
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:FUBP3-ABL1 (FusionGDB2 ID:31757) |
Fusion Gene Summary for FUBP3-ABL1 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: FUBP3-ABL1 | Fusion gene ID: 31757 | Hgene | Tgene | Gene symbol | FUBP3 | ABL1 | Gene ID | 8939 | 25 |
| Gene name | far upstream element binding protein 3 | ABL proto-oncogene 1, non-receptor tyrosine kinase | |
| Synonyms | FBP3 | ABL|BCR-ABL|CHDSKM|JTK7|bcr/abl|c-ABL|c-ABL1|p150|v-abl | |
| Cytomap | 9q34.11-q34.12 | 9q34.12 | |
| Type of gene | protein-coding | protein-coding | |
| Description | far upstream element-binding protein 3FUSE-binding protein 3far upstream element (FUSE) binding protein 3 | tyrosine-protein kinase ABL1ABL protooncogene 1 nonreceptor tyrosine kinaseAbelson tyrosine-protein kinase 1bcr/c-abl oncogene proteinc-abl oncogene 1, receptor tyrosine kinaseproto-oncogene c-Ablproto-oncogene tyrosine-protein kinase ABL1truncated | |
| Modification date | 20200313 | 20200327 | |
| UniProtAcc | Q96I24 | P00519 | |
| Ensembl transtripts involved in fusion gene | ENST00000467100, ENST00000319725, | ENST00000318560, | |
| Fusion gene scores | * DoF score | 5 X 4 X 4=80 | 21 X 149 X 8=25032 |
| # samples | 6 | 154 | |
| ** MAII score | log2(6/80*10)=-0.415037499278844 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(154/25032*10)=-4.02277130765866 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: FUBP3 [Title/Abstract] AND ABL1 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | ABL1(133589842)-FUBP3(133493184), # samples:4 FUBP3(133455151)-ABL1(133738149), # samples:2 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | FUBP3 | GO:0006351 | transcription, DNA-templated | 8940189 |
| Hgene | FUBP3 | GO:0010628 | positive regulation of gene expression | 19087307 |
| Hgene | FUBP3 | GO:0045893 | positive regulation of transcription, DNA-templated | 8940189 |
| Hgene | FUBP3 | GO:0045944 | positive regulation of transcription by RNA polymerase II | 8940183|8940189 |
| Tgene | ABL1 | GO:0006974 | cellular response to DNA damage stimulus | 15657060 |
| Tgene | ABL1 | GO:0006975 | DNA damage induced protein phosphorylation | 18280240 |
| Tgene | ABL1 | GO:0018108 | peptidyl-tyrosine phosphorylation | 7590236|9144171|10713049|11121037 |
| Tgene | ABL1 | GO:0038083 | peptidyl-tyrosine autophosphorylation | 10518561 |
| Tgene | ABL1 | GO:0042770 | signal transduction in response to DNA damage | 9037071|15657060 |
| Tgene | ABL1 | GO:0043065 | positive regulation of apoptotic process | 9037071 |
| Tgene | ABL1 | GO:0046777 | protein autophosphorylation | 10713049 |
| Tgene | ABL1 | GO:0050731 | positive regulation of peptidyl-tyrosine phosphorylation | 15657060 |
| Tgene | ABL1 | GO:0051353 | positive regulation of oxidoreductase activity | 12893824 |
| Tgene | ABL1 | GO:0051444 | negative regulation of ubiquitin-protein transferase activity | 20823226 |
| Tgene | ABL1 | GO:0070301 | cellular response to hydrogen peroxide | 10713049 |
| Tgene | ABL1 | GO:0071103 | DNA conformation change | 9558345 |
| Tgene | ABL1 | GO:0071901 | negative regulation of protein serine/threonine kinase activity | 11121037 |
| Tgene | ABL1 | GO:1990051 | activation of protein kinase C activity | 10713049 |
| Tgene | ABL1 | GO:2001020 | regulation of response to DNA damage stimulus | 9461559 |
 Fusion gene breakpoints across FUBP3 (5'-gene) Fusion gene breakpoints across FUBP3 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
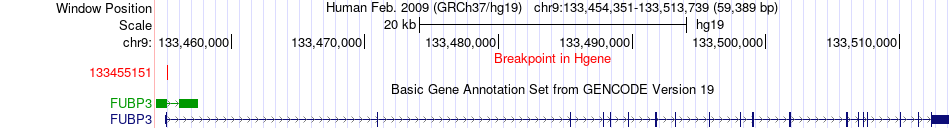 |
 Fusion gene breakpoints across ABL1 (3'-gene) Fusion gene breakpoints across ABL1 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
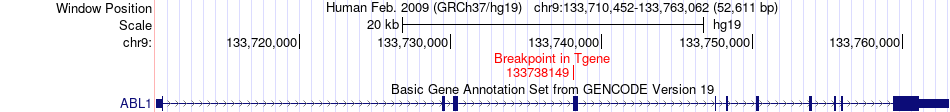 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | ESCA | TCGA-LN-A4A8 | FUBP3 | chr9 | 133455151 | + | ABL1 | chr9 | 133738149 | + |
Top |
Fusion Gene ORF analysis for FUBP3-ABL1 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 3UTR-3CDS | ENST00000467100 | ENST00000318560 | FUBP3 | chr9 | 133455151 | + | ABL1 | chr9 | 133738149 | + |
| In-frame | ENST00000319725 | ENST00000318560 | FUBP3 | chr9 | 133455151 | + | ABL1 | chr9 | 133738149 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000319725 | FUBP3 | chr9 | 133455151 | + | ENST00000318560 | ABL1 | chr9 | 133738149 | + | 4995 | 159 | 75 | 3002 | 975 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000319725 | ENST00000318560 | FUBP3 | chr9 | 133455151 | + | ABL1 | chr9 | 133738149 | + | 0.001717061 | 0.9982829 |
Top |
Fusion Genomic Features for FUBP3-ABL1 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| FUBP3 | chr9 | 133455151 | + | ABL1 | chr9 | 133738149 | + | 1.44E-10 | 1 |
| FUBP3 | chr9 | 133455151 | + | ABL1 | chr9 | 133738149 | + | 1.44E-10 | 1 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
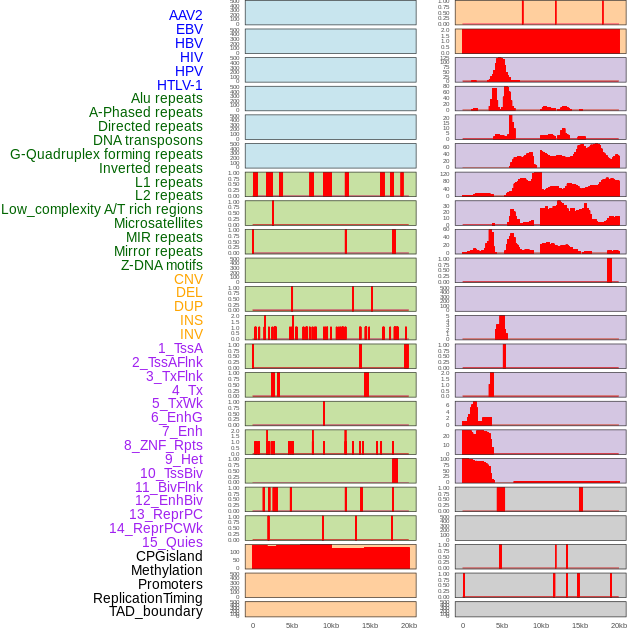 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
 |
Top |
Fusion Protein Features for FUBP3-ABL1 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr9:133589842/chr9:133493184) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| FUBP3 | ABL1 |
| FUNCTION: May interact with single-stranded DNA from the far-upstream element (FUSE). May activate gene expression. | FUNCTION: Non-receptor tyrosine-protein kinase that plays a role in many key processes linked to cell growth and survival such as cytoskeleton remodeling in response to extracellular stimuli, cell motility and adhesion, receptor endocytosis, autophagy, DNA damage response and apoptosis. Coordinates actin remodeling through tyrosine phosphorylation of proteins controlling cytoskeleton dynamics like WASF3 (involved in branch formation); ANXA1 (involved in membrane anchoring); DBN1, DBNL, CTTN, RAPH1 and ENAH (involved in signaling); or MAPT and PXN (microtubule-binding proteins). Phosphorylation of WASF3 is critical for the stimulation of lamellipodia formation and cell migration. Involved in the regulation of cell adhesion and motility through phosphorylation of key regulators of these processes such as BCAR1, CRK, CRKL, DOK1, EFS or NEDD9. Phosphorylates multiple receptor tyrosine kinases and more particularly promotes endocytosis of EGFR, facilitates the formation of neuromuscular synapses through MUSK, inhibits PDGFRB-mediated chemotaxis and modulates the endocytosis of activated B-cell receptor complexes. Other substrates which are involved in endocytosis regulation are the caveolin (CAV1) and RIN1. Moreover, ABL1 regulates the CBL family of ubiquitin ligases that drive receptor down-regulation and actin remodeling. Phosphorylation of CBL leads to increased EGFR stability. Involved in late-stage autophagy by regulating positively the trafficking and function of lysosomal components. ABL1 targets to mitochondria in response to oxidative stress and thereby mediates mitochondrial dysfunction and cell death. In response to oxidative stress, phosphorylates serine/threonine kinase PRKD2 at 'Tyr-717' (PubMed:28428613). ABL1 is also translocated in the nucleus where it has DNA-binding activity and is involved in DNA-damage response and apoptosis. Many substrates are known mediators of DNA repair: DDB1, DDB2, ERCC3, ERCC6, RAD9A, RAD51, RAD52 or WRN. Activates the proapoptotic pathway when the DNA damage is too severe to be repaired. Phosphorylates TP73, a primary regulator for this type of damage-induced apoptosis. Phosphorylates the caspase CASP9 on 'Tyr-153' and regulates its processing in the apoptotic response to DNA damage. Phosphorylates PSMA7 that leads to an inhibition of proteasomal activity and cell cycle transition blocks. ABL1 acts also as a regulator of multiple pathological signaling cascades during infection. Several known tyrosine-phosphorylated microbial proteins have been identified as ABL1 substrates. This is the case of A36R of Vaccinia virus, Tir (translocated intimin receptor) of pathogenic E.coli and possibly Citrobacter, CagA (cytotoxin-associated gene A) of H.pylori, or AnkA (ankyrin repeat-containing protein A) of A.phagocytophilum. Pathogens can highjack ABL1 kinase signaling to reorganize the host actin cytoskeleton for multiple purposes, like facilitating intracellular movement and host cell exit. Finally, functions as its own regulator through autocatalytic activity as well as through phosphorylation of its inhibitor, ABI1. Regulates T-cell differentiation in a TBX21-dependent manner. Phosphorylates TBX21 on tyrosine residues leading to an enhancement of its transcriptional activator activity (By similarity). {ECO:0000250|UniProtKB:P00520, ECO:0000269|PubMed:10391250, ECO:0000269|PubMed:11971963, ECO:0000269|PubMed:12379650, ECO:0000269|PubMed:12531427, ECO:0000269|PubMed:12672821, ECO:0000269|PubMed:15031292, ECO:0000269|PubMed:15556646, ECO:0000269|PubMed:15657060, ECO:0000269|PubMed:15886098, ECO:0000269|PubMed:16424036, ECO:0000269|PubMed:16678104, ECO:0000269|PubMed:16943190, ECO:0000269|PubMed:17306540, ECO:0000269|PubMed:17623672, ECO:0000269|PubMed:18328268, ECO:0000269|PubMed:18945674, ECO:0000269|PubMed:19891780, ECO:0000269|PubMed:20357770, ECO:0000269|PubMed:20417104, ECO:0000269|PubMed:28428613, ECO:0000269|PubMed:9037071, ECO:0000269|PubMed:9144171, ECO:0000269|PubMed:9461559}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | ABL1 | chr9:133455151 | chr9:133738149 | ENST00000318560 | 2 | 11 | 605_609 | 183 | 1131.0 | Compositional bias | Note=Poly-Lys | |
| Tgene | ABL1 | chr9:133455151 | chr9:133738149 | ENST00000318560 | 2 | 11 | 782_1019 | 183 | 1131.0 | Compositional bias | Note=Pro-rich | |
| Tgene | ABL1 | chr9:133455151 | chr9:133738149 | ENST00000318560 | 2 | 11 | 897_903 | 183 | 1131.0 | Compositional bias | Note=Poly-Pro | |
| Tgene | ABL1 | chr9:133455151 | chr9:133738149 | ENST00000318560 | 2 | 11 | 242_493 | 183 | 1131.0 | Domain | Protein kinase | |
| Tgene | ABL1 | chr9:133455151 | chr9:133738149 | ENST00000318560 | 2 | 11 | 1090_1100 | 183 | 1131.0 | Motif | Nuclear export signal | |
| Tgene | ABL1 | chr9:133455151 | chr9:133738149 | ENST00000318560 | 2 | 11 | 381_405 | 183 | 1131.0 | Motif | Note=Kinase activation loop | |
| Tgene | ABL1 | chr9:133455151 | chr9:133738149 | ENST00000318560 | 2 | 11 | 709_715 | 183 | 1131.0 | Motif | Nuclear localization signal 2 | |
| Tgene | ABL1 | chr9:133455151 | chr9:133738149 | ENST00000318560 | 2 | 11 | 762_769 | 183 | 1131.0 | Motif | Nuclear localization signal 3 | |
| Tgene | ABL1 | chr9:133455151 | chr9:133738149 | ENST00000318560 | 2 | 11 | 248_256 | 183 | 1131.0 | Nucleotide binding | Note=ATP | |
| Tgene | ABL1 | chr9:133455151 | chr9:133738149 | ENST00000318560 | 2 | 11 | 316_322 | 183 | 1131.0 | Nucleotide binding | Note=ATP | |
| Tgene | ABL1 | chr9:133455151 | chr9:133738149 | ENST00000318560 | 2 | 11 | 869_968 | 183 | 1131.0 | Region | DNA-binding | |
| Tgene | ABL1 | chr9:133455151 | chr9:133738149 | ENST00000318560 | 2 | 11 | 953_1130 | 183 | 1131.0 | Region | Note=F-actin-binding |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | FUBP3 | chr9:133455151 | chr9:133738149 | ENST00000319725 | + | 1 | 19 | 162_228 | 28 | 573.0 | Domain | KH 2 |
| Hgene | FUBP3 | chr9:133455151 | chr9:133738149 | ENST00000319725 | + | 1 | 19 | 253_317 | 28 | 573.0 | Domain | KH 3 |
| Hgene | FUBP3 | chr9:133455151 | chr9:133738149 | ENST00000319725 | + | 1 | 19 | 354_421 | 28 | 573.0 | Domain | KH 4 |
| Hgene | FUBP3 | chr9:133455151 | chr9:133738149 | ENST00000319725 | + | 1 | 19 | 77_141 | 28 | 573.0 | Domain | KH 1 |
| Tgene | ABL1 | chr9:133455151 | chr9:133738149 | ENST00000318560 | 2 | 11 | 18_22 | 183 | 1131.0 | Compositional bias | Note=Poly-Ser | |
| Tgene | ABL1 | chr9:133455151 | chr9:133738149 | ENST00000318560 | 2 | 11 | 127_217 | 183 | 1131.0 | Domain | SH2 | |
| Tgene | ABL1 | chr9:133455151 | chr9:133738149 | ENST00000318560 | 2 | 11 | 61_121 | 183 | 1131.0 | Domain | SH3 | |
| Tgene | ABL1 | chr9:133455151 | chr9:133738149 | ENST00000318560 | 2 | 11 | 1_60 | 183 | 1131.0 | Region | Note=CAP |
Top |
Fusion Gene Sequence for FUBP3-ABL1 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >31757_31757_1_FUBP3-ABL1_FUBP3_chr9_133455151_ENST00000319725_ABL1_chr9_133738149_ENST00000318560_length(transcript)=4995nt_BP=159nt CGGGAGGCCGGACCGGGGAGCCGAGCGGCGGCGTCGGCGGCGTCGGCGGCGGCGGCGACGGCGGCGGGGGCGGTAATGGCGGAGCTGGTG CAGGGGCAGAGCGCTCCTGTGGGGATGAAGGCCGAGGGCTTCGTGGATGCCCTGCACCGGGTCCGGCAGCTCTACGTCTCCTCCGAGAGC CGCTTCAACACCCTGGCCGAGTTGGTTCATCATCATTCAACGGTGGCCGACGGGCTCATCACCACGCTCCATTATCCAGCCCCAAAGCGC AACAAGCCCACTGTCTATGGTGTGTCCCCCAACTACGACAAGTGGGAGATGGAACGCACGGACATCACCATGAAGCACAAGCTGGGCGGG GGCCAGTACGGGGAGGTGTACGAGGGCGTGTGGAAGAAATACAGCCTGACGGTGGCCGTGAAGACCTTGAAGGAGGACACCATGGAGGTG GAAGAGTTCTTGAAAGAAGCTGCAGTCATGAAAGAGATCAAACACCCTAACCTGGTGCAGCTCCTTGGGGTCTGCACCCGGGAGCCCCCG TTCTATATCATCACTGAGTTCATGACCTACGGGAACCTCCTGGACTACCTGAGGGAGTGCAACCGGCAGGAGGTGAACGCCGTGGTGCTG CTGTACATGGCCACTCAGATCTCGTCAGCCATGGAGTACCTGGAGAAGAAAAACTTCATCCACAGAGATCTTGCTGCCCGAAACTGCCTG GTAGGGGAGAACCACTTGGTGAAGGTAGCTGATTTTGGCCTGAGCAGGTTGATGACAGGGGACACCTACACAGCCCATGCTGGAGCCAAG TTCCCCATCAAATGGACTGCACCCGAGAGCCTGGCCTACAACAAGTTCTCCATCAAGTCCGACGTCTGGGCATTTGGAGTATTGCTTTGG GAAATTGCTACCTATGGCATGTCCCCTTACCCGGGAATTGACCTGTCCCAGGTGTATGAGCTGCTAGAGAAGGACTACCGCATGGAGCGC CCAGAAGGCTGCCCAGAGAAGGTCTATGAACTCATGCGAGCATGTTGGCAGTGGAATCCCTCTGACCGGCCCTCCTTTGCTGAAATCCAC CAAGCCTTTGAAACAATGTTCCAGGAATCCAGTATCTCAGACGAAGTGGAAAAGGAGCTGGGGAAACAAGGCGTCCGTGGGGCTGTGAGT ACCTTGCTGCAGGCCCCAGAGCTGCCCACCAAGACGAGGACCTCCAGGAGAGCTGCAGAGCACAGAGACACCACTGACGTGCCTGAGATG CCTCACTCCAAGGGCCAGGGAGAGAGCGATCCTCTGGACCATGAGCCTGCCGTGTCTCCATTGCTCCCTCGAAAAGAGCGAGGTCCCCCG GAGGGCGGCCTGAATGAAGATGAGCGCCTTCTCCCCAAAGACAAAAAGACCAACTTGTTCAGCGCCTTGATCAAGAAGAAGAAGAAGACA GCCCCAACCCCTCCCAAACGCAGCAGCTCCTTCCGGGAGATGGACGGCCAGCCGGAGCGCAGAGGGGCCGGCGAGGAAGAGGGCCGAGAC ATCAGCAACGGGGCACTGGCTTTCACCCCCTTGGACACAGCTGACCCAGCCAAGTCCCCAAAGCCCAGCAATGGGGCTGGGGTCCCCAAT GGAGCCCTCCGGGAGTCCGGGGGCTCAGGCTTCCGGTCTCCCCACCTGTGGAAGAAGTCCAGCACGCTGACCAGCAGCCGCCTAGCCACC GGCGAGGAGGAGGGCGGTGGCAGCTCCAGCAAGCGCTTCCTGCGCTCTTGCTCCGCCTCCTGCGTTCCCCATGGGGCCAAGGACACGGAG TGGAGGTCAGTCACGCTGCCTCGGGACTTGCAGTCCACGGGAAGACAGTTTGACTCGTCCACATTTGGAGGGCACAAAAGTGAGAAGCCG GCTCTGCCTCGGAAGAGGGCAGGGGAGAACAGGTCTGACCAGGTGACCCGAGGCACAGTAACGCCTCCCCCCAGGCTGGTGAAAAAGAAT GAGGAAGCTGCTGATGAGGTCTTCAAAGACATCATGGAGTCCAGCCCGGGCTCCAGCCCGCCCAACCTGACTCCAAAACCCCTCCGGCGG CAGGTCACCGTGGCCCCTGCCTCGGGCCTCCCCCACAAGGAAGAAGCTGGAAAGGGCAGTGCCTTAGGGACCCCTGCTGCAGCTGAGCCA GTGACCCCCACCAGCAAAGCAGGCTCAGGTGCACCAGGGGGCACCAGCAAGGGCCCCGCCGAGGAGTCCAGAGTGAGGAGGCACAAGCAC TCCTCTGAGTCGCCAGGGAGGGACAAGGGGAAATTGTCCAGGCTCAAACCTGCCCCGCCGCCCCCACCAGCAGCCTCTGCAGGGAAGGCT GGAGGAAAGCCCTCGCAGAGCCCGAGCCAGGAGGCGGCCGGGGAGGCAGTCCTGGGCGCAAAGACAAAAGCCACGAGTCTGGTTGATGCT GTGAACAGTGACGCTGCCAAGCCCAGCCAGCCGGGAGAGGGCCTCAAAAAGCCCGTGCTCCCGGCCACTCCAAAGCCACAGTCCGCCAAG CCGTCGGGGACCCCCATCAGCCCAGCCCCCGTTCCCTCCACGTTGCCATCAGCATCCTCGGCCCTGGCAGGGGACCAGCCGTCTTCCACC GCCTTCATCCCTCTCATATCAACCCGAGTGTCTCTTCGGAAAACCCGCCAGCCTCCAGAGCGGATCGCCAGCGGCGCCATCACCAAGGGC GTGGTCCTGGACAGCACCGAGGCGCTGTGCCTCGCCATCTCTAGGAACTCCGAGCAGATGGCCAGCCACAGCGCAGTGCTGGAGGCCGGC AAAAACCTCTACACGTTCTGCGTGAGCTATGTGGATTCCATCCAGCAAATGAGGAACAAGTTTGCCTTCCGAGAGGCCATCAACAAACTG GAGAATAATCTCCGGGAGCTTCAGATCTGCCCGGCGACAGCAGGCAGTGGTCCAGCGGCCACTCAGGACTTCAGCAAGCTCCTCAGTTCG GTGAAGGAAATCAGTGACATAGTGCAGAGGTAGCAGCAGTCAGGGGTCAGGTGTCAGGCCCGTCGGAGCTGCCTGCAGCACATGCGGGCT CGCCCATACCCGTGACAGTGGCTGACAAGGGACTAGTGAGTCAGCACCTTGGCCCAGGAGCTCTGCGCCAGGCAGAGCTGAGGGCCCTGT GGAGTCCAGCTCTACTACCTACGTTTGCACCGCCTGCCCTCCCGCACCTTCCTCCTCCCCGCTCCGTCTCTGTCCTCGAATTTTATCTGT GGAGTTCCTGCTCCGTGGACTGCAGTCGGCATGCCAGGACCCGCCAGCCCCGCTCCCACCTAGTGCCCCAGACTGAGCTCTCCAGGCCAG GTGGGAACGGCTGATGTGGACTGTCTTTTTCATTTTTTTCTCTCTGGAGCCCCTCCTCCCCCGGCTGGGCCTCCTTCTTCCACTTCTCCA AGAATGGAAGCCTGAACTGAGGCCTTGTGTGTCAGGCCCTCTGCCTGCACTCCCTGGCCTTGCCCGTCGTGTGCTGAAGACATGTTTCAA GAACCGCATTTCGGGAAGGGCATGCACGGGCATGCACACGGCTGGTCACTCTGCCCTCTGCTGCTGCCCGGGGTGGGGTGCACTCGCCAT TTCCTCACGTGCAGGACAGCTCTTGATTTGGGTGGAAAACAGGGTGCTAAAGCCAACCAGCCTTTGGGTCCTGGGCAGGTGGGAGCTGAA AAGGATCGAGGCATGGGGCATGTCCTTTCCATCTGTCCACATCCCCAGAGCCCAGCTCTTGCTCTCTTGTGACGTGCACTGTGAATCCTG GCAAGAAAGCTTGAGTCTCAAGGGTGGCAGGTCACTGTCACTGCCGACATCCCTCCCCCAGCAGAATGGAGGCAGGGGACAAGGGAGGCA GTGGCTAGTGGGGTGAACAGCTGGTGCCAAATAGCCCCAGACTGGGCCCAGGCAGGTCTGCAAGGGCCCAGAGTGAACCGTCCTTTCACA CATCTGGGTGCCCTGAAAGGGCCCTTCCCCTCCCCCACTCCTCTAAGACAAAGTAGATTCTTACAAGGCCCTTTCCTTTGGAACAAGACA GCCTTCACTTTTCTGAGTTCTTGAAGCATTTCAAAGCCCTGCCTCTGTGTAGCCGCCCTGAGAGAGAATAGAGCTGCCACTGGGCACCTG CGCACAGGTGGGAGGAAAGGGCCTGGCCAGTCCTGGTCCTGGCTGCACTCTTGAACTGGGCGAATGTCTTATTTAATTACCGTGAGTGAC ATAGCCTCATGTTCTGTGGGGGTCATCAGGGAGGGTTAGGAAAACCACAAACGGAGCCCCTGAAAGCCTCACGTATTTCACAGAGCACGC CTGCCATCTTCTCCCCGAGGCTGCCCCAGGCCGGAGCCCAGATACGGGGGCTGTGACTCTGGGCAGGGACCCGGGGTCTCCTGGACCTTG ACAGAGCAGCTAACTCCGAGAGCAGTGGGCAGGTGGCCGCCCCTGAGGCTTCACGCCGGGAGAAGCCACCTTCCCACCCCTTCATACCGC CTCGTGCCAGCAGCCTCGCACAGGCCCTAGCTTTACGCTCATCACCTAAACTTGTACTTTATTTTTCTGATAGAAATGGTTTCCTCTGGA TCGTTTTATGCGGTTCTTACAGCACATCACCTCTTTGCCCCCGACGGCTGTGACGCAGCCGGAGGGAGGCACTAGTCACCGACAGCGGCC TTGAAGACAGAGCAAAGCGCCCACCCAGGTCCCCCGACTGCCTGTCTCCATGAGGTACTGGTCCCTTCCTTTTGTTAACGTGATGTGCCA CTATATTTTACACGTATCTCTTGGTATGCATCTTTTATAGACGCTCTTTTCTAAGTGGCGTGTGCATAGCGTCCTGCCCTGCCCCCTCGG GGGCCTGTGGTGGCTCCCCCTCTGCTTCTCGGGGTCCAGTGCATTTTGTTTCTGTATATGATTCTCTGTGGTTTTTTTTGAATCCAAATC >31757_31757_1_FUBP3-ABL1_FUBP3_chr9_133455151_ENST00000319725_ABL1_chr9_133738149_ENST00000318560_length(amino acids)=975AA_BP=0 MAELVQGQSAPVGMKAEGFVDALHRVRQLYVSSESRFNTLAELVHHHSTVADGLITTLHYPAPKRNKPTVYGVSPNYDKWEMERTDITMK HKLGGGQYGEVYEGVWKKYSLTVAVKTLKEDTMEVEEFLKEAAVMKEIKHPNLVQLLGVCTREPPFYIITEFMTYGNLLDYLRECNRQEV NAVVLLYMATQISSAMEYLEKKNFIHRDLAARNCLVGENHLVKVADFGLSRLMTGDTYTAHAGAKFPIKWTAPESLAYNKFSIKSDVWAF GVLLWEIATYGMSPYPGIDLSQVYELLEKDYRMERPEGCPEKVYELMRACWQWNPSDRPSFAEIHQAFETMFQESSISDEVEKELGKQGV RGAVSTLLQAPELPTKTRTSRRAAEHRDTTDVPEMPHSKGQGESDPLDHEPAVSPLLPRKERGPPEGGLNEDERLLPKDKKTNLFSALIK KKKKTAPTPPKRSSSFREMDGQPERRGAGEEEGRDISNGALAFTPLDTADPAKSPKPSNGAGVPNGALRESGGSGFRSPHLWKKSSTLTS SRLATGEEEGGGSSSKRFLRSCSASCVPHGAKDTEWRSVTLPRDLQSTGRQFDSSTFGGHKSEKPALPRKRAGENRSDQVTRGTVTPPPR LVKKNEEAADEVFKDIMESSPGSSPPNLTPKPLRRQVTVAPASGLPHKEEAGKGSALGTPAAAEPVTPTSKAGSGAPGGTSKGPAEESRV RRHKHSSESPGRDKGKLSRLKPAPPPPPAASAGKAGGKPSQSPSQEAAGEAVLGAKTKATSLVDAVNSDAAKPSQPGEGLKKPVLPATPK PQSAKPSGTPISPAPVPSTLPSASSALAGDQPSSTAFIPLISTRVSLRKTRQPPERIASGAITKGVVLDSTEALCLAISRNSEQMASHSA -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for FUBP3-ABL1 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for FUBP3-ABL1 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for FUBP3-ABL1 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
