGACCCTGGGTTCCCCATTAGAGCCCACCTTCTGAGC AGCAGCCTCAGTGGGAGGTGGAGGCAGGTAGTGATGCTGGGTGCCAGGTGGGAGTGGAGAGGGGACTGCTCTCCTCCAAGTGTGCACTTT CCTCATTATTCTCAGGGGCGCAGATGCTCAGGCTGGCGCAGGGGAGAGGCTAGGGAGGGAGCCATGGTGCCCAGAAGGCCTGGCGACCAG CCCCTGCTGAGAGATGGAGCTAACATCCTGTGTTTACGGCCAACGGGGTTGCCGCTAGGCTGGTGCAGCTGTCAGTGAGCTGGCGTGCTG CAGACCACCTGAGAGCTGGCCCTAGGGTCTCAGGCAGACTGGGGAGTGGGGATCCACAGTGGGAAACCTGTGTTTTGGCAGTAGACTCCT >31316_31316_5_FRMD3-BRD4_FRMD3_chr9_85987828_ENST00000376438_BRD4_chr19_15365073_ENST00000360016_length(amino acids)=246AA_BP=135 MSRAQVSRTVRASVCVRAGEHRAGPEPPARSPRPRCMFASCHCVPRGRRTMKMIHFRSSSVKSLSQEMRCTIRLLDDSEISCHIQRETKG QFLIDHICNYYSLLEKDYFGIRYVDPEKQRHWLEPNKSIFKQMKTEKVDVIAGSSKMKGFSSSESESSSESSSSDSEDSETAFCTSGDFV -------------------------------------------------------------- >31316_31316_6_FRMD3-BRD4_FRMD3_chr9_85987828_ENST00000376438_BRD4_chr19_15365073_ENST00000371835_length(transcript)=2955nt_BP=610nt CGCGAGCCCGCCGGGGGTCTCAGGCTCTGGGCGTCCTCGGCGAGCGAGCCCGGGCAGAGGGAGGCGCACGAGCGCGCGCGACAGAAGGAG GCGGGGAAAGGAGGGGGCGAGGCGGAGGCGAGCGAACAGAGGGAGGGACCCGCCCGCCGCGCCCCGGCCGCTGGGCATGTGTGTCCGCAG GCGCCCGACGCTGCCGATGTCCCGGGGCTGAGCCGCGCCCAGGTGTCCCGGACAGTGCGTGCGAGCGTGTGTGTCCGCGCAGGCGAGCAC CGCGCCGGCCCTGAGCCTCCCGCTCGCTCCCCACGGCCGCGGTGCATGTTCGCCTCCTGCCACTGTGTGCCGAGAGGCAGGAGGACCATG AAAATGATCCACTTTCGGAGCTCCAGCGTCAAATCGCTCAGCCAGGAGATGAGATGCACCATCCGGCTGCTGGACGACTCGGAGATCTCC TGCCACATCCAGAGGGAAACCAAAGGGCAGTTTCTCATTGACCACATCTGCAACTACTACAGCCTGCTGGAGAAGGACTACTTTGGCATT CGCTATGTGGACCCAGAGAAGCAAAGGCACTGGCTTGAACCTAACAAGTCCATCTTCAAGCAAATGAAAACTGAGAAAGTTGATGTGATT GCCGGCTCCTCCAAGATGAAGGGCTTCTCGTCCTCAGAGTCGGAGAGCTCCAGTGAGTCCAGCTCCTCTGACAGCGAAGACTCCGAAACA GGTCCTGCCTAATCATTGGACACGGACTCTTAATAAAACGGTCTTCAGTTCCAGATTCCTTCCCAGCAAGCTATAGCTTAAGTCCATTTT CTTCCGTGAAAGGGACAGGACTCCATCAAGTTATGGAATTCCTCAGAGCCCTGGGCCTGTCCCCCGGGGTGGATTAGTCATGTCCAGCAG CACACGCCTAGTCCCGCCTTCGGGAAGGCTGCCTGCCTGGCCAGCCGCCCAGGCCTCTCTGTGTAAAGACTGCCTGGCTGTCCTGCCCAG CCTTCCTGGTTCTCTGGGGTCCTCTGGGTGGGTGGCATCTCCTGGAGGGTGATGACAATCCCCAACACATGCATTCATGTGGTGCTACTC TGTGTGCAAAGCCAGACCCCAAGTATGTTTTCTCTCTTTGTCCCATCCCTCTTTTTCTGGGACTTTGGACCCTAACTACTTCCCTCCTGA ACCTTGCAGTGACATCAGTCCAGGAGAGCTCTCGTTCAGTGTGCGGAAGAACACTCTGACCTCTAGAGCTGTCCTAGATAAGGAGTGGGA GCTTTAGAGGCAAGGCCTCTAGACCCTGGAAGGCTCAGTGAGGCTCTTCCCACAGCATGCTTCTCACTGGTGCCCTGTAAGGCTCGAGCC ACCGCTGACTCTGAGCCTTTTGGAGTCTTTCCTCCTTCGTCTCCATTGTTCCCGTGCATTTCCAAAAGCTTAAGTTGCCTGGTGGGCATT TCCCCAGTTTCTTTGGCCTCCGTCTTCTCAAGTCACATAGGGAAAGTACCTCCTGGAACCAGGCTGCAGTATGCAGGACCTGCCAGGCAG GCACTGGTGAAGGGCCTTGGGCCTATCATCCCCCCAACCCCACCTCACCCCACCCGCCTCCTCTAGTGGGGTGAGTCTGGGCTGGTGGAC CAGAGAGGGTGTCACAGACCCTCAGGGACTGCCCCATGGACACCTCTGACTGGTGTTAACAGTGTGAACATTTTCCCCGTCTTCAGTCCC TTAGAATGACGACAGCCCCTGGGGTTGGGGCAGGCGAGTGTGGCCACATCATCCAAGCCCTCCCAGAGACACAAATAGGCTTTTTTGCTC TAAAAATAAATACCAGCCCTTTTTTGGTCACAAATCCAGCATCTCAGCAGAAAACTGCCTGACATGAAAAGTCCCCTGAGGAACTGCATC TGCGTTTCAGGGGCTTTTCATTTTTTCTCCTTTTTTAAAGTGTAGATTGTGGGTGCTTCCTAGAGGCCTGCCTTCTTCTGGAACTGGAAG TGGGCTATCACCATGGGCAAGCCCTTGGGTGCAGGCTCCCCACCTGCCTGGGAACTCTGGCAGCTCTCCTCAGCTCCTTGGGCTTGAGCA GCTGCAACTGCCCCAGATTTGCTGTGGAAGCAGGGGCTAGCCCTGGCCTCACCAGGGCCTCCCGGGGCCCTGCATTGATGCTCAGGAGTT CCTGGGCTGCTCTTGATCCTTTCTGGGCATCCAGCTTCCAGTTAAGCTCTGTTTGCCAAACAAACTATTCTCAGCTGCCCTTTGGCCTGC GCCTGATGTGTTCCTGTTGCAGTCCCGCCTGCCTGAGACAGGAGCAGGCAGGAGAGCCTTCATGCCCAGATTCCCACAGGACAATTGGGG AGCTGCTGGCATTGTCTTTCTGGGAAGATTCTGCTTTCTTGGACCAAATGGCAGCCTGATTACCAGTGTCGGGCCTGCATGCTGCCCCCG ACACACGCACGCACGCGCACACACGTGTGCACATGGGCCATAGCCACAAGCCAGCTCTCCTCCAGGGTCCTTTCAACCTCGCTGTCCAGG GACCCTGTCCTTCTTGCCCGTGGGGCTTCCATCTGGCAGAGAACGTTCAGGGCTTGTTGAACTTGAAAGCTCATTAGACTTAAGCTGTCA CCTGTGCTTGGTGCCCCAGGAACAGCCAGAGAGGACAGTGCCCACTCACTTCTTGTTGGCAGCCTCCTGTGCAGGAAGTGCCAGCCGGGC CTCGACGCACCAGCTGGCTGTGGGTCCTGAGGAGGGGCGGGAGGCGGCCGCTCAGTGCAGATGGGGACTCCTCTCCTCTGCCCTGACCTT ACCCTCCATTACCTCCTTCACTGGAGTGGGGCTGGGGGGTGGGTGGAATCAGTGTTTTAATCGGATTTTTAAAAAACATTTTATTTCTTT >31316_31316_6_FRMD3-BRD4_FRMD3_chr9_85987828_ENST00000376438_BRD4_chr19_15365073_ENST00000371835_length(amino acids)=201AA_BP= MLEDGLVRFKPVPLLLWVHIANAKVVLLQQAVVVADVVNEKLPFGFPLDVAGDLRVVQQPDGASHLLAERFDAGAPKVDHFHGPPASRHT VAGGEHAPRPWGASGRLRAGAVLACADTHARTHCPGHLGAAQPRDIGSVGRLRTHMPSGRGAAGGSLPLFARLRLAPSFPRLLLSRALVR -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for FRMD3-BRD4 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for FRMD3-BRD4 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for FRMD3-BRD4 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
