
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:FUBP3-EIF4E2 (FusionGDB2 ID:31758) |
Fusion Gene Summary for FUBP3-EIF4E2 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: FUBP3-EIF4E2 | Fusion gene ID: 31758 | Hgene | Tgene | Gene symbol | FUBP3 | EIF4E2 | Gene ID | 8939 | 9470 |
| Gene name | far upstream element binding protein 3 | eukaryotic translation initiation factor 4E family member 2 | |
| Synonyms | FBP3 | 4E-LP|4EHP|EIF4EL3|IF4e|h4EHP | |
| Cytomap | 9q34.11-q34.12 | 2q37.1 | |
| Type of gene | protein-coding | protein-coding | |
| Description | far upstream element-binding protein 3FUSE-binding protein 3far upstream element (FUSE) binding protein 3 | eukaryotic translation initiation factor 4E type 2eIF-4E type 2eIF4E-like cap-binding proteineIF4E-like protein 4E-LPeukaryotic translation initiation factor 4E homologous proteineukaryotic translation initiation factor 4E-like 3mRNA cap-binding pro | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | Q96I24 | O60573 | |
| Ensembl transtripts involved in fusion gene | ENST00000467100, ENST00000319725, | ENST00000258416, ENST00000409098, ENST00000409167, ENST00000409322, ENST00000409394, ENST00000409495, ENST00000409514, ENST00000479834, | |
| Fusion gene scores | * DoF score | 5 X 4 X 4=80 | 6 X 9 X 4=216 |
| # samples | 6 | 9 | |
| ** MAII score | log2(6/80*10)=-0.415037499278844 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(9/216*10)=-1.26303440583379 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: FUBP3 [Title/Abstract] AND EIF4E2 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | FUBP3(133455151)-EIF4E2(233428957), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | FUBP3 | GO:0006351 | transcription, DNA-templated | 8940189 |
| Hgene | FUBP3 | GO:0010628 | positive regulation of gene expression | 19087307 |
| Hgene | FUBP3 | GO:0045893 | positive regulation of transcription, DNA-templated | 8940189 |
| Hgene | FUBP3 | GO:0045944 | positive regulation of transcription by RNA polymerase II | 8940183|8940189 |
 Fusion gene breakpoints across FUBP3 (5'-gene) Fusion gene breakpoints across FUBP3 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
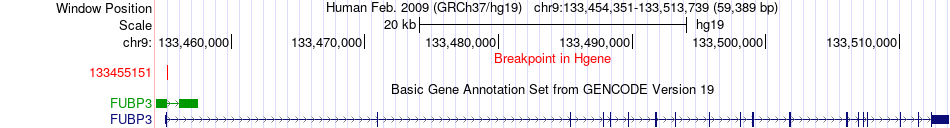 |
 Fusion gene breakpoints across EIF4E2 (3'-gene) Fusion gene breakpoints across EIF4E2 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
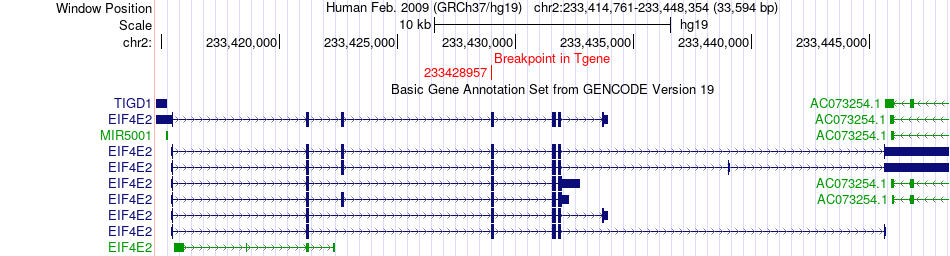 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | BLCA | TCGA-E7-A4XJ-01A | FUBP3 | chr9 | 133455151 | + | EIF4E2 | chr2 | 233428957 | + |
Top |
Fusion Gene ORF analysis for FUBP3-EIF4E2 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 3UTR-3CDS | ENST00000467100 | ENST00000258416 | FUBP3 | chr9 | 133455151 | + | EIF4E2 | chr2 | 233428957 | + |
| 3UTR-3CDS | ENST00000467100 | ENST00000409098 | FUBP3 | chr9 | 133455151 | + | EIF4E2 | chr2 | 233428957 | + |
| 3UTR-3CDS | ENST00000467100 | ENST00000409167 | FUBP3 | chr9 | 133455151 | + | EIF4E2 | chr2 | 233428957 | + |
| 3UTR-3CDS | ENST00000467100 | ENST00000409322 | FUBP3 | chr9 | 133455151 | + | EIF4E2 | chr2 | 233428957 | + |
| 3UTR-3CDS | ENST00000467100 | ENST00000409394 | FUBP3 | chr9 | 133455151 | + | EIF4E2 | chr2 | 233428957 | + |
| 3UTR-3CDS | ENST00000467100 | ENST00000409495 | FUBP3 | chr9 | 133455151 | + | EIF4E2 | chr2 | 233428957 | + |
| 3UTR-3CDS | ENST00000467100 | ENST00000409514 | FUBP3 | chr9 | 133455151 | + | EIF4E2 | chr2 | 233428957 | + |
| 3UTR-intron | ENST00000467100 | ENST00000479834 | FUBP3 | chr9 | 133455151 | + | EIF4E2 | chr2 | 233428957 | + |
| 5CDS-intron | ENST00000319725 | ENST00000479834 | FUBP3 | chr9 | 133455151 | + | EIF4E2 | chr2 | 233428957 | + |
| In-frame | ENST00000319725 | ENST00000258416 | FUBP3 | chr9 | 133455151 | + | EIF4E2 | chr2 | 233428957 | + |
| In-frame | ENST00000319725 | ENST00000409098 | FUBP3 | chr9 | 133455151 | + | EIF4E2 | chr2 | 233428957 | + |
| In-frame | ENST00000319725 | ENST00000409167 | FUBP3 | chr9 | 133455151 | + | EIF4E2 | chr2 | 233428957 | + |
| In-frame | ENST00000319725 | ENST00000409322 | FUBP3 | chr9 | 133455151 | + | EIF4E2 | chr2 | 233428957 | + |
| In-frame | ENST00000319725 | ENST00000409394 | FUBP3 | chr9 | 133455151 | + | EIF4E2 | chr2 | 233428957 | + |
| In-frame | ENST00000319725 | ENST00000409495 | FUBP3 | chr9 | 133455151 | + | EIF4E2 | chr2 | 233428957 | + |
| In-frame | ENST00000319725 | ENST00000409514 | FUBP3 | chr9 | 133455151 | + | EIF4E2 | chr2 | 233428957 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000319725 | FUBP3 | chr9 | 133455151 | + | ENST00000409514 | EIF4E2 | chr2 | 233428957 | + | 3372 | 159 | 75 | 599 | 174 |
| ENST00000319725 | FUBP3 | chr9 | 133455151 | + | ENST00000409098 | EIF4E2 | chr2 | 233428957 | + | 3295 | 159 | 75 | 593 | 172 |
| ENST00000319725 | FUBP3 | chr9 | 133455151 | + | ENST00000409394 | EIF4E2 | chr2 | 233428957 | + | 607 | 159 | 75 | 593 | 172 |
| ENST00000319725 | FUBP3 | chr9 | 133455151 | + | ENST00000258416 | EIF4E2 | chr2 | 233428957 | + | 820 | 159 | 75 | 626 | 183 |
| ENST00000319725 | FUBP3 | chr9 | 133455151 | + | ENST00000409495 | EIF4E2 | chr2 | 233428957 | + | 867 | 159 | 265 | 858 | 197 |
| ENST00000319725 | FUBP3 | chr9 | 133455151 | + | ENST00000409167 | EIF4E2 | chr2 | 233428957 | + | 1360 | 159 | 265 | 858 | 197 |
| ENST00000319725 | FUBP3 | chr9 | 133455151 | + | ENST00000409322 | EIF4E2 | chr2 | 233428957 | + | 788 | 159 | 75 | 626 | 183 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000319725 | ENST00000409514 | FUBP3 | chr9 | 133455151 | + | EIF4E2 | chr2 | 233428957 | + | 0.0027292 | 0.99727076 |
| ENST00000319725 | ENST00000409098 | FUBP3 | chr9 | 133455151 | + | EIF4E2 | chr2 | 233428957 | + | 0.001886812 | 0.99811316 |
| ENST00000319725 | ENST00000409394 | FUBP3 | chr9 | 133455151 | + | EIF4E2 | chr2 | 233428957 | + | 0.003658253 | 0.99634176 |
| ENST00000319725 | ENST00000258416 | FUBP3 | chr9 | 133455151 | + | EIF4E2 | chr2 | 233428957 | + | 0.008640088 | 0.9913599 |
| ENST00000319725 | ENST00000409495 | FUBP3 | chr9 | 133455151 | + | EIF4E2 | chr2 | 233428957 | + | 0.4091638 | 0.59083617 |
| ENST00000319725 | ENST00000409167 | FUBP3 | chr9 | 133455151 | + | EIF4E2 | chr2 | 233428957 | + | 0.6096648 | 0.39033526 |
| ENST00000319725 | ENST00000409322 | FUBP3 | chr9 | 133455151 | + | EIF4E2 | chr2 | 233428957 | + | 0.008267183 | 0.99173284 |
Top |
Fusion Genomic Features for FUBP3-EIF4E2 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| FUBP3 | chr9 | 133455151 | + | EIF4E2 | chr2 | 233428956 | + | 2.37E-10 | 1 |
| FUBP3 | chr9 | 133455151 | + | EIF4E2 | chr2 | 233428956 | + | 2.37E-10 | 1 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
 |
Top |
Fusion Protein Features for FUBP3-EIF4E2 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr9:133455151/chr2:233428957) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| FUBP3 | EIF4E2 |
| FUNCTION: May interact with single-stranded DNA from the far-upstream element (FUSE). May activate gene expression. | FUNCTION: Recognizes and binds the 7-methylguanosine-containing mRNA cap during an early step in the initiation (PubMed:17368478, PubMed:25624349, PubMed:9582349). Acts as a repressor of translation initiation (PubMed:22751931). In contrast to EIF4E, it is unable to bind eIF4G (EIF4G1, EIF4G2 or EIF4G3), suggesting that it acts by competing with EIF4E and block assembly of eIF4F at the cap (By similarity). In P-bodies, component of a complex that promotes miRNA-mediated translational repression (PubMed:28487484). {ECO:0000250|UniProtKB:Q8BMB3, ECO:0000269|PubMed:17368478, ECO:0000269|PubMed:22751931, ECO:0000269|PubMed:25624349, ECO:0000269|PubMed:28487484, ECO:0000269|PubMed:9582349}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | EIF4E2 | chr9:133455151 | chr2:233428957 | ENST00000258416 | 2 | 7 | 124_125 | 90 | 246.0 | Region | 7-methylguanosine-containing mRNA cap binding | |
| Tgene | EIF4E2 | chr9:133455151 | chr2:233428957 | ENST00000258416 | 2 | 7 | 150_157 | 90 | 246.0 | Region | EIF4EBP1/2/3 binding | |
| Tgene | EIF4E2 | chr9:133455151 | chr2:233428957 | ENST00000258416 | 2 | 7 | 174_179 | 90 | 246.0 | Region | 7-methylguanosine-containing mRNA cap binding | |
| Tgene | EIF4E2 | chr9:133455151 | chr2:233428957 | ENST00000258416 | 2 | 7 | 222_224 | 90 | 246.0 | Region | 7-methylguanosine-containing mRNA cap binding | |
| Tgene | EIF4E2 | chr9:133455151 | chr2:233428957 | ENST00000258416 | 2 | 7 | 95_99 | 90 | 246.0 | Region | EIF4EBP1/2/3 binding |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | FUBP3 | chr9:133455151 | chr2:233428957 | ENST00000319725 | + | 1 | 19 | 162_228 | 28 | 573.0 | Domain | KH 2 |
| Hgene | FUBP3 | chr9:133455151 | chr2:233428957 | ENST00000319725 | + | 1 | 19 | 253_317 | 28 | 573.0 | Domain | KH 3 |
| Hgene | FUBP3 | chr9:133455151 | chr2:233428957 | ENST00000319725 | + | 1 | 19 | 354_421 | 28 | 573.0 | Domain | KH 4 |
| Hgene | FUBP3 | chr9:133455151 | chr2:233428957 | ENST00000319725 | + | 1 | 19 | 77_141 | 28 | 573.0 | Domain | KH 1 |
| Tgene | EIF4E2 | chr9:133455151 | chr2:233428957 | ENST00000258416 | 2 | 7 | 54_57 | 90 | 246.0 | Region | EIF4EBP1/2/3 binding | |
| Tgene | EIF4E2 | chr9:133455151 | chr2:233428957 | ENST00000258416 | 2 | 7 | 78_79 | 90 | 246.0 | Region | 7-methylguanosine-containing mRNA cap binding |
Top |
Fusion Gene Sequence for FUBP3-EIF4E2 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >31758_31758_1_FUBP3-EIF4E2_FUBP3_chr9_133455151_ENST00000319725_EIF4E2_chr2_233428957_ENST00000258416_length(transcript)=820nt_BP=159nt CGGGAGGCCGGACCGGGGAGCCGAGCGGCGGCGTCGGCGGCGTCGGCGGCGGCGGCGACGGCGGCGGGGGCGGTAATGGCGGAGCTGGTG CAGGGGCAGAGCGCTCCTGTGGGGATGAAGGCCGAGGGCTTCGTGGATGCCCTGCACCGGGTCCGGCAGGTGGAGCAGTTCTGGAGGTTT TATAGCCACATGGTACGTCCTGGGGACCTGACAGGCCACAGTGACTTCCATCTCTTCAAAGAAGGAATTAAACCCATGTGGGAGGATGAT GCAAATAAAAATGGTGGCAAGTGGATTATTCGGCTGCGGAAGGGCTTGGCCTCCCGTTGCTGGGAGAATCTCATTTTGGCCATGCTGGGG GAACAGTTCATGGTTGGGGAGGAGATCTGTGGGGCTGTGGTGTCTGTCCGCTTTCAGGAAGACATTATTTCAATATGGAATAAGACTGCC AGTGACCAAGCAACCACAGCCCGAATCCGGGACACACTTCGGCGAGTGCTTAACCTACCTCCCAACACCATTATGGAATACAAAACTCAC ACCGACAGCATCAAAATGCCAGGCAGGCTGGGCCCCCAAAGGCTCCTTTTTCAAAACCTCTGGAAGCCGCGGTTGAATGTGCCATGACCC TCTCCCTCTCTGGATGGCACCATCATTGAAGCTGGCGTCATCGGAGTCTCTTGTTCTGTTGGCGTGCTACCTGGAAGATCCTTCTGTCCT GGACAAGAGGAATTGGAAGAGCATTTTATGTTTTAAGAACAGGCTGACACGCAGCAGCTACAACAACAGCTGAGATCACTTAATAAATGG >31758_31758_1_FUBP3-EIF4E2_FUBP3_chr9_133455151_ENST00000319725_EIF4E2_chr2_233428957_ENST00000258416_length(amino acids)=183AA_BP=27 MAELVQGQSAPVGMKAEGFVDALHRVRQVEQFWRFYSHMVRPGDLTGHSDFHLFKEGIKPMWEDDANKNGGKWIIRLRKGLASRCWENLI LAMLGEQFMVGEEICGAVVSVRFQEDIISIWNKTASDQATTARIRDTLRRVLNLPPNTIMEYKTHTDSIKMPGRLGPQRLLFQNLWKPRL -------------------------------------------------------------- >31758_31758_2_FUBP3-EIF4E2_FUBP3_chr9_133455151_ENST00000319725_EIF4E2_chr2_233428957_ENST00000409098_length(transcript)=3295nt_BP=159nt CGGGAGGCCGGACCGGGGAGCCGAGCGGCGGCGTCGGCGGCGTCGGCGGCGGCGGCGACGGCGGCGGGGGCGGTAATGGCGGAGCTGGTG CAGGGGCAGAGCGCTCCTGTGGGGATGAAGGCCGAGGGCTTCGTGGATGCCCTGCACCGGGTCCGGCAGGTGGAGCAGTTCTGGAGGTTT TATAGCCACATGGTACGTCCTGGGGACCTGACAGGCCACAGTGACTTCCATCTCTTCAAAGAAGGAATTAAACCCATGTGGGAGGATGAT GCAAATAAAAATGGTGGCAAGTGGATTATTCGGCTGCGGAAGGGCTTGGCCTCCCGTTGCTGGGAGAATCTCATTTTGGCCATGCTGGGG GAACAGTTCATGGTTGGGGAGGAGATCTGTGGGGCTGTGGTGTCTGTCCGCTTTCAGGAAGACATTATTTCAATATGGAATAAGACTGCC AGTGACCAAGCAACCACAGCCCGAATCCGGGACACACTTCGGCGAGTGCTTAACCTACCTCCCAACACCATTATGGAATACAAAACTCAC ACCGACAGCATCAAGGACAATTCAAGCTTTCGAAATACAAAAATCACATTGTGAGAGTACACAAGCTGGCAATAATCCTTTTCTGTATGT GTGTGTTTGTCCGAAATGGATGGGAGGCCTCCAGCCAGTCCTTCCATTGCTCACTGAAGGGACGTCCCTGAGCCGTGCGCTCTCCTTTTG CACTCATTCCTGGAGGACGGATTCCGCTAGACACCCTCCAGCATCGCTGACTTTATAAAGCGGAAAAACGGAAAACGTGACTTTGTAATA GACAAACATTGTTTTTTAATGGGGTTCCATGGGGGGGCGTTCAGCCTTTCTGTTTGCTGTGTGCTGTCCAGATGCCTCTTGGGCGTCCTG TTGTTCTCTTCCCGTGTTGTACGAAGGGTACCGTGGCCACGTGTACATGCCAGAGCTGTTGATGAGAGTTACCAGTATTATGAATGTCTG TGCATCCAGGAAAAGTGTTCTGTTGGAGAGTCCCAAAATAGCTGTAAATGCTCTCTTCTAGCTCTGCCATTAAATTATTGGGACATTTTG TTTCTTTCTCTCCCCTCCTCCAAGCCCACCACTTTCTGAAGCTGTGTTACTGATGTGATGACTGTCCAGGATTTTGTATCATCCCCTGCC TTATGCCCTCCGGTCTCCTCTCCTGTGGGACTCTCTCCTTCTTAGGCACAGCTGCACTTAGAATTGTAATCCAGTGGCTCTCACTGGTTT CACTGAGGCAGCTTAGAGGGAATGCCTTAGTAAACTGAACTGGAGCAGAATCTAGTTTGATTAGGAGCTAGATTATAATCTAGGTTCCAA GTACAGAATTTGGAGCTAAGGACTGTGACTGAAATCATTTTCCCATATGAGCAGACCCTGTGTGTCAGGCCTGTTTCCCATATGAGCAGA GCCTGTGTGCAAGTCTGTTTCTGGCATGTCCCTCATTGAGGAAGGGAAGCAAAAGCTGGTTATTGCCAGGCCTATTAACACTTAATATGC AAATTCTATCATCCTGAAACTGGGGCATCTGAGGAAAAGGTGACCTTGCTGGATGGCTTTATTTGCATGGCTCTGCCTGTCTGCAGTGGT TGAGTCCTCATCACCTGGTATGTGTATGAGCAAATGTGTGCTGATTGTGATGCCCAGGCAGAAACCTCTTGAAGACTGCTGCAGGCATGC TTTAAAAATGACCAGTCACTCATCAGAGAAGCTGGGTGATCTGACTCCAGAAGGACTGAAGTCAGAGAAGTCACAAGAGCACCTAGGATT CAAATAAATAGCGTCAGAGTCCTATAGCAACCTCCAAGTAGCACCGTCTTACTTGGCTCTTGTGAGCAAAGACTGCAGTACCTTAAATTA AGGCCTCTCTTTAAAACATATGTGGAAGACTAGGGGATCCTTGGCCACCTGGTCTCAGAGAAATCATATGAGAGTAACAGGCATTTCCTT ATTGTATTTGTACTACACTCTTCCTACTTTTCCATTCCTGAACACCCTCTAATTACCACTGTTTTGGGGATGCTTTTTTTCTGAAAGAAC GGGGAGTACAGGGGCCAAAAGGGAGGTGCTTCTATTACAGGGCAAGTTAGATCAGATAAGAAGATCTAGCAGTGATTTAAACTCCAGGAA TATGAAGAGTGCATTCTGGTGTCCAGACAGTTGTGAAGGGCTCGGCAAATATAAGAGCACCATTGGCTACATGGAGAGCAAAGGCTGTCT TTGAAGACCCCAGGGAGGTCTTCACTTTTGCCTAAATTCAGATTTGCCGTGAAAATTCCAAAGAGAGCAGATATTTGGATTTGCCCTCCT TTGGGCATCTACCTGACTGTTGTGTGTGTGGGAAAGTCAGTGTGTATGTGTAGAGTGTGCTTAGGAGTGAGTGAGTGGTCAGGCCTCCTG GCTAGGTGTGTTGTCCATAGTTTTGTTGTTGTTGTTGTTGTTGTTCAGAGTTTTGAATCTTACGTTTTCTAGAGCTGCTCATGTTTTCCC TTCCTTTTTGTCGTTGTAGTATTTGACAGTTGTCTTTTCCATCAAAAACATACTGGCCCTAGGCTGGTTAGAGCAAGGGGTTATGCCTGT TAGGCAGCATCTTACGCCCAGTGTTCTCCCAGATATTCTGCCTAACAGGTTTAAGTAGGAGATTAAATAGTCAGTTTATCTAAACCCTCT ATTTTTCCAAACTAGCTTGAGAGGTCTTTTCATCTATTTTTACTCCATGGGCCTCTGATATGCTGAGATGTGGCACGGTTATGATTATGG TATCACCGTTACAGAGGCAAAGGGATAAAGGTCTACCAGGGCCAACGTAACCAGAATGTGAGGAGAGTGAAGAGTCGGTCTCCGCTTTGC ACAGTACCAGAATGTGAGGTTGCCACTGGAGAGTGGAGAGTCGGTCTCCACTTTGCACAGTTGGAGCTGTTCTCCTCTAACTCCTTTGCT GGGTTTTCTGTTTGAAATTGGCCCCTACTCCTCTCCAGCCCTTTACTGGGTTTTCTGTTTGAAATTGGCCCCTACTCCTCTCCAGTGTCT GCTTTTTGAATCCTTCTGTTCGTGTGTGTGTGTGTGTGTGTGTGTGTTCCCTATGATACTGGCAGTGGCAATAATTTTCCACCCAGAGAG AAGCTTGTGGTCCACAATGCTCGAAGAATGAATTTCCAAGTATTTGCCAGTGGAAACGGAGCAGAGGCTATGACAAGAGTGATTTAGTGT >31758_31758_2_FUBP3-EIF4E2_FUBP3_chr9_133455151_ENST00000319725_EIF4E2_chr2_233428957_ENST00000409098_length(amino acids)=172AA_BP=27 MAELVQGQSAPVGMKAEGFVDALHRVRQVEQFWRFYSHMVRPGDLTGHSDFHLFKEGIKPMWEDDANKNGGKWIIRLRKGLASRCWENLI -------------------------------------------------------------- >31758_31758_3_FUBP3-EIF4E2_FUBP3_chr9_133455151_ENST00000319725_EIF4E2_chr2_233428957_ENST00000409167_length(transcript)=1360nt_BP=159nt CGGGAGGCCGGACCGGGGAGCCGAGCGGCGGCGTCGGCGGCGTCGGCGGCGGCGGCGACGGCGGCGGGGGCGGTAATGGCGGAGCTGGTG CAGGGGCAGAGCGCTCCTGTGGGGATGAAGGCCGAGGGCTTCGTGGATGCCCTGCACCGGGTCCGGCAGGTGGAGCAGTTCTGGAGGTTT TATAGCCACATGGTACGTCCTGGGGACCTGACAGGCCACAGTGACTTCCATCTCTTCAAAGAAGGAATTAAACCCATGTGGGAGGATGAT GCAAATAAAAATGGTGGCAAGTGGATTATTCGGCTGCGGAAGGGCTTGGCCTCCCGTTGCTGGGAGAATCTCATTTTGGCCATGCTGGGG GAACAGTTCATGGTTGGGGAGGAGATCTGTGGGGCTGTGGTGTCTGTCCGCTTTCAGGAAGACATTATTTCAATATGGAATAAGACTGCC AGTGACCAAGCAACCACAGCCCGAATCCGGGACACACTTCGGCGAGTGCTTAACCTACCTCCCAACACCATTATGGAATACAAAACTCAC ACCGACAGCATCAAGTACGTGTTGGGGGGTTATGGGAGGACGTGTCCCTAAGCTTAGTATAAGTAGATCTGTAGTTGGGCCCAGAGGAAT ATGGCCGGACAGGAGACTTACTTGTGGAGAAGGGACAGACCAGGCCTTCCCTCTAGGGCTTCTGGCTACTTGCTGCAGTTCTTTGTCAGC GAGGCTCCCTGCCACCTATACTACCAGGTGCTTTGTAGCAGAGAGAGCCCATCTGCAGAAACCCAGTTGTACTACCTGGGCTTGCTTAAT TTCTACAGGCTGTCTTGTCTTTCATATGCTAACCACTATCTTAAAATGAAAGACAAGTGCTTTTCTTTTTCTTGCTGTTTTTTAAGATAA TTTCACCCTGCCCACCCTCACCCCACTTCTGCATGTCTTTTTAACTTGTTAGCTTCAGTCTATTTTCTTTGTCCTGGGATACTGACTACT TATCAGGAATGACTCCATTTCCTTGTGTATAGAGCTGACTCTTAGAAGAAGGGGGATAGTGTGTGTGTGAGAGAGCAGAATCAAAGGGAA GCATGAAGGCCACAGGGAAAATAAGCTTGGCAATCTCCCTAGGACTCAGGGAAATAGAGCTCTCTGTCCACAGTGAGAATCACTGTTAGG CTTGCTCCTCAGTGATTGAGTGTCAGAAGTAAGCTCAGGCTATTGTGAAAAAGTGTGGGGAGTGGCTGTCAGATTGTCTCAAAGCTGGAG GGAGAGCTAGCCAGCTAAGAGTTAGGACTCTTAGGTTTAGTTCTCTCATATGGCCCCATACCAATATGTGCCTTTGACAAGTGTCTTTGC >31758_31758_3_FUBP3-EIF4E2_FUBP3_chr9_133455151_ENST00000319725_EIF4E2_chr2_233428957_ENST00000409167_length(amino acids)=197AA_BP= MMQIKMVASGLFGCGRAWPPVAGRISFWPCWGNSSWLGRRSVGLWCLSAFRKTLFQYGIRLPVTKQPQPESGTHFGECLTYLPTPLWNTK LTPTASSTCWGVMGGRVPKLSISRSVVGPRGIWPDRRLTCGEGTDQAFPLGLLATCCSSLSARLPATYTTRCFVAERAHLQKPSCTTWAC -------------------------------------------------------------- >31758_31758_4_FUBP3-EIF4E2_FUBP3_chr9_133455151_ENST00000319725_EIF4E2_chr2_233428957_ENST00000409322_length(transcript)=788nt_BP=159nt CGGGAGGCCGGACCGGGGAGCCGAGCGGCGGCGTCGGCGGCGTCGGCGGCGGCGGCGACGGCGGCGGGGGCGGTAATGGCGGAGCTGGTG CAGGGGCAGAGCGCTCCTGTGGGGATGAAGGCCGAGGGCTTCGTGGATGCCCTGCACCGGGTCCGGCAGGTGGAGCAGTTCTGGAGGTTT TATAGCCACATGGTACGTCCTGGGGACCTGACAGGCCACAGTGACTTCCATCTCTTCAAAGAAGGAATTAAACCCATGTGGGAGGATGAT GCAAATAAAAATGGTGGCAAGTGGATTATTCGGCTGCGGAAGGGCTTGGCCTCCCGTTGCTGGGAGAATCTCATTTTGGCCATGCTGGGG GAACAGTTCATGGTTGGGGAGGAGATCTGTGGGGCTGTGGTGTCTGTCCGCTTTCAGGAAGACATTATTTCAATATGGAATAAGACTGCC AGTGACCAAGCAACCACAGCCCGAATCCGGGACACACTTCGGCGAGTGCTTAACCTACCTCCCAACACCATTATGGAATACAAAACTCAC ACCGACAGCATCAAAATGCCAGGCAGGCTGGGCCCCCAAAGGCTCCTTTTTCAAAACCTCTGGAAGCCGCGGTTGAATGTGCCATGACCC TCTCCCTCTCTGGATGGCACCATCATTGAAGCTGGCGTCATCGGAGTCTCTTGTTCTGTTGGCGTGCTACCTGGAAGATCCTTCTGTCCT >31758_31758_4_FUBP3-EIF4E2_FUBP3_chr9_133455151_ENST00000319725_EIF4E2_chr2_233428957_ENST00000409322_length(amino acids)=183AA_BP=27 MAELVQGQSAPVGMKAEGFVDALHRVRQVEQFWRFYSHMVRPGDLTGHSDFHLFKEGIKPMWEDDANKNGGKWIIRLRKGLASRCWENLI LAMLGEQFMVGEEICGAVVSVRFQEDIISIWNKTASDQATTARIRDTLRRVLNLPPNTIMEYKTHTDSIKMPGRLGPQRLLFQNLWKPRL -------------------------------------------------------------- >31758_31758_5_FUBP3-EIF4E2_FUBP3_chr9_133455151_ENST00000319725_EIF4E2_chr2_233428957_ENST00000409394_length(transcript)=607nt_BP=159nt CGGGAGGCCGGACCGGGGAGCCGAGCGGCGGCGTCGGCGGCGTCGGCGGCGGCGGCGACGGCGGCGGGGGCGGTAATGGCGGAGCTGGTG CAGGGGCAGAGCGCTCCTGTGGGGATGAAGGCCGAGGGCTTCGTGGATGCCCTGCACCGGGTCCGGCAGGTGGAGCAGTTCTGGAGGTTT TATAGCCACATGGTACGTCCTGGGGACCTGACAGGCCACAGTGACTTCCATCTCTTCAAAGAAGGAATTAAACCCATGTGGGAGGATGAT GCAAATAAAAATGGTGGCAAGTGGATTATTCGGCTGCGGAAGGGCTTGGCCTCCCGTTGCTGGGAGAATCTCATTTTGGCCATGCTGGGG GAACAGTTCATGGTTGGGGAGGAGATCTGTGGGGCTGTGGTGTCTGTCCGCTTTCAGGAAGACATTATTTCAATATGGAATAAGACTGCC AGTGACCAAGCAACCACAGCCCGAATCCGGGACACACTTCGGCGAGTGCTTAACCTACCTCCCAACACCATTATGGAATACAAAACTCAC >31758_31758_5_FUBP3-EIF4E2_FUBP3_chr9_133455151_ENST00000319725_EIF4E2_chr2_233428957_ENST00000409394_length(amino acids)=172AA_BP=27 MAELVQGQSAPVGMKAEGFVDALHRVRQVEQFWRFYSHMVRPGDLTGHSDFHLFKEGIKPMWEDDANKNGGKWIIRLRKGLASRCWENLI -------------------------------------------------------------- >31758_31758_6_FUBP3-EIF4E2_FUBP3_chr9_133455151_ENST00000319725_EIF4E2_chr2_233428957_ENST00000409495_length(transcript)=867nt_BP=159nt CGGGAGGCCGGACCGGGGAGCCGAGCGGCGGCGTCGGCGGCGTCGGCGGCGGCGGCGACGGCGGCGGGGGCGGTAATGGCGGAGCTGGTG CAGGGGCAGAGCGCTCCTGTGGGGATGAAGGCCGAGGGCTTCGTGGATGCCCTGCACCGGGTCCGGCAGGTGGAGCAGTTCTGGAGGTTT TATAGCCACATGGTACGTCCTGGGGACCTGACAGGCCACAGTGACTTCCATCTCTTCAAAGAAGGAATTAAACCCATGTGGGAGGATGAT GCAAATAAAAATGGTGGCAAGTGGATTATTCGGCTGCGGAAGGGCTTGGCCTCCCGTTGCTGGGAGAATCTCATTTTGGCCATGCTGGGG GAACAGTTCATGGTTGGGGAGGAGATCTGTGGGGCTGTGGTGTCTGTCCGCTTTCAGGAAGACATTATTTCAATATGGAATAAGACTGCC AGTGACCAAGCAACCACAGCCCGAATCCGGGACACACTTCGGCGAGTGCTTAACCTACCTCCCAACACCATTATGGAATACAAAACTCAC ACCGACAGCATCAAGTACGTGTTGGGGGGTTATGGGAGGACGTGTCCCTAAGCTTAGTATAAGTAGATCTGTAGTTGGGCCCAGAGGAAT ATGGCCGGACAGGAGACTTACTTGTGGAGAAGGGACAGACCAGGCCTTCCCTCTAGGGCTTCTGGCTACTTGCTGCAGTTCTTTGTCAGC GAGGCTCCCTGCCACCTATACTACCAGGTGCTTTGTAGCAGAGAGAGCCCATCTGCAGAAACCCAGTTGTACTACCTGGGCTTGCTTAAT >31758_31758_6_FUBP3-EIF4E2_FUBP3_chr9_133455151_ENST00000319725_EIF4E2_chr2_233428957_ENST00000409495_length(amino acids)=197AA_BP= MMQIKMVASGLFGCGRAWPPVAGRISFWPCWGNSSWLGRRSVGLWCLSAFRKTLFQYGIRLPVTKQPQPESGTHFGECLTYLPTPLWNTK LTPTASSTCWGVMGGRVPKLSISRSVVGPRGIWPDRRLTCGEGTDQAFPLGLLATCCSSLSARLPATYTTRCFVAERAHLQKPSCTTWAC -------------------------------------------------------------- >31758_31758_7_FUBP3-EIF4E2_FUBP3_chr9_133455151_ENST00000319725_EIF4E2_chr2_233428957_ENST00000409514_length(transcript)=3372nt_BP=159nt CGGGAGGCCGGACCGGGGAGCCGAGCGGCGGCGTCGGCGGCGTCGGCGGCGGCGGCGACGGCGGCGGGGGCGGTAATGGCGGAGCTGGTG CAGGGGCAGAGCGCTCCTGTGGGGATGAAGGCCGAGGGCTTCGTGGATGCCCTGCACCGGGTCCGGCAGGTGGAGCAGTTCTGGAGGTTT TATAGCCACATGGTACGTCCTGGGGACCTGACAGGCCACAGTGACTTCCATCTCTTCAAAGAAGGAATTAAACCCATGTGGGAGGATGAT GCAAATAAAAATGGTGGCAAGTGGATTATTCGGCTGCGGAAGGGCTTGGCCTCCCGTTGCTGGGAGAATCTCATTTTGGCCATGCTGGGG GAACAGTTCATGGTTGGGGAGGAGATCTGTGGGGCTGTGGTGTCTGTCCGCTTTCAGGAAGACATTATTTCAATATGGAATAAGACTGCC AGTGACCAAGCAACCACAGCCCGAATCCGGGACACACTTCGGCGAGTGCTTAACCTACCTCCCAACACCATTATGGAATACAAAACTCAC ACCGACAGCATCAAGGCCTGGGAGGAGTTTCATGGCCTGGTGAACAGCAGCGGCCGCTGATCGGACGCTCCCCCTCTGTCTCTCACACTC AGGGGACAATTCAAGCTTTCGAAATACAAAAATCACATTGTGAGAGTACACAAGCTGGCAATAATCCTTTTCTGTATGTGTGTGTTTGTC CGAAATGGATGGGAGGCCTCCAGCCAGTCCTTCCATTGCTCACTGAAGGGACGTCCCTGAGCCGTGCGCTCTCCTTTTGCACTCATTCCT GGAGGACGGATTCCGCTAGACACCCTCCAGCATCGCTGACTTTATAAAGCGGAAAAACGGAAAACGTGACTTTGTAATAGACAAACATTG TTTTTTAATGGGGTTCCATGGGGGGGCGTTCAGCCTTTCTGTTTGCTGTGTGCTGTCCAGATGCCTCTTGGGCGTCCTGTTGTTCTCTTC CCGTGTTGTACGAAGGGTACCGTGGCCACGTGTACATGCCAGAGCTGTTGATGAGAGTTACCAGTATTATGAATGTCTGTGCATCCAGGA AAAGTGTTCTGTTGGAGAGTCCCAAAATAGCTGTAAATGCTCTCTTCTAGCTCTGCCATTAAATTATTGGGACATTTTGTTTCTTTCTCT CCCCTCCTCCAAGCCCACCACTTTCTGAAGCTGTGTTACTGATGTGATGACTGTCCAGGATTTTGTATCATCCCCTGCCTTATGCCCTCC GGTCTCCTCTCCTGTGGGACTCTCTCCTTCTTAGGCACAGCTGCACTTAGAATTGTAATCCAGTGGCTCTCACTGGTTTCACTGAGGCAG CTTAGAGGGAATGCCTTAGTAAACTGAACTGGAGCAGAATCTAGTTTGATTAGGAGCTAGATTATAATCTAGGTTCCAAGTACAGAATTT GGAGCTAAGGACTGTGACTGAAATCATTTTCCCATATGAGCAGACCCTGTGTGTCAGGCCTGTTTCCCATATGAGCAGAGCCTGTGTGCA AGTCTGTTTCTGGCATGTCCCTCATTGAGGAAGGGAAGCAAAAGCTGGTTATTGCCAGGCCTATTAACACTTAATATGCAAATTCTATCA TCCTGAAACTGGGGCATCTGAGGAAAAGGTGACCTTGCTGGATGGCTTTATTTGCATGGCTCTGCCTGTCTGCAGTGGTTGAGTCCTCAT CACCTGGTATGTGTATGAGCAAATGTGTGCTGATTGTGATGCCCAGGCAGAAACCTCTTGAAGACTGCTGCAGGCATGCTTTAAAAATGA CCAGTCACTCATCAGAGAAGCTGGGTGATCTGACTCCAGAAGGACTGAAGTCAGAGAAGTCACAAGAGCACCTAGGATTCAAATAAATAG CGTCAGAGTCCTATAGCAACCTCCAAGTAGCACCGTCTTACTTGGCTCTTGTGAGCAAAGACTGCAGTACCTTAAATTAAGGCCTCTCTT TAAAACATATGTGGAAGACTAGGGGATCCTTGGCCACCTGGTCTCAGAGAAATCATATGAGAGTAACAGGCATTTCCTTATTGTATTTGT ACTACACTCTTCCTACTTTTCCATTCCTGAACACCCTCTAATTACCACTGTTTTGGGGATGCTTTTTTTCTGAAAGAACGGGGAGTACAG GGGCCAAAAGGGAGGTGCTTCTATTACAGGGCAAGTTAGATCAGATAAGAAGATCTAGCAGTGATTTAAACTCCAGGAATATGAAGAGTG CATTCTGGTGTCCAGACAGTTGTGAAGGGCTCGGCAAATATAAGAGCACCATTGGCTACATGGAGAGCAAAGGCTGTCTTTGAAGACCCC AGGGAGGTCTTCACTTTTGCCTAAATTCAGATTTGCCGTGAAAATTCCAAAGAGAGCAGATATTTGGATTTGCCCTCCTTTGGGCATCTA CCTGACTGTTGTGTGTGTGGGAAAGTCAGTGTGTATGTGTAGAGTGTGCTTAGGAGTGAGTGAGTGGTCAGGCCTCCTGGCTAGGTGTGT TGTCCATAGTTTTGTTGTTGTTGTTGTTGTTGTTCAGAGTTTTGAATCTTACGTTTTCTAGAGCTGCTCATGTTTTCCCTTCCTTTTTGT CGTTGTAGTATTTGACAGTTGTCTTTTCCATCAAAAACATACTGGCCCTAGGCTGGTTAGAGCAAGGGGTTATGCCTGTTAGGCAGCATC TTACGCCCAGTGTTCTCCCAGATATTCTGCCTAACAGGTTTAAGTAGGAGATTAAATAGTCAGTTTATCTAAACCCTCTATTTTTCCAAA CTAGCTTGAGAGGTCTTTTCATCTATTTTTACTCCATGGGCCTCTGATATGCTGAGATGTGGCACGGTTATGATTATGGTATCACCGTTA CAGAGGCAAAGGGATAAAGGTCTACCAGGGCCAACGTAACCAGAATGTGAGGAGAGTGAAGAGTCGGTCTCCGCTTTGCACAGTACCAGA ATGTGAGGTTGCCACTGGAGAGTGGAGAGTCGGTCTCCACTTTGCACAGTTGGAGCTGTTCTCCTCTAACTCCTTTGCTGGGTTTTCTGT TTGAAATTGGCCCCTACTCCTCTCCAGCCCTTTACTGGGTTTTCTGTTTGAAATTGGCCCCTACTCCTCTCCAGTGTCTGCTTTTTGAAT CCTTCTGTTCGTGTGTGTGTGTGTGTGTGTGTGTGTTCCCTATGATACTGGCAGTGGCAATAATTTTCCACCCAGAGAGAAGCTTGTGGT CCACAATGCTCGAAGAATGAATTTCCAAGTATTTGCCAGTGGAAACGGAGCAGAGGCTATGACAAGAGTGATTTAGTGTTGCACTTTCAG >31758_31758_7_FUBP3-EIF4E2_FUBP3_chr9_133455151_ENST00000319725_EIF4E2_chr2_233428957_ENST00000409514_length(amino acids)=174AA_BP=27 MAELVQGQSAPVGMKAEGFVDALHRVRQVEQFWRFYSHMVRPGDLTGHSDFHLFKEGIKPMWEDDANKNGGKWIIRLRKGLASRCWENLI -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for FUBP3-EIF4E2 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for FUBP3-EIF4E2 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for FUBP3-EIF4E2 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
