
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:FRMD4A-MCM10 (FusionGDB2 ID:31329) |
Fusion Gene Summary for FRMD4A-MCM10 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: FRMD4A-MCM10 | Fusion gene ID: 31329 | Hgene | Tgene | Gene symbol | FRMD4A | MCM10 | Gene ID | 55691 | 55388 |
| Gene name | FERM domain containing 4A | minichromosome maintenance 10 replication initiation factor | |
| Synonyms | CCAFCA|FRMD4|bA295P9.4 | CNA43|DNA43|PRO2249 | |
| Cytomap | 10p13 | 10p13 | |
| Type of gene | protein-coding | protein-coding | |
| Description | FERM domain-containing protein 4AFERM domain containing 4 | protein MCM10 homologMCM10 minichromosome maintenance deficient 10homolog of yeast MCM10hsMCM10minichromosome maintenance complex component 10 | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | Q9P2Q2 | Q7L590 | |
| Ensembl transtripts involved in fusion gene | ENST00000342409, ENST00000358621, ENST00000378503, ENST00000357447, ENST00000475141, ENST00000492155, ENST00000493380, | ENST00000378694, ENST00000378714, ENST00000484800, | |
| Fusion gene scores | * DoF score | 16 X 15 X 7=1680 | 2 X 2 X 2=8 |
| # samples | 17 | 2 | |
| ** MAII score | log2(17/1680*10)=-3.30485458152842 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(2/8*10)=1.32192809488736 | |
| Context | PubMed: FRMD4A [Title/Abstract] AND MCM10 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | FRMD4A(13900847)-MCM10(13214376), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | FRMD4A-MCM10 seems lost the major protein functional domain in Hgene partner, which is a essential gene due to the frame-shifted ORF. FRMD4A-MCM10 seems lost the major protein functional domain in Tgene partner, which is a essential gene due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
 Fusion gene breakpoints across FRMD4A (5'-gene) Fusion gene breakpoints across FRMD4A (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
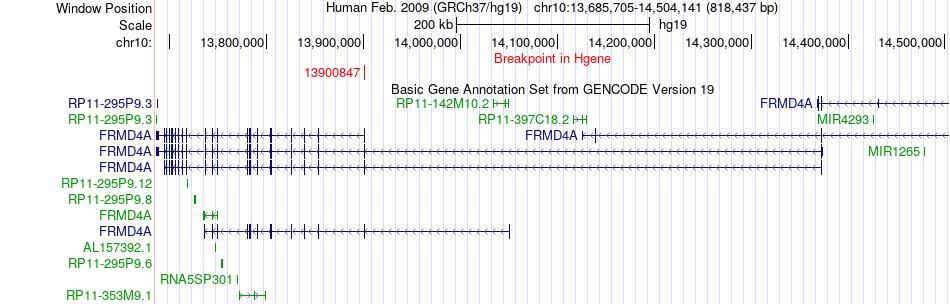 |
 Fusion gene breakpoints across MCM10 (3'-gene) Fusion gene breakpoints across MCM10 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
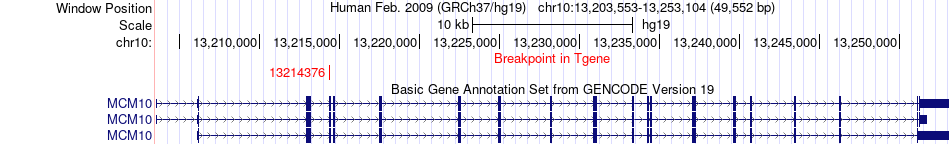 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | PRAD | TCGA-EJ-5525-01A | FRMD4A | chr10 | 13900847 | - | MCM10 | chr10 | 13214376 | + |
Top |
Fusion Gene ORF analysis for FRMD4A-MCM10 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| Frame-shift | ENST00000342409 | ENST00000378694 | FRMD4A | chr10 | 13900847 | - | MCM10 | chr10 | 13214376 | + |
| Frame-shift | ENST00000342409 | ENST00000378714 | FRMD4A | chr10 | 13900847 | - | MCM10 | chr10 | 13214376 | + |
| Frame-shift | ENST00000342409 | ENST00000484800 | FRMD4A | chr10 | 13900847 | - | MCM10 | chr10 | 13214376 | + |
| Frame-shift | ENST00000358621 | ENST00000378694 | FRMD4A | chr10 | 13900847 | - | MCM10 | chr10 | 13214376 | + |
| Frame-shift | ENST00000358621 | ENST00000378714 | FRMD4A | chr10 | 13900847 | - | MCM10 | chr10 | 13214376 | + |
| Frame-shift | ENST00000358621 | ENST00000484800 | FRMD4A | chr10 | 13900847 | - | MCM10 | chr10 | 13214376 | + |
| Frame-shift | ENST00000378503 | ENST00000378694 | FRMD4A | chr10 | 13900847 | - | MCM10 | chr10 | 13214376 | + |
| Frame-shift | ENST00000378503 | ENST00000378714 | FRMD4A | chr10 | 13900847 | - | MCM10 | chr10 | 13214376 | + |
| Frame-shift | ENST00000378503 | ENST00000484800 | FRMD4A | chr10 | 13900847 | - | MCM10 | chr10 | 13214376 | + |
| In-frame | ENST00000357447 | ENST00000378694 | FRMD4A | chr10 | 13900847 | - | MCM10 | chr10 | 13214376 | + |
| In-frame | ENST00000357447 | ENST00000378714 | FRMD4A | chr10 | 13900847 | - | MCM10 | chr10 | 13214376 | + |
| In-frame | ENST00000357447 | ENST00000484800 | FRMD4A | chr10 | 13900847 | - | MCM10 | chr10 | 13214376 | + |
| intron-3CDS | ENST00000475141 | ENST00000378694 | FRMD4A | chr10 | 13900847 | - | MCM10 | chr10 | 13214376 | + |
| intron-3CDS | ENST00000475141 | ENST00000378714 | FRMD4A | chr10 | 13900847 | - | MCM10 | chr10 | 13214376 | + |
| intron-3CDS | ENST00000475141 | ENST00000484800 | FRMD4A | chr10 | 13900847 | - | MCM10 | chr10 | 13214376 | + |
| intron-3CDS | ENST00000492155 | ENST00000378694 | FRMD4A | chr10 | 13900847 | - | MCM10 | chr10 | 13214376 | + |
| intron-3CDS | ENST00000492155 | ENST00000378714 | FRMD4A | chr10 | 13900847 | - | MCM10 | chr10 | 13214376 | + |
| intron-3CDS | ENST00000492155 | ENST00000484800 | FRMD4A | chr10 | 13900847 | - | MCM10 | chr10 | 13214376 | + |
| intron-3CDS | ENST00000493380 | ENST00000378694 | FRMD4A | chr10 | 13900847 | - | MCM10 | chr10 | 13214376 | + |
| intron-3CDS | ENST00000493380 | ENST00000378714 | FRMD4A | chr10 | 13900847 | - | MCM10 | chr10 | 13214376 | + |
| intron-3CDS | ENST00000493380 | ENST00000484800 | FRMD4A | chr10 | 13900847 | - | MCM10 | chr10 | 13214376 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000357447 | FRMD4A | chr10 | 13900847 | - | ENST00000378714 | MCM10 | chr10 | 13214376 | + | 4550 | 480 | 494 | 2755 | 753 |
| ENST00000357447 | FRMD4A | chr10 | 13900847 | - | ENST00000484800 | MCM10 | chr10 | 13214376 | + | 3185 | 480 | 494 | 2758 | 754 |
| ENST00000357447 | FRMD4A | chr10 | 13900847 | - | ENST00000378694 | MCM10 | chr10 | 13214376 | + | 4643 | 480 | 494 | 2698 | 734 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000357447 | ENST00000378714 | FRMD4A | chr10 | 13900847 | - | MCM10 | chr10 | 13214376 | + | 0.00063766 | 0.9993624 |
| ENST00000357447 | ENST00000484800 | FRMD4A | chr10 | 13900847 | - | MCM10 | chr10 | 13214376 | + | 0.003960487 | 0.9960395 |
| ENST00000357447 | ENST00000378694 | FRMD4A | chr10 | 13900847 | - | MCM10 | chr10 | 13214376 | + | 0.000666906 | 0.9993331 |
Top |
Fusion Genomic Features for FRMD4A-MCM10 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| FRMD4A | chr10 | 13900846 | - | MCM10 | chr10 | 13214375 | + | 6.49E-06 | 0.99999356 |
| FRMD4A | chr10 | 13900846 | - | MCM10 | chr10 | 13214375 | + | 6.49E-06 | 0.99999356 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
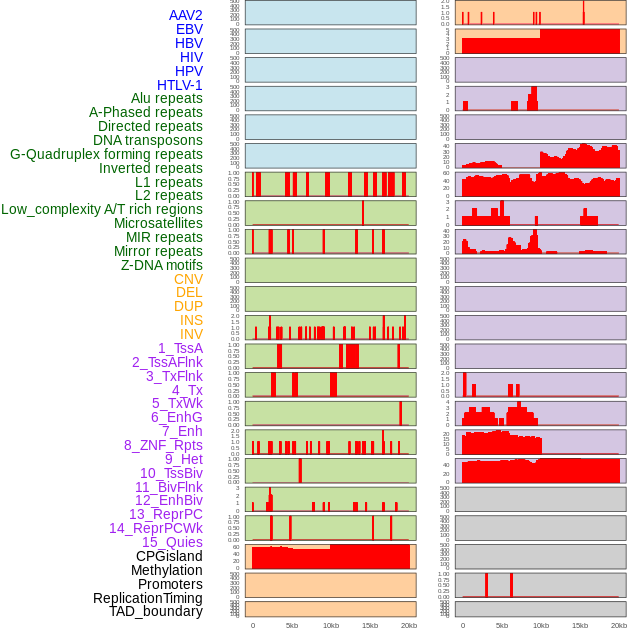 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
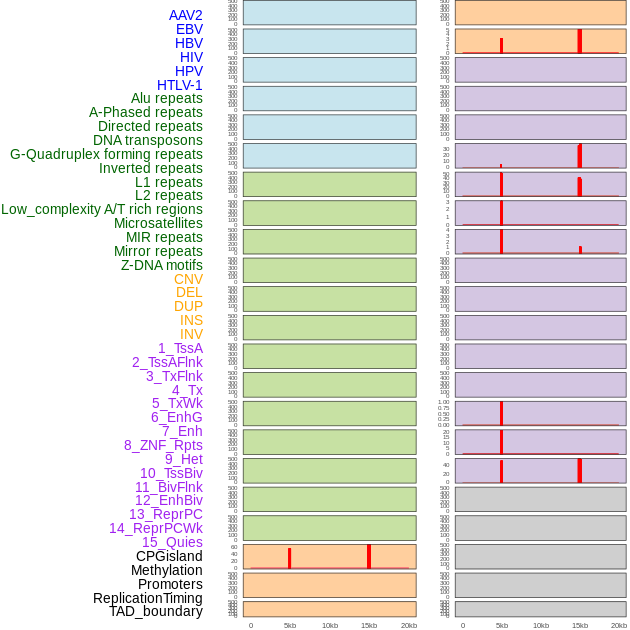 |
Top |
Fusion Protein Features for FRMD4A-MCM10 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr10:13900847/chr10:13214376) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| FRMD4A | MCM10 |
| FUNCTION: Scaffolding protein that regulates epithelial cell polarity by connecting ARF6 activation with the PAR3 complex (By similarity). Plays a redundant role with FRMD4B in epithelial polarization (By similarity). May regulate MAPT secretion by activating ARF6-signaling (PubMed:27044754). {ECO:0000250|UniProtKB:Q8BIE6, ECO:0000269|PubMed:27044754}. | FUNCTION: Acts as a replication initiation factor that brings together the MCM2-7 helicase and the DNA polymerase alpha/primase complex in order to initiate DNA replication. Additionally, plays a role in preventing DNA damage during replication. Key effector of the RBBP6 and ZBTB38-mediated regulation of DNA-replication and common fragile sites stability; acts as a direct target of transcriptional repression by ZBTB38 (PubMed:24726359). {ECO:0000269|PubMed:11095689, ECO:0000269|PubMed:15136575, ECO:0000269|PubMed:17699597, ECO:0000269|PubMed:19608746, ECO:0000269|PubMed:24726359}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | MCM10 | chr10:13900847 | chr10:13214376 | ENST00000378714 | 2 | 20 | 238_389 | 116 | 875.0 | Region | OB-fold domain | |
| Tgene | MCM10 | chr10:13900847 | chr10:13214376 | ENST00000378714 | 2 | 20 | 390_415 | 116 | 875.0 | Region | Note=Zinc finger-like 1 | |
| Tgene | MCM10 | chr10:13900847 | chr10:13214376 | ENST00000378714 | 2 | 20 | 783_802 | 116 | 875.0 | Region | Zinc finger-like 2 | |
| Tgene | MCM10 | chr10:13900847 | chr10:13214376 | ENST00000378714 | 2 | 20 | 816_836 | 116 | 875.0 | Region | Zinc finger-like 3 | |
| Tgene | MCM10 | chr10:13900847 | chr10:13214376 | ENST00000484800 | 2 | 20 | 238_389 | 116 | 876.0 | Region | OB-fold domain | |
| Tgene | MCM10 | chr10:13900847 | chr10:13214376 | ENST00000484800 | 2 | 20 | 390_415 | 116 | 876.0 | Region | Note=Zinc finger-like 1 | |
| Tgene | MCM10 | chr10:13900847 | chr10:13214376 | ENST00000484800 | 2 | 20 | 783_802 | 116 | 876.0 | Region | Zinc finger-like 2 | |
| Tgene | MCM10 | chr10:13900847 | chr10:13214376 | ENST00000484800 | 2 | 20 | 816_836 | 116 | 876.0 | Region | Zinc finger-like 3 |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | FRMD4A | chr10:13900847 | chr10:13214376 | ENST00000357447 | - | 3 | 25 | 382_416 | 37 | 2034.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | FRMD4A | chr10:13900847 | chr10:13214376 | ENST00000378503 | - | 2 | 23 | 382_416 | 37 | 1040.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | FRMD4A | chr10:13900847 | chr10:13214376 | ENST00000357447 | - | 3 | 25 | 627_756 | 37 | 2034.0 | Compositional bias | Note=Ser-rich |
| Hgene | FRMD4A | chr10:13900847 | chr10:13214376 | ENST00000357447 | - | 3 | 25 | 935_1007 | 37 | 2034.0 | Compositional bias | Note=Ser-rich |
| Hgene | FRMD4A | chr10:13900847 | chr10:13214376 | ENST00000378503 | - | 2 | 23 | 627_756 | 37 | 1040.0 | Compositional bias | Note=Ser-rich |
| Hgene | FRMD4A | chr10:13900847 | chr10:13214376 | ENST00000378503 | - | 2 | 23 | 935_1007 | 37 | 1040.0 | Compositional bias | Note=Ser-rich |
| Hgene | FRMD4A | chr10:13900847 | chr10:13214376 | ENST00000357447 | - | 3 | 25 | 20_322 | 37 | 2034.0 | Domain | FERM |
| Hgene | FRMD4A | chr10:13900847 | chr10:13214376 | ENST00000378503 | - | 2 | 23 | 20_322 | 37 | 1040.0 | Domain | FERM |
| Tgene | MCM10 | chr10:13900847 | chr10:13214376 | ENST00000378714 | 2 | 20 | 104_142 | 116 | 875.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | MCM10 | chr10:13900847 | chr10:13214376 | ENST00000484800 | 2 | 20 | 104_142 | 116 | 876.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | MCM10 | chr10:13900847 | chr10:13214376 | ENST00000378714 | 2 | 20 | 1_156 | 116 | 875.0 | Region | N-terminal domain | |
| Tgene | MCM10 | chr10:13900847 | chr10:13214376 | ENST00000484800 | 2 | 20 | 1_156 | 116 | 876.0 | Region | N-terminal domain |
Top |
Fusion Gene Sequence for FRMD4A-MCM10 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >31329_31329_1_FRMD4A-MCM10_FRMD4A_chr10_13900847_ENST00000357447_MCM10_chr10_13214376_ENST00000378694_length(transcript)=4643nt_BP=480nt GAACGCTCACAGAACAGGCAGTGCAATTCCATGTTCCTCTTAAGTATGTTAGCCCTACCGGGAGCTGAGCTGGCCAGTCTACTTGGAGAG GAAAAGTAGATCTGGGGAAGGTGGAAGGGTCAGTTCCTAAGTGACTTCCTCCTCGGGGATGGTAAGGGCATTTGCTGATCTCCAGTGACT GCCTGGTGCCTCATGGTCAGACTCGGCTGTCTCACTCCCAGATATCTGATTTTGCAAAAAGGGACACACCTATCTGCAGCAAAGAAGACA CTGACCAGATTGCGAGCGGTGCTTTTGGATGCTCTGTAGCCACCCGGGGCCCAGGAGGACTGACTCGGCAGCAGGATTCGTGCATGGGAA TCGGAGACCATGGCAGTGCAGCTGGTGCCCGACTCAGCTCTCGGCCTGCTGATGATGACGGAGGGCCGCCGATGTCAAGTACATCTTCTT GATGACAGGAAGCTGGAACTCCTAGTACAGAGGAATTAAGGAATTTGCAAGAGCAAATGAAGGCCTTACAAGAGCAGCTAAAAGTAACAA CAATTAAACAGACAGCAAGCCCAGCCCGTCTGCAAAAATCCCCTGAGAAGTCTCCCCGGCCACCTCTTAAGGAGAGGAGAGTTCAGAGAA TTCAGGAGTCAACATGCTTTTCTGCGGAGCTTGATGTCCCTGCGCTACCAAGAACCAAGAGGGTGGCTCGAACACCAAAGGCTTCACCTC CAGATCCCAAAAGCTCATCTTCAAGGATGACAAGTGCACCCTCCCAACCCCTACAGACGATTTCTCGGAACAAACCTAGTGGGATAACTA GAGGTCAAATTGTGGGGACCCCAGGAAGTTCTGGGGAAACGACTCAACCCATCTGTGTGGAAGCCTTCTCTGGTCTGCGGCTCAGGCGGC CTCGAGTATCCTCCACAGAAATGAACAAGAAAATGACCGGCCGAAAACTGATCAGACTGTCTCAGATCAAGGAAAAGATGGCCAGAGAGA AGCTGGAAGAAATAGATTGGGTGACATTTGGGGTTATATTGAAGAAGGTTACGCCACAGAGTGTGAATAGTGGAAAAACCTTCAGCATAT GGAAACTGAATGATCTTCGTGACCTGACACAATGTGTGTCCTTGTTCTTATTTGGAGAAGTTCACAAAGCGCTCTGGAAGACGGAGCAGG GGACTGTCGTAGGGATCCTCAATGCCAACCCCATGAAGCCCAAGGATGGTTCAGAGGAGGTGTGTTTATCTATCGATCATCCTCAGAAGG TCTTAATTATGGGTGAAGCTCTTGACCTGGGAACCTGTAAAGCCAAGAAGAAGAATGGAGAGCCGTGCACGCAGACTGTGAATTTGCGTG ACTGTGAGTACTGTCAGTACCATGTCCAGGCTCAGTACAAGAAGCTCAGCGCAAAGCGTGCGGATCTGCAGTCCACCTTCTCTGGAGGAC GAATTCCAAAGAAGTTTGCCCGCAGAGGCACCAGCCTCAAAGAACGGCTGTGCCAAGATGGCTTTTACTACGGAGGGGTTTCTTCTGCCT CGTATGCAGCTTCAATTGCAGCAGCTGTGGCTCCTAAGAAGAAGATTCAAACCACTCTGAGTAATCTGGTTGTTAAGGGCACAAACTTGA TCATCCAGGAAACACGGCAAAAACTCGGAATACCCCAGAAGAGCCTGTCTTGCTCTGAGGAGTTCAAGGAACTGATGGACCTGCCGACGT GTGGAGCCAGGAACTTAAAACAACATTTAGCCAAAGCCACAGCTTCAGGGATTATGGGGAGCCCAAAACCAGCCATCAAGTCCATCTCGG CCTCAGCACTCTTGAAGCAACAGAAGCAGCGGATGTTGGAGATGAGGAGAAGGAAATCAGAAGAAATACAGAAGCGATTTCTGCAGAGCT CAAGTGAAGTTGAGAGCCCAGCTGTGCCATCTTCATCAAGACAGCCCCCTGCTCAGCCTCCACGGACAGGATCCGAGTTCCCCAGGCTGG AGGGAGCCCCGGCCACAATGACGCCCAAGCTGGGGCGAGGTGTCTTGGAAGGAGATGATGTTCTCTTTTATGATGAGTCACCACCACCAA GACCAAAACTGAGTGCTTTAGCAGAAGCCAAAAAGTTAGCTGCTATCACCAAATTAAGGGCAAAAGGCCAGGTTCTTACAAAAACAAACC CAAACAGCATTAAGAAGAAACAAAAGGACCCTCAGGACATCCTGGAGGTGAAGGAACGTGTAGAAAAAAACACCATGTTTTCTTCTCAAG CTGAGGATGAATTGGAGCCTGCCAGGAAAAAAAGGAGAGAACAACTTGCCTATCTGGAATCTGAGGAATTTCAGAAAATCCTAAAAGCAA AATCAAAACACACAGGCATCCTGAAAGAGGCCGAGGCTGAGATGCAGGAGCGCTACTTTGAGCCACTGGTGAAAAAAGAACAAATGGAAG AAAAGATGAGAAACATCAGAGAAGTGAAGTGCCGTGTCGTGACATGCAAGACGTGCGCCTATACCCACTTCAAGCTGCTGGAGACCTGCG TCAGTGAGCAGCATGAATACCACTGGCATGATGGTGTGAAGAGGTTTTTCAAATGTCCCTGTGGAAACAGAAGCATCTCCTTGGACAGAC TCCCGAACAAGCACTGCAGTAACTGTGGCCTCTACAAATGGGAACGGGACGGAATGCTAAAGGTATGCCATTTGCGTACTAATTTTTGAC TCCTTTTAGTGACCCATGCTAATAATGTGGAACCATCTCCTATTAAAATATTTTCATTTTTCTAGGAAAAGACTGGTCCAAAGATAGGAG GAGAAACTCTGTTACCAAGAGGAGAAGAACATGCTAAATTTCTGAACAGCCTTAAATAACCCGAACTTCAGACATTTTCCCACAGACTTC CTGGCCTCCTGTGACTCTGGAAAGCAAAGGATTGGCTGTGTATTGTCCATTGATTCCTGATTGACGCCGTCAAAAACAAATGCTTGTTAA GCCCATAAGCTTTGCCTGCTTACTTTCTGCCATTGGGTTGGTTTGATACCACATTTAACATTGACATTTAAGTGGAAAACCAAGTTATCA TTGTCTTTTCTAAGCTCAGTGTGGATGATTGCATTACTTCATTCACTGAAGTTTTTGCCCAAAAATTGGAAGGTAAACAGAGAGCTATGT TTCTGTATCTTTTGGTTATAGAGTGTTCACTTCTTTATCATAACAAAATTCTAGTGTTTATACGAACACCCAGAGGCAAAAGAATTTGGC TTAATTCTCACTCCAGGTAAGTAGCTTAACTTCTGGGCTTCAGTTTTCTCATCTGTAAAATCAGGAAGATTGGACTAAGTGATCCTGAAA TGTATTTTTTAGCACTGGATTTCTACAAATAATAAAACTTTCCCATCTAGATAATGATGATCACATAGTCTTGATGTACGGACATTAAAA GCCAGATTTCTTCATTCAATTCTGTTATCTCTGTTTTACTCTTTGAAATTGATCAAGCCACTGAATCACTTTGCATTTCAGTTTATATAT ATAGAGAGAAAGAAGGTGTCTGCTCTTACATTATTGTGGAGCCCTGTGATAGAAATATGTAAAATCTCATATTATTTTTTTTTTAATTTT TTTATTTTTTATGACAGGGTCTCACTATGTCACCCTGGCTGGAGTGCAGTAGTGCGATCGCGGCACACTGCAGCCTTGGCTTCCCTGGGC TCAAGCAGTCCTCCCACCTCAGTCTCCCAAATAGCTAGGACTACAGGCGTGCGTGACCAAGCCCAGCTAATTTTTGCATTTTTTGTAGAG ATGGGGTTTTGCCATGTTGCTCAGGCTGGTCTCAAACTCCTGAGCACTAGCAATCCACCCACCTCTGTTTCCAAAAAAAAAAAAAAAATG AAAGGTCAACCCCTATGCAAATTACCACAGCAAAGGTTTCATTCAGGAGATTCTTCCATCTGGGCAACCTGGTTTTCCAAATATCATTTG ACCTAAGTGAATGTTGATACTAGCTAAAGATTGGGTAAATTGGTTGAATTATTGTATTGAAGCTTGAGCTGTAGCTAAAAGTAATTTAGG TTTCCCCTAAGATGTTATTATGTTAGGGACATAACACTTTTGGGAGGTTGTTGTGGGAGATGGTTGATTTAGGTTTTCAAAAGCTAGAAA TAAAATTTACATGCCTTAGATTTCATAAAATTCTGCTCTAATTGGGTGGAAGGTGCTGTATCTAACTTGTGTTCCTCCTAAGGTTATGTC CTAATAACTATTCTTTTAGGAGTATACTTCTACTTTATAGAAGGTTGCTTTTCTTTTTAATTTTTTCTAACAAAGAAAAGAATAAAGTAT TTATTAATAAGAACCAGAAAGCACTTGAAACTGATGTTTTTAATGGCTCATTTAGGGTAGATTTATTTATCTCATTAACTTAAAACAGCT ATGTGTATGAAATAGGTCACAACAGAACTTGAACACCAGGTTGGTGTCTGAGCAATCCCTTTCTTATGGGAAAAACAATGTTCTTGTTTG AACAGAGGGTATCATTGCAGTCAGTATTCACGTGTATATTGTTATATAAGTTGTATAATATGCTTGTAAAGGCTGAGGGTGAGCTGTATC >31329_31329_1_FRMD4A-MCM10_FRMD4A_chr10_13900847_ENST00000357447_MCM10_chr10_13214376_ENST00000378694_length(amino acids)=734AA_BP= MQEQMKALQEQLKVTTIKQTASPARLQKSPEKSPRPPLKERRVQRIQESTCFSAELDVPALPRTKRVARTPKASPPDPKSSSSRMTSAPS QPLQTISRNKPSGITRGQIVGTPGSSGETTQPICVEAFSGLRLRRPRVSSTEMNKKMTGRKLIRLSQIKEKMAREKLEEIDWVTFGVILK KVTPQSVNSGKTFSIWKLNDLRDLTQCVSLFLFGEVHKALWKTEQGTVVGILNANPMKPKDGSEEVCLSIDHPQKVLIMGEALDLGTCKA KKKNGEPCTQTVNLRDCEYCQYHVQAQYKKLSAKRADLQSTFSGGRIPKKFARRGTSLKERLCQDGFYYGGVSSASYAASIAAAVAPKKK IQTTLSNLVVKGTNLIIQETRQKLGIPQKSLSCSEEFKELMDLPTCGARNLKQHLAKATASGIMGSPKPAIKSISASALLKQQKQRMLEM RRRKSEEIQKRFLQSSSEVESPAVPSSSRQPPAQPPRTGSEFPRLEGAPATMTPKLGRGVLEGDDVLFYDESPPPRPKLSALAEAKKLAA ITKLRAKGQVLTKTNPNSIKKKQKDPQDILEVKERVEKNTMFSSQAEDELEPARKKRREQLAYLESEEFQKILKAKSKHTGILKEAEAEM QERYFEPLVKKEQMEEKMRNIREVKCRVVTCKTCAYTHFKLLETCVSEQHEYHWHDGVKRFFKCPCGNRSISLDRLPNKHCSNCGLYKWE -------------------------------------------------------------- >31329_31329_2_FRMD4A-MCM10_FRMD4A_chr10_13900847_ENST00000357447_MCM10_chr10_13214376_ENST00000378714_length(transcript)=4550nt_BP=480nt GAACGCTCACAGAACAGGCAGTGCAATTCCATGTTCCTCTTAAGTATGTTAGCCCTACCGGGAGCTGAGCTGGCCAGTCTACTTGGAGAG GAAAAGTAGATCTGGGGAAGGTGGAAGGGTCAGTTCCTAAGTGACTTCCTCCTCGGGGATGGTAAGGGCATTTGCTGATCTCCAGTGACT GCCTGGTGCCTCATGGTCAGACTCGGCTGTCTCACTCCCAGATATCTGATTTTGCAAAAAGGGACACACCTATCTGCAGCAAAGAAGACA CTGACCAGATTGCGAGCGGTGCTTTTGGATGCTCTGTAGCCACCCGGGGCCCAGGAGGACTGACTCGGCAGCAGGATTCGTGCATGGGAA TCGGAGACCATGGCAGTGCAGCTGGTGCCCGACTCAGCTCTCGGCCTGCTGATGATGACGGAGGGCCGCCGATGTCAAGTACATCTTCTT GATGACAGGAAGCTGGAACTCCTAGTACAGAGGAATTAAGGAATTTGCAAGAGCAAATGAAGGCCTTACAAGAGCAGCTAAAAGTAACAA CAATTAAACAGACAGCAAGCCCAGCCCGTCTGCAAAAATCCCCTGAGAAGTCTCCCCGGCCACCTCTTAAGGAGAGGAGAGTTCAGAGAA TTCAGGAGTCAACATGCTTTTCTGCGGAGCTTGATGTCCCTGCGCTACCAAGAACCAAGAGGGTGGCTCGAACACCAAAGGCTTCACCTC CAGATCCCAAAAGCTCATCTTCAAGGATGACAAGTGCACCCTCCCAACCCCTACAGACGATTTCTCGGAACAAACCTAGTGGGATAACTA GAGGTCAAATTGTGGGGACCCCAGGAAGTTCTGGGGAAACGACTCAACCCATCTGTGTGGAAGCCTTCTCTGGTCTGCGGCTCAGGCGGC CTCGAGTATCCTCCACAGAAATGAACAAGAAAATGACCGGCCGAAAACTGATCAGACTGTCTCAGATCAAGGAAAAGATGGCCAGAGAGA AGCTGGAAGAAATAGATTGGGTGACATTTGGGGTTATATTGAAGAAGGTTACGCCACAGAGTGTGAATAGTGGAAAAACCTTCAGCATAT GGAAACTGAATGATCTTCGTGACCTGACACAATGTGTGTCCTTGTTCTTATTTGGAGAAGTTCACAAAGCGCTCTGGAAGACGGAGCAGG GGACTGTCGTAGGGATCCTCAATGCCAACCCCATGAAGCCCAAGGATGGTTCAGAGGAGGTGTGTTTATCTATCGATCATCCTCAGAAGG TCTTAATTATGGGTGAAGCTCTTGACCTGGGAACCTGTAAAGCCAAGAAGAAGAATGGAGAGCCGTGCACGCAGACTGTGAATTTGCGTG ACTGTGAGTACTGTCAGTACCATGTCCAGGCTCAGTACAAGAAGCTCAGCGCAAAGCGTGCGGATCTGCAGTCCACCTTCTCTGGAGGAC GAATTCCAAAGAAGTTTGCCCGCAGAGGCACCAGCCTCAAAGAACGGCTGTGCCAAGATGGCTTTTACTACGGAGGGGTTTCTTCTGCCT CGTATGCAGCTTCAATTGCAGCAGCTGTGGCTCCTAAGAAGAAGATTCAAACCACTCTGAGTAATCTGGTTGTTAAGGGCACAAACTTGA TCATCCAGGAAACACGGCAAAAACTCGGAATACCCCAGAAGAGCCTGTCTTGCTCTGAGGAGTTCAAGGAACTGATGGACCTGCCGACGT GTGGAGCCAGGAACTTAAAACAACATTTAGCCAAAGCCACAGCTTCAGGGATTATGGGGAGCCCAAAACCAGCCATCAAGTCCATCTCGG CCTCAGCACTCTTGAAGCAACAGAAGCAGCGGATGTTGGAGATGAGGAGAAGGAAATCAGAAGAAATACAGAAGCGATTTCTGCAGAGCT CAAGTGAAGTTGAGAGCCCAGCTGTGCCATCTTCATCAAGACAGCCCCCTGCTCAGCCTCCACGGACAGGATCCGAGTTCCCCAGGCTGG AGGGAGCCCCGGCCACAATGACGCCCAAGCTGGGGCGAGGTGTCTTGGAAGGAGATGATGTTCTCTTTTATGATGAGTCACCACCACCAA GACCAAAACTGAGTGCTTTAGCAGAAGCCAAAAAGTTAGCTGCTATCACCAAATTAAGGGCAAAAGGCCAGGTTCTTACAAAAACAAACC CAAACAGCATTAAGAAGAAACAAAAGGACCCTCAGGACATCCTGGAGGTGAAGGAACGTGTAGAAAAAAACACCATGTTTTCTTCTCAAG CTGAGGATGAATTGGAGCCTGCCAGGAAAAAAAGGAGAGAACAACTTGCCTATCTGGAATCTGAGGAATTTCAGAAAATCCTAAAAGCAA AATCAAAACACACAGGCATCCTGAAAGAGGCCGAGGCTGAGATGCAGGAGCGCTACTTTGAGCCACTGGTGAAAAAAGAACAAATGGAAG AAAAGATGAGAAACATCAGAGAAGTGAAGTGCCGTGTCGTGACATGCAAGACGTGCGCCTATACCCACTTCAAGCTGCTGGAGACCTGCG TCAGTGAGCAGCATGAATACCACTGGCATGATGGTGTGAAGAGGTTTTTCAAATGTCCCTGTGGAAACAGAAGCATCTCCTTGGACAGAC TCCCGAACAAGCACTGCAGTAACTGTGGCCTCTACAAATGGGAACGGGACGGAATGCTAAAGGAAAAGACTGGTCCAAAGATAGGAGGAG AAACTCTGTTACCAAGAGGAGAAGAACATGCTAAATTTCTGAACAGCCTTAAATAACCCGAACTTCAGACATTTTCCCACAGACTTCCTG GCCTCCTGTGACTCTGGAAAGCAAAGGATTGGCTGTGTATTGTCCATTGATTCCTGATTGACGCCGTCAAAAACAAATGCTTGTTAAGCC CATAAGCTTTGCCTGCTTACTTTCTGCCATTGGGTTGGTTTGATACCACATTTAACATTGACATTTAAGTGGAAAACCAAGTTATCATTG TCTTTTCTAAGCTCAGTGTGGATGATTGCATTACTTCATTCACTGAAGTTTTTGCCCAAAAATTGGAAGGTAAACAGAGAGCTATGTTTC TGTATCTTTTGGTTATAGAGTGTTCACTTCTTTATCATAACAAAATTCTAGTGTTTATACGAACACCCAGAGGCAAAAGAATTTGGCTTA ATTCTCACTCCAGGTAAGTAGCTTAACTTCTGGGCTTCAGTTTTCTCATCTGTAAAATCAGGAAGATTGGACTAAGTGATCCTGAAATGT ATTTTTTAGCACTGGATTTCTACAAATAATAAAACTTTCCCATCTAGATAATGATGATCACATAGTCTTGATGTACGGACATTAAAAGCC AGATTTCTTCATTCAATTCTGTTATCTCTGTTTTACTCTTTGAAATTGATCAAGCCACTGAATCACTTTGCATTTCAGTTTATATATATA GAGAGAAAGAAGGTGTCTGCTCTTACATTATTGTGGAGCCCTGTGATAGAAATATGTAAAATCTCATATTATTTTTTTTTTAATTTTTTT ATTTTTTATGACAGGGTCTCACTATGTCACCCTGGCTGGAGTGCAGTAGTGCGATCGCGGCACACTGCAGCCTTGGCTTCCCTGGGCTCA AGCAGTCCTCCCACCTCAGTCTCCCAAATAGCTAGGACTACAGGCGTGCGTGACCAAGCCCAGCTAATTTTTGCATTTTTTGTAGAGATG GGGTTTTGCCATGTTGCTCAGGCTGGTCTCAAACTCCTGAGCACTAGCAATCCACCCACCTCTGTTTCCAAAAAAAAAAAAAAAATGAAA GGTCAACCCCTATGCAAATTACCACAGCAAAGGTTTCATTCAGGAGATTCTTCCATCTGGGCAACCTGGTTTTCCAAATATCATTTGACC TAAGTGAATGTTGATACTAGCTAAAGATTGGGTAAATTGGTTGAATTATTGTATTGAAGCTTGAGCTGTAGCTAAAAGTAATTTAGGTTT CCCCTAAGATGTTATTATGTTAGGGACATAACACTTTTGGGAGGTTGTTGTGGGAGATGGTTGATTTAGGTTTTCAAAAGCTAGAAATAA AATTTACATGCCTTAGATTTCATAAAATTCTGCTCTAATTGGGTGGAAGGTGCTGTATCTAACTTGTGTTCCTCCTAAGGTTATGTCCTA ATAACTATTCTTTTAGGAGTATACTTCTACTTTATAGAAGGTTGCTTTTCTTTTTAATTTTTTCTAACAAAGAAAAGAATAAAGTATTTA TTAATAAGAACCAGAAAGCACTTGAAACTGATGTTTTTAATGGCTCATTTAGGGTAGATTTATTTATCTCATTAACTTAAAACAGCTATG TGTATGAAATAGGTCACAACAGAACTTGAACACCAGGTTGGTGTCTGAGCAATCCCTTTCTTATGGGAAAAACAATGTTCTTGTTTGAAC AGAGGGTATCATTGCAGTCAGTATTCACGTGTATATTGTTATATAAGTTGTATAATATGCTTGTAAAGGCTGAGGGTGAGCTGTATCTGG >31329_31329_2_FRMD4A-MCM10_FRMD4A_chr10_13900847_ENST00000357447_MCM10_chr10_13214376_ENST00000378714_length(amino acids)=753AA_BP= MQEQMKALQEQLKVTTIKQTASPARLQKSPEKSPRPPLKERRVQRIQESTCFSAELDVPALPRTKRVARTPKASPPDPKSSSSRMTSAPS QPLQTISRNKPSGITRGQIVGTPGSSGETTQPICVEAFSGLRLRRPRVSSTEMNKKMTGRKLIRLSQIKEKMAREKLEEIDWVTFGVILK KVTPQSVNSGKTFSIWKLNDLRDLTQCVSLFLFGEVHKALWKTEQGTVVGILNANPMKPKDGSEEVCLSIDHPQKVLIMGEALDLGTCKA KKKNGEPCTQTVNLRDCEYCQYHVQAQYKKLSAKRADLQSTFSGGRIPKKFARRGTSLKERLCQDGFYYGGVSSASYAASIAAAVAPKKK IQTTLSNLVVKGTNLIIQETRQKLGIPQKSLSCSEEFKELMDLPTCGARNLKQHLAKATASGIMGSPKPAIKSISASALLKQQKQRMLEM RRRKSEEIQKRFLQSSSEVESPAVPSSSRQPPAQPPRTGSEFPRLEGAPATMTPKLGRGVLEGDDVLFYDESPPPRPKLSALAEAKKLAA ITKLRAKGQVLTKTNPNSIKKKQKDPQDILEVKERVEKNTMFSSQAEDELEPARKKRREQLAYLESEEFQKILKAKSKHTGILKEAEAEM QERYFEPLVKKEQMEEKMRNIREVKCRVVTCKTCAYTHFKLLETCVSEQHEYHWHDGVKRFFKCPCGNRSISLDRLPNKHCSNCGLYKWE -------------------------------------------------------------- >31329_31329_3_FRMD4A-MCM10_FRMD4A_chr10_13900847_ENST00000357447_MCM10_chr10_13214376_ENST00000484800_length(transcript)=3185nt_BP=480nt GAACGCTCACAGAACAGGCAGTGCAATTCCATGTTCCTCTTAAGTATGTTAGCCCTACCGGGAGCTGAGCTGGCCAGTCTACTTGGAGAG GAAAAGTAGATCTGGGGAAGGTGGAAGGGTCAGTTCCTAAGTGACTTCCTCCTCGGGGATGGTAAGGGCATTTGCTGATCTCCAGTGACT GCCTGGTGCCTCATGGTCAGACTCGGCTGTCTCACTCCCAGATATCTGATTTTGCAAAAAGGGACACACCTATCTGCAGCAAAGAAGACA CTGACCAGATTGCGAGCGGTGCTTTTGGATGCTCTGTAGCCACCCGGGGCCCAGGAGGACTGACTCGGCAGCAGGATTCGTGCATGGGAA TCGGAGACCATGGCAGTGCAGCTGGTGCCCGACTCAGCTCTCGGCCTGCTGATGATGACGGAGGGCCGCCGATGTCAAGTACATCTTCTT GATGACAGGAAGCTGGAACTCCTAGTACAGAGGAATTAAGGAATTTGCAAGAGCAAATGAAGGCCTTACAAGAGCAGCTAAAAGTAACAA CAATTAAACAGACAGCAAGCCCAGCCCGTCTGCAAAAATCCCCTGTAGAGAAGTCTCCCCGGCCACCTCTTAAGGAGAGGAGAGTTCAGA GAATTCAGGAGTCAACATGCTTTTCTGCGGAGCTTGATGTCCCTGCGCTACCAAGAACCAAGAGGGTGGCTCGAACACCAAAGGCTTCAC CTCCAGATCCCAAAAGCTCATCTTCAAGGATGACAAGTGCACCCTCCCAACCCCTACAGACGATTTCTCGGAACAAACCTAGTGGGATAA CTAGAGGTCAAATTGTGGGGACCCCAGGAAGTTCTGGGGAAACGACTCAACCCATCTGTGTGGAAGCCTTCTCTGGTCTGCGGCTCAGGC GGCCTCGAGTATCCTCCACAGAAATGAACAAGAAAATGACCGGCCGAAAACTGATCAGACTGTCTCAGATCAAGGAAAAGATGGCCAGAG AGAAGCTGGAAGAAATAGATTGGGTGACATTTGGGGTTATATTGAAGAAGGTTACGCCACAGAGTGTGAATAGTGGAAAAACCTTCAGCA TATGGAAACTGAATGATCTTCGTGACCTGACACAATGTGTGTCCTTGTTCTTATTTGGAGAAGTTCACAAAGCGCTCTGGAAGACGGAGC AGGGGACTGTCGTAGGGATCCTCAATGCCAACCCCATGAAGCCCAAGGATGGTTCAGAGGAGGTGTGTTTATCTATCGATCATCCTCAGA AGGTCTTAATTATGGGTGAAGCTCTTGACCTGGGAACCTGTAAAGCCAAGAAGAAGAATGGAGAGCCGTGCACGCAGACTGTGAATTTGC GTGACTGTGAGTACTGTCAGTACCATGTCCAGGCTCAGTACAAGAAGCTCAGCGCAAAGCGTGCGGATCTGCAGTCCACCTTCTCTGGAG GACGAATTCCAAAGAAGTTTGCCCGCAGAGGCACCAGCCTCAAAGAACGGCTGTGCCAAGATGGCTTTTACTACGGAGGGGTTTCTTCTG CCTCGTATGCAGCTTCAATTGCAGCAGCTGTGGCTCCTAAGAAGAAGATTCAAACCACTCTGAGTAATCTGGTTGTTAAGGGCACAAACT TGATCATCCAGGAAACACGGCAAAAACTCGGAATACCCCAGAAGAGCCTGTCTTGCTCTGAGGAGTTCAAGGAACTGATGGACCTGCCGA CGTGTGGAGCCAGGAACTTAAAACAACATTTAGCCAAAGCCACAGCTTCAGGGATTATGGGGAGCCCAAAACCAGCCATCAAGTCCATCT CGGCCTCAGCACTCTTGAAGCAACAGAAGCAGCGGATGTTGGAGATGAGGAGAAGGAAATCAGAAGAAATACAGAAGCGATTTCTGCAGA GCTCAAGTGAAGTTGAGAGCCCAGCTGTGCCATCTTCATCAAGACAGCCCCCTGCTCAGCCTCCACGGACAGGATCCGAGTTCCCCAGGC TGGAGGGAGCCCCGGCCACAATGACGCCCAAGCTGGGGCGAGGTGTCTTGGAAGGAGATGATGTTCTCTTTTATGATGAGTCACCACCAC CAAGACCAAAACTGAGTGCTTTAGCAGAAGCCAAAAAGTTAGCTGCTATCACCAAATTAAGGGCAAAAGGCCAGGTTCTTACAAAAACAA ACCCAAACAGCATTAAGAAGAAACAAAAGGACCCTCAGGACATCCTGGAGGTGAAGGAACGTGTAGAAAAAAACACCATGTTTTCTTCTC AAGCTGAGGATGAATTGGAGCCTGCCAGGAAAAAAAGGAGAGAACAACTTGCCTATCTGGAATCTGAGGAATTTCAGAAAATCCTAAAAG CAAAATCAAAACACACAGGCATCCTGAAAGAGGCCGAGGCTGAGATGCAGGAGCGCTACTTTGAGCCACTGGTGAAAAAAGAACAAATGG AAGAAAAGATGAGAAACATCAGAGAAGTGAAGTGCCGTGTCGTGACATGCAAGACGTGCGCCTATACCCACTTCAAGCTGCTGGAGACCT GCGTCAGTGAGCAGCATGAATACCACTGGCATGATGGTGTGAAGAGGTTTTTCAAATGTCCCTGTGGAAACAGAAGCATCTCCTTGGACA GACTCCCGAACAAGCACTGCAGTAACTGTGGCCTCTACAAATGGGAACGGGACGGAATGCTAAAGGAAAAGACTGGTCCAAAGATAGGAG GAGAAACTCTGTTACCAAGAGGAGAAGAACATGCTAAATTTCTGAACAGCCTTAAATAACCCGAACTTCAGACATTTTCCCACAGACTTC CTGGCCTCCTGTGACTCTGGAAAGCAAAGGATTGGCTGTGTATTGTCCATTGATTCCTGATTGACGCCGTCAAAAACAAATGCTTGTTAA GCCCATAAGCTTTGCCTGCTTACTTTCTGCCATTGGGTTGGTTTGATACCACATTTAACATTGACATTTAAGTGGAAAACCAAGTTATCA TTGTCTTTTCTAAGCTCAGTGTGGATGATTGCATTACTTCATTCACTGAAGTTTTTGCCCAAAAATTGGAAGGTAAACAGAGAGCTATGT TTCTGTATCTTTTGGTTATAGAGTGTTCACTTCTTTATCATAACAAAATTCTAGTGTTTATACGAACACCCAGAGGCAAAAGAATTTGGC >31329_31329_3_FRMD4A-MCM10_FRMD4A_chr10_13900847_ENST00000357447_MCM10_chr10_13214376_ENST00000484800_length(amino acids)=754AA_BP= MQEQMKALQEQLKVTTIKQTASPARLQKSPVEKSPRPPLKERRVQRIQESTCFSAELDVPALPRTKRVARTPKASPPDPKSSSSRMTSAP SQPLQTISRNKPSGITRGQIVGTPGSSGETTQPICVEAFSGLRLRRPRVSSTEMNKKMTGRKLIRLSQIKEKMAREKLEEIDWVTFGVIL KKVTPQSVNSGKTFSIWKLNDLRDLTQCVSLFLFGEVHKALWKTEQGTVVGILNANPMKPKDGSEEVCLSIDHPQKVLIMGEALDLGTCK AKKKNGEPCTQTVNLRDCEYCQYHVQAQYKKLSAKRADLQSTFSGGRIPKKFARRGTSLKERLCQDGFYYGGVSSASYAASIAAAVAPKK KIQTTLSNLVVKGTNLIIQETRQKLGIPQKSLSCSEEFKELMDLPTCGARNLKQHLAKATASGIMGSPKPAIKSISASALLKQQKQRMLE MRRRKSEEIQKRFLQSSSEVESPAVPSSSRQPPAQPPRTGSEFPRLEGAPATMTPKLGRGVLEGDDVLFYDESPPPRPKLSALAEAKKLA AITKLRAKGQVLTKTNPNSIKKKQKDPQDILEVKERVEKNTMFSSQAEDELEPARKKRREQLAYLESEEFQKILKAKSKHTGILKEAEAE MQERYFEPLVKKEQMEEKMRNIREVKCRVVTCKTCAYTHFKLLETCVSEQHEYHWHDGVKRFFKCPCGNRSISLDRLPNKHCSNCGLYKW -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for FRMD4A-MCM10 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for FRMD4A-MCM10 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for FRMD4A-MCM10 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
