
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:FUCA2-FIP1L1 (FusionGDB2 ID:31765) |
Fusion Gene Summary for FUCA2-FIP1L1 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: FUCA2-FIP1L1 | Fusion gene ID: 31765 | Hgene | Tgene | Gene symbol | FUCA2 | FIP1L1 | Gene ID | 2519 | 81608 |
| Gene name | alpha-L-fucosidase 2 | factor interacting with PAPOLA and CPSF1 | |
| Synonyms | dJ20N2.5 | FIP1|Rhe|hFip1 | |
| Cytomap | 6q24.2 | 4q12 | |
| Type of gene | protein-coding | protein-coding | |
| Description | plasma alpha-L-fucosidaseI+/--L-fucosidase 2a-L-fucosidase 2alpha-L-fucoside fucohydrolase 2fucosidase, alpha-L- 2, plasmaplasma fucosidase | pre-mRNA 3'-end-processing factor FIP1FIP1 like 1FIP1-like 1 proteinFIP1L1 cleavage and polyadenylation specific factor subunitfactor interacting with PAPrearranged in hypereosinophilia | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | Q9BTY2 | Q6UN15 | |
| Ensembl transtripts involved in fusion gene | ENST00000002165, ENST00000438118, ENST00000367585, | ENST00000510668, ENST00000306932, ENST00000337488, ENST00000358575, ENST00000507166, ENST00000507922, | |
| Fusion gene scores | * DoF score | 5 X 5 X 3=75 | 17 X 15 X 11=2805 |
| # samples | 5 | 23 | |
| ** MAII score | log2(5/75*10)=-0.584962500721156 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(23/2805*10)=-3.60829500455178 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: FUCA2 [Title/Abstract] AND FIP1L1 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | FUCA2(143828374)-FIP1L1(54265897), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | FUCA2-FIP1L1 seems lost the major protein functional domain in Tgene partner, which is a CGC due to the frame-shifted ORF. FUCA2-FIP1L1 seems lost the major protein functional domain in Tgene partner, which is a essential gene due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | FUCA2 | GO:0006004 | fucose metabolic process | 19666478 |
| Hgene | FUCA2 | GO:0016139 | glycoside catabolic process | 19666478 |
 Fusion gene breakpoints across FUCA2 (5'-gene) Fusion gene breakpoints across FUCA2 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
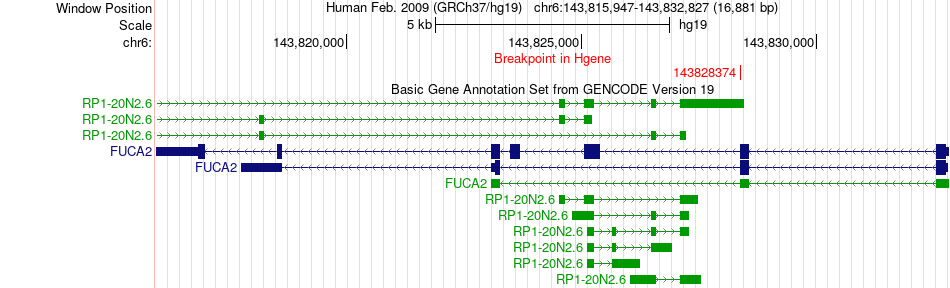 |
 Fusion gene breakpoints across FIP1L1 (3'-gene) Fusion gene breakpoints across FIP1L1 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
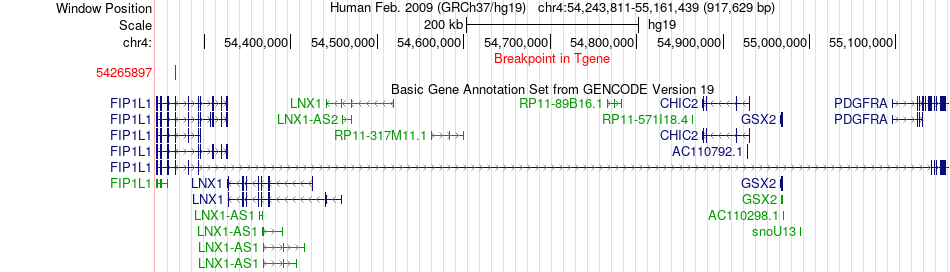 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | STAD | TCGA-HU-A4HB-01A | FUCA2 | chr6 | 143828374 | - | FIP1L1 | chr4 | 54265897 | + |
Top |
Fusion Gene ORF analysis for FUCA2-FIP1L1 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-intron | ENST00000002165 | ENST00000510668 | FUCA2 | chr6 | 143828374 | - | FIP1L1 | chr4 | 54265897 | + |
| 5CDS-intron | ENST00000438118 | ENST00000510668 | FUCA2 | chr6 | 143828374 | - | FIP1L1 | chr4 | 54265897 | + |
| 5UTR-3CDS | ENST00000367585 | ENST00000306932 | FUCA2 | chr6 | 143828374 | - | FIP1L1 | chr4 | 54265897 | + |
| 5UTR-3CDS | ENST00000367585 | ENST00000337488 | FUCA2 | chr6 | 143828374 | - | FIP1L1 | chr4 | 54265897 | + |
| 5UTR-3CDS | ENST00000367585 | ENST00000358575 | FUCA2 | chr6 | 143828374 | - | FIP1L1 | chr4 | 54265897 | + |
| 5UTR-3CDS | ENST00000367585 | ENST00000507166 | FUCA2 | chr6 | 143828374 | - | FIP1L1 | chr4 | 54265897 | + |
| 5UTR-3CDS | ENST00000367585 | ENST00000507922 | FUCA2 | chr6 | 143828374 | - | FIP1L1 | chr4 | 54265897 | + |
| 5UTR-intron | ENST00000367585 | ENST00000510668 | FUCA2 | chr6 | 143828374 | - | FIP1L1 | chr4 | 54265897 | + |
| Frame-shift | ENST00000002165 | ENST00000306932 | FUCA2 | chr6 | 143828374 | - | FIP1L1 | chr4 | 54265897 | + |
| Frame-shift | ENST00000002165 | ENST00000337488 | FUCA2 | chr6 | 143828374 | - | FIP1L1 | chr4 | 54265897 | + |
| Frame-shift | ENST00000002165 | ENST00000358575 | FUCA2 | chr6 | 143828374 | - | FIP1L1 | chr4 | 54265897 | + |
| Frame-shift | ENST00000002165 | ENST00000507166 | FUCA2 | chr6 | 143828374 | - | FIP1L1 | chr4 | 54265897 | + |
| Frame-shift | ENST00000002165 | ENST00000507922 | FUCA2 | chr6 | 143828374 | - | FIP1L1 | chr4 | 54265897 | + |
| In-frame | ENST00000438118 | ENST00000306932 | FUCA2 | chr6 | 143828374 | - | FIP1L1 | chr4 | 54265897 | + |
| In-frame | ENST00000438118 | ENST00000337488 | FUCA2 | chr6 | 143828374 | - | FIP1L1 | chr4 | 54265897 | + |
| In-frame | ENST00000438118 | ENST00000358575 | FUCA2 | chr6 | 143828374 | - | FIP1L1 | chr4 | 54265897 | + |
| In-frame | ENST00000438118 | ENST00000507166 | FUCA2 | chr6 | 143828374 | - | FIP1L1 | chr4 | 54265897 | + |
| In-frame | ENST00000438118 | ENST00000507922 | FUCA2 | chr6 | 143828374 | - | FIP1L1 | chr4 | 54265897 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000438118 | FUCA2 | chr6 | 143828374 | - | ENST00000507166 | FIP1L1 | chr4 | 54265897 | + | 2285 | 440 | 434 | 2284 | 617 |
| ENST00000438118 | FUCA2 | chr6 | 143828374 | - | ENST00000358575 | FIP1L1 | chr4 | 54265897 | + | 1766 | 440 | 434 | 1546 | 370 |
| ENST00000438118 | FUCA2 | chr6 | 143828374 | - | ENST00000337488 | FIP1L1 | chr4 | 54265897 | + | 1739 | 440 | 434 | 1519 | 361 |
| ENST00000438118 | FUCA2 | chr6 | 143828374 | - | ENST00000507922 | FIP1L1 | chr4 | 54265897 | + | 947 | 440 | 16 | 501 | 161 |
| ENST00000438118 | FUCA2 | chr6 | 143828374 | - | ENST00000306932 | FIP1L1 | chr4 | 54265897 | + | 1584 | 440 | 434 | 1411 | 325 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000438118 | ENST00000507166 | FUCA2 | chr6 | 143828374 | - | FIP1L1 | chr4 | 54265897 | + | 0.003163563 | 0.9968364 |
| ENST00000438118 | ENST00000358575 | FUCA2 | chr6 | 143828374 | - | FIP1L1 | chr4 | 54265897 | + | 0.016735017 | 0.983265 |
| ENST00000438118 | ENST00000337488 | FUCA2 | chr6 | 143828374 | - | FIP1L1 | chr4 | 54265897 | + | 0.022038277 | 0.9779618 |
| ENST00000438118 | ENST00000507922 | FUCA2 | chr6 | 143828374 | - | FIP1L1 | chr4 | 54265897 | + | 0.04438797 | 0.955612 |
| ENST00000438118 | ENST00000306932 | FUCA2 | chr6 | 143828374 | - | FIP1L1 | chr4 | 54265897 | + | 0.016110793 | 0.9838892 |
Top |
Fusion Genomic Features for FUCA2-FIP1L1 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| FUCA2 | chr6 | 143828373 | - | FIP1L1 | chr4 | 54265896 | + | 1.65E-05 | 0.99998343 |
| FUCA2 | chr6 | 143828373 | - | FIP1L1 | chr4 | 54265896 | + | 1.65E-05 | 0.99998343 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
 |
Top |
Fusion Protein Features for FUCA2-FIP1L1 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr6:143828374/chr4:54265897) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| FUCA2 | FIP1L1 |
| FUNCTION: Alpha-L-fucosidase is responsible for hydrolyzing the alpha-1,6-linked fucose joined to the reducing-end N-acetylglucosamine of the carbohydrate moieties of glycoproteins. | FUNCTION: Component of the cleavage and polyadenylation specificity factor (CPSF) complex that plays a key role in pre-mRNA 3'-end formation, recognizing the AAUAAA signal sequence and interacting with poly(A) polymerase and other factors to bring about cleavage and poly(A) addition. FIP1L1 contributes to poly(A) site recognition and stimulates poly(A) addition. Binds to U-rich RNA sequence elements surrounding the poly(A) site. May act to tether poly(A) polymerase to the CPSF complex. {ECO:0000269|PubMed:14749727}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | FIP1L1 | chr6:143828374 | chr4:54265897 | ENST00000306932 | 6 | 15 | 356_406 | 197 | 521.0 | Compositional bias | Note=Pro-rich | |
| Tgene | FIP1L1 | chr6:143828374 | chr4:54265897 | ENST00000306932 | 6 | 15 | 456_562 | 197 | 521.0 | Compositional bias | Note=Arg-rich | |
| Tgene | FIP1L1 | chr6:143828374 | chr4:54265897 | ENST00000306932 | 6 | 15 | 478_594 | 197 | 521.0 | Compositional bias | Note=Glu-rich | |
| Tgene | FIP1L1 | chr6:143828374 | chr4:54265897 | ENST00000337488 | 8 | 18 | 356_406 | 235 | 595.0 | Compositional bias | Note=Pro-rich | |
| Tgene | FIP1L1 | chr6:143828374 | chr4:54265897 | ENST00000337488 | 8 | 18 | 456_562 | 235 | 595.0 | Compositional bias | Note=Arg-rich | |
| Tgene | FIP1L1 | chr6:143828374 | chr4:54265897 | ENST00000337488 | 8 | 18 | 478_594 | 235 | 595.0 | Compositional bias | Note=Glu-rich | |
| Tgene | FIP1L1 | chr6:143828374 | chr4:54265897 | ENST00000358575 | 7 | 18 | 356_406 | 220 | 589.0 | Compositional bias | Note=Pro-rich | |
| Tgene | FIP1L1 | chr6:143828374 | chr4:54265897 | ENST00000358575 | 7 | 18 | 456_562 | 220 | 589.0 | Compositional bias | Note=Arg-rich | |
| Tgene | FIP1L1 | chr6:143828374 | chr4:54265897 | ENST00000358575 | 7 | 18 | 478_594 | 220 | 589.0 | Compositional bias | Note=Glu-rich | |
| Tgene | FIP1L1 | chr6:143828374 | chr4:54265897 | ENST00000507166 | 8 | 24 | 356_406 | 235 | 850.0 | Compositional bias | Note=Pro-rich | |
| Tgene | FIP1L1 | chr6:143828374 | chr4:54265897 | ENST00000507166 | 8 | 24 | 456_562 | 235 | 850.0 | Compositional bias | Note=Arg-rich | |
| Tgene | FIP1L1 | chr6:143828374 | chr4:54265897 | ENST00000507166 | 8 | 24 | 478_594 | 235 | 850.0 | Compositional bias | Note=Glu-rich | |
| Tgene | FIP1L1 | chr6:143828374 | chr4:54265897 | ENST00000507922 | 7 | 12 | 356_406 | 220 | 379.0 | Compositional bias | Note=Pro-rich | |
| Tgene | FIP1L1 | chr6:143828374 | chr4:54265897 | ENST00000507922 | 7 | 12 | 456_562 | 220 | 379.0 | Compositional bias | Note=Arg-rich | |
| Tgene | FIP1L1 | chr6:143828374 | chr4:54265897 | ENST00000507922 | 7 | 12 | 478_594 | 220 | 379.0 | Compositional bias | Note=Glu-rich | |
| Tgene | FIP1L1 | chr6:143828374 | chr4:54265897 | ENST00000306932 | 6 | 15 | 457_490 | 197 | 521.0 | Region | Arg/Asp/Glu-rich domain | |
| Tgene | FIP1L1 | chr6:143828374 | chr4:54265897 | ENST00000337488 | 8 | 18 | 457_490 | 235 | 595.0 | Region | Arg/Asp/Glu-rich domain | |
| Tgene | FIP1L1 | chr6:143828374 | chr4:54265897 | ENST00000358575 | 7 | 18 | 457_490 | 220 | 589.0 | Region | Arg/Asp/Glu-rich domain | |
| Tgene | FIP1L1 | chr6:143828374 | chr4:54265897 | ENST00000507166 | 8 | 24 | 457_490 | 235 | 850.0 | Region | Arg/Asp/Glu-rich domain | |
| Tgene | FIP1L1 | chr6:143828374 | chr4:54265897 | ENST00000507922 | 7 | 12 | 457_490 | 220 | 379.0 | Region | Arg/Asp/Glu-rich domain |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | FIP1L1 | chr6:143828374 | chr4:54265897 | ENST00000306932 | 6 | 15 | 1_356 | 197 | 521.0 | Region | Note=Necessary for stimulating PAPOLA activity | |
| Tgene | FIP1L1 | chr6:143828374 | chr4:54265897 | ENST00000337488 | 8 | 18 | 1_356 | 235 | 595.0 | Region | Note=Necessary for stimulating PAPOLA activity | |
| Tgene | FIP1L1 | chr6:143828374 | chr4:54265897 | ENST00000358575 | 7 | 18 | 1_356 | 220 | 589.0 | Region | Note=Necessary for stimulating PAPOLA activity | |
| Tgene | FIP1L1 | chr6:143828374 | chr4:54265897 | ENST00000507166 | 8 | 24 | 1_356 | 235 | 850.0 | Region | Note=Necessary for stimulating PAPOLA activity | |
| Tgene | FIP1L1 | chr6:143828374 | chr4:54265897 | ENST00000507922 | 7 | 12 | 1_356 | 220 | 379.0 | Region | Note=Necessary for stimulating PAPOLA activity |
Top |
Fusion Gene Sequence for FUCA2-FIP1L1 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >31765_31765_1_FUCA2-FIP1L1_FUCA2_chr6_143828374_ENST00000438118_FIP1L1_chr4_54265897_ENST00000306932_length(transcript)=1584nt_BP=440nt AGAGAGGACAGCCGGCCTGCGCCGGGACATGCGGCCCCAGGAGCTCCCCAGGCTCGCGTTCCCGTTGCTGCTGTTGCTGTTGCTGCTGCT GCCGCCGCCGCCGTGCCCTGCCCACAGCGCCACGCGCTTCGACCCCACCTGGGAGTCCCTGGACGCCCGCCAGCTGCCCGCGTGGTTTGA CCAGGCCAAGTTCGGCATCTTCATCCACTGGGGAGTGTTTTCCGTGCCCAGCTTCGGTAGCGAGTGGTTCTGGTGGTATTGGCAAAAGGA AAAGATACCGAAGTATGTGGAATTTATGAAAGATAATTACCCTCCTAGTTTCAAATATGAAGATTTTGGACCACTATTTACAGCAAAATT TTTTAATGCCAACCAGTGGGCAGATATTTTTCAGGCCTCTGGTGCCAAATACATTGTCTTAACTTCCAAACATCATGAAGGTACAGCAGG GAAGAACTGGAAACTCAGAGAAAGAAACTGCCCTTCCATCTACAAAAGCTGAGTTTACTTCTCCTCCTTCTTTGTTCAAGACTGGGCTTC CACCGAGCAGGAGATTACCTGGGGCAATTGATGTTATCGGTCAGACTATAACTATCAGCCGAGTAGAAGGCAGGCGACGGGCAAATGAGA ACAGCAACATACAGGTCCTTTCTGAAAGATCTGCTACTGAAGTAGACAACAATTTTAGCAAACCACCTCCGTTTTTCCCTCCAGGAGCTC CTCCCACTCACCTTCCACCTCCTCCATTTCTTCCACCTCCTCCGACTGTCAGCACTGCTCCACCTCTGATTCCACCACCGGGTTTTCCTC CTCCACCAGGCGCTCCACCTCCATCTCTTATACCAACAATAGAAAGTGGACATTCCTCTGGTTATGATAGTCGTTCTGCACGTGCATTTC CATATGGCAATGTTGCCTTTCCCCATCTTCCTGGTTCTGCTCCTTCGTGGCCTAGTCTTGTGGACACCAGCAAGCAGTGGGACTATTATG CCAGAAGAGAGAAAGACCGAGATAGAGAGAGAGACAGAGACAGAGAGCGAGACCGTGATCGGGACAGAGAAAGAGAACGCACCAGAGAGA GAGAGAGGGAGCGTGATCACAGTCCTACACCAAGTGTTTTCAACAGCGATGAAGAACGATACAGATACAGGGAATATGCAGAAAGAGGTT ATGAGCGTCACAGAGCAAGTCGAGAAAAAGAAGAACGACATAGAGAAAGACGACACAGGGAGAAAGAGGAAACCAGACATAAGTCTTCTC GAAGTAATAGTAGACGTCGCCATGAAAGTGAAGAAGGAGATAGTCACAGGAGACACAAACACAAAAAATCTAAAAGAAGCAAAGAAGGAA AAGAAGCGGGCAGTGAGCCTGCCCCTGAACAGGAGAGCACCGAAGCTACACCTGCAGAATAGGCATGGTTTTGGCCTTTTGTGTATATTA GTACCAGAAGTAGATACTATAAATCTTGTTATTTTTCTGGATAATGTTTAAGAAATTTACCTTAAATCTTGTTCTGTTTGTTAGTATGAA >31765_31765_1_FUCA2-FIP1L1_FUCA2_chr6_143828374_ENST00000438118_FIP1L1_chr4_54265897_ENST00000306932_length(amino acids)=325AA_BP=1 MKVQQGRTGNSEKETALPSTKAEFTSPPSLFKTGLPPSRRLPGAIDVIGQTITISRVEGRRRANENSNIQVLSERSATEVDNNFSKPPPF FPPGAPPTHLPPPPFLPPPPTVSTAPPLIPPPGFPPPPGAPPPSLIPTIESGHSSGYDSRSARAFPYGNVAFPHLPGSAPSWPSLVDTSK QWDYYARREKDRDRERDRDRERDRDRDRERERTRERERERDHSPTPSVFNSDEERYRYREYAERGYERHRASREKEERHRERRHREKEET -------------------------------------------------------------- >31765_31765_2_FUCA2-FIP1L1_FUCA2_chr6_143828374_ENST00000438118_FIP1L1_chr4_54265897_ENST00000337488_length(transcript)=1739nt_BP=440nt AGAGAGGACAGCCGGCCTGCGCCGGGACATGCGGCCCCAGGAGCTCCCCAGGCTCGCGTTCCCGTTGCTGCTGTTGCTGTTGCTGCTGCT GCCGCCGCCGCCGTGCCCTGCCCACAGCGCCACGCGCTTCGACCCCACCTGGGAGTCCCTGGACGCCCGCCAGCTGCCCGCGTGGTTTGA CCAGGCCAAGTTCGGCATCTTCATCCACTGGGGAGTGTTTTCCGTGCCCAGCTTCGGTAGCGAGTGGTTCTGGTGGTATTGGCAAAAGGA AAAGATACCGAAGTATGTGGAATTTATGAAAGATAATTACCCTCCTAGTTTCAAATATGAAGATTTTGGACCACTATTTACAGCAAAATT TTTTAATGCCAACCAGTGGGCAGATATTTTTCAGGCCTCTGGTGCCAAATACATTGTCTTAACTTCCAAACATCATGAAGGTACAGCAGG GAAGAACTGGAAACTCAGAGAAAGAAACTGCCCTTCCATCTACAAAAGCTGAGTTTACTTCTCCTCCTTCTTTGTTCAAGACTGGGCTTC CACCGAGCAGAAACAGCACTTCTTCTCAGTCTCAGACAAGTACTGCCTCCAGAAAAGCCAATTCAAGCGTTGGGAAGTGGCAGGATCGAT ATGGGAGGGCCGAATCACCTGATCTAAGGAGATTACCTGGGGCAATTGATGTTATCGGTCAGACTATAACTATCAGCCGAGTAGAAGGCA GGCGACGGGCAAATGAGAACAGCAACATACAGGTCCTTTCTGAAAGATCTGCTACTGAAGTAGACAACAATTTTAGCAAACCACCTCCGT TTTTCCCTCCAGGAGCTCCTCCCACTCACCTTCCACCTCCTCCATTTCTTCCACCTCCTCCGACTGTCAGCACTGCTCCACCTCTGATTC CACCACCGGGTTTTCCTCCTCCACCAGGCGCTCCACCTCCATCTCTTATACCAACAATAGAAAGTGGACATTCCTCTGGTTATGATAGTC GTTCTGCACGTGCATTTCCATATGGCAATGTTGCCTTTCCCCATCTTCCTGGTTCTGCTCCTTCGTGGCCTAGTCTTGTGGACACCAGCA AGCAGTGGGACTATTATGCCAGAAGAGAGAAAGACCGAGATAGAGAGAGAGACAGAGACAGAGAGCGAGACCGTGATCGGGACAGAGAAA GAGAACGCACCAGAGAGAGAGAGAGGGAGCGTGATCACAGTCCTACACCAAGTGTTTTCAACAGCGATGAAGAACGATACAGATACAGGG AATATGCAGAAAGAGGTTATGAGCGTCACAGAGCAAGTCGAGAAAAAGAAGAACGACATAGAGAAAGACGACACAGGGAGAAAGAGGAAA CCAGACATAAGTCTTCTCGAAGTAATAGTAGACGTCGCCATGAAAGTGAAGAAGGAGATAGTCACAGGAGACACAAACACAAAAAATCTA AAAGAAGCAAAGAAGGAAAAGAAGCGGGCAGTGAGCCTGCCCCTGAACAGGAGAGCACCGAAGCTACACCTGCAGAATAGGCATGGTTTT GGCCTTTTGTGTATATTAGTACCAGAAGTAGATACTATAAATCTTGTTATTTTTCTGGATAATGTTTAAGAAATTTACCTTAAATCTTGT TCTGTTTGTTAGTATGAAAAGTTAACTTTTTTTCCAAAATAAAAGAGTGAATTTTTCATGTTAAGTTAAAAATCTTTGTCTTGTACTATT >31765_31765_2_FUCA2-FIP1L1_FUCA2_chr6_143828374_ENST00000438118_FIP1L1_chr4_54265897_ENST00000337488_length(amino acids)=361AA_BP=1 MKVQQGRTGNSEKETALPSTKAEFTSPPSLFKTGLPPSRNSTSSQSQTSTASRKANSSVGKWQDRYGRAESPDLRRLPGAIDVIGQTITI SRVEGRRRANENSNIQVLSERSATEVDNNFSKPPPFFPPGAPPTHLPPPPFLPPPPTVSTAPPLIPPPGFPPPPGAPPPSLIPTIESGHS SGYDSRSARAFPYGNVAFPHLPGSAPSWPSLVDTSKQWDYYARREKDRDRERDRDRERDRDRDRERERTRERERERDHSPTPSVFNSDEE RYRYREYAERGYERHRASREKEERHRERRHREKEETRHKSSRSNSRRRHESEEGDSHRRHKHKKSKRSKEGKEAGSEPAPEQESTEATPA -------------------------------------------------------------- >31765_31765_3_FUCA2-FIP1L1_FUCA2_chr6_143828374_ENST00000438118_FIP1L1_chr4_54265897_ENST00000358575_length(transcript)=1766nt_BP=440nt AGAGAGGACAGCCGGCCTGCGCCGGGACATGCGGCCCCAGGAGCTCCCCAGGCTCGCGTTCCCGTTGCTGCTGTTGCTGTTGCTGCTGCT GCCGCCGCCGCCGTGCCCTGCCCACAGCGCCACGCGCTTCGACCCCACCTGGGAGTCCCTGGACGCCCGCCAGCTGCCCGCGTGGTTTGA CCAGGCCAAGTTCGGCATCTTCATCCACTGGGGAGTGTTTTCCGTGCCCAGCTTCGGTAGCGAGTGGTTCTGGTGGTATTGGCAAAAGGA AAAGATACCGAAGTATGTGGAATTTATGAAAGATAATTACCCTCCTAGTTTCAAATATGAAGATTTTGGACCACTATTTACAGCAAAATT TTTTAATGCCAACCAGTGGGCAGATATTTTTCAGGCCTCTGGTGCCAAATACATTGTCTTAACTTCCAAACATCATGAAGGTACAGCAGG GAAGAACTGGAAACTCAGAGAAAGAAACTGCCCTTCCATCTACAAAAGCTGAGTTTACTTCTCCTCCTTCTTTGTTCAAGACTGGGCTTC CACCGAGCAGAAACAGCACTTCTTCTCAGTCTCAGACAAGTACTGCCTCCAGAAAAGCCAATTCAAGCGTTGGGAAGTGGCAGGATCGAT ATGGGAGGGCCGAATCACCTGATCTAAGGAGATTACCTGGGGCAATTGATGTTATCGGTCAGACTATAACTATCAGCCGAGTAGAAGGCA GGCGACGGGCAAATGAGAACAGCAACATACAGGTCCTTTCTGAAAGATCTGCTACTGAAGTAGACAACAATTTTAGCAAACCACCTCCGT TTTTCCCTCCAGGAGCTCCTCCCACTCACCTTCCACCTCCTCCATTTCTTCCACCTCCTCCGACTGTCAGCACTGCTCCACCTCTGATTC CACCACCGGGTATTCCAATAACTGTACCACCTCCAGGTTTTCCTCCTCCACCAGGCGCTCCACCTCCATCTCTTATACCAACAATAGAAA GTGGACATTCCTCTGGTTATGATAGTCGTTCTGCACGTGCATTTCCATATGGCAATGTTGCCTTTCCCCATCTTCCTGGTTCTGCTCCTT CGTGGCCTAGTCTTGTGGACACCAGCAAGCAGTGGGACTATTATGCCAGAAGAGAGAAAGACCGAGATAGAGAGAGAGACAGAGACAGAG AGCGAGACCGTGATCGGGACAGAGAAAGAGAACGCACCAGAGAGAGAGAGAGGGAGCGTGATCACAGTCCTACACCAAGTGTTTTCAACA GCGATGAAGAACGATACAGATACAGGGAATATGCAGAAAGAGGTTATGAGCGTCACAGAGCAAGTCGAGAAAAAGAAGAACGACATAGAG AAAGACGACACAGGGAGAAAGAGGAAACCAGACATAAGTCTTCTCGAAGTAATAGTAGACGTCGCCATGAAAGTGAAGAAGGAGATAGTC ACAGGAGACACAAACACAAAAAATCTAAAAGAAGCAAAGAAGGAAAAGAAGCGGGCAGTGAGCCTGCCCCTGAACAGGAGAGCACCGAAG CTACACCTGCAGAATAGGCATGGTTTTGGCCTTTTGTGTATATTAGTACCAGAAGTAGATACTATAAATCTTGTTATTTTTCTGGATAAT GTTTAAGAAATTTACCTTAAATCTTGTTCTGTTTGTTAGTATGAAAAGTTAACTTTTTTTCCAAAATAAAAGAGTGAATTTTTCATGTTA >31765_31765_3_FUCA2-FIP1L1_FUCA2_chr6_143828374_ENST00000438118_FIP1L1_chr4_54265897_ENST00000358575_length(amino acids)=370AA_BP=1 MKVQQGRTGNSEKETALPSTKAEFTSPPSLFKTGLPPSRNSTSSQSQTSTASRKANSSVGKWQDRYGRAESPDLRRLPGAIDVIGQTITI SRVEGRRRANENSNIQVLSERSATEVDNNFSKPPPFFPPGAPPTHLPPPPFLPPPPTVSTAPPLIPPPGIPITVPPPGFPPPPGAPPPSL IPTIESGHSSGYDSRSARAFPYGNVAFPHLPGSAPSWPSLVDTSKQWDYYARREKDRDRERDRDRERDRDRDRERERTRERERERDHSPT PSVFNSDEERYRYREYAERGYERHRASREKEERHRERRHREKEETRHKSSRSNSRRRHESEEGDSHRRHKHKKSKRSKEGKEAGSEPAPE -------------------------------------------------------------- >31765_31765_4_FUCA2-FIP1L1_FUCA2_chr6_143828374_ENST00000438118_FIP1L1_chr4_54265897_ENST00000507166_length(transcript)=2285nt_BP=440nt AGAGAGGACAGCCGGCCTGCGCCGGGACATGCGGCCCCAGGAGCTCCCCAGGCTCGCGTTCCCGTTGCTGCTGTTGCTGTTGCTGCTGCT GCCGCCGCCGCCGTGCCCTGCCCACAGCGCCACGCGCTTCGACCCCACCTGGGAGTCCCTGGACGCCCGCCAGCTGCCCGCGTGGTTTGA CCAGGCCAAGTTCGGCATCTTCATCCACTGGGGAGTGTTTTCCGTGCCCAGCTTCGGTAGCGAGTGGTTCTGGTGGTATTGGCAAAAGGA AAAGATACCGAAGTATGTGGAATTTATGAAAGATAATTACCCTCCTAGTTTCAAATATGAAGATTTTGGACCACTATTTACAGCAAAATT TTTTAATGCCAACCAGTGGGCAGATATTTTTCAGGCCTCTGGTGCCAAATACATTGTCTTAACTTCCAAACATCATGAAGGTACAGCAGG GAAGAACTGGAAACTCAGAGAAAGAAACTGCCCTTCCATCTACAAAAGCTGAGTTTACTTCTCCTCCTTCTTTGTTCAAGACTGGGCTTC CACCGAGCAGAAACAGCACTTCTTCTCAGTCTCAGACAAGTACTGCCTCCAGAAAAGCCAATTCAAGCGTTGGGAAGTGGCAGGATCGAT ATGGGAGGGCCGAATCACCTGATCTAAGGAGATTACCTGGGGCAATTGATGTTATCGGTCAGACTATAACTATCAGCCGAGTAGAAGGCA GGCGACGGGCAAATGAGAACAGCAACATACAGCTGCCTTATGACTCAAGATGGGAGTTTCCAAGAGATGGACTAGTGCTTGGTCGGGTCT TGGGGTCTGGAGCGTTTGGGAAGGTGGTTGAAGGAACAGCCTATGGATTAAGCCGGTCCCAACCTGTCATGAAAGTTGCAGTGAAGATGC TAAAACCCACGGCCAGATCCAGTGAAAAACAAGCTCTCATGTCTGAACTGAAGATAATGACTCACCTGGGGCCACATTTGAACATTGTAA ACTTGCTGGGAGCCTGCACCAAGTCAGGCCCCATTTACATCATCACAGAGTATTGCTTCTATGGAGATTTGGTCAACTATTTGCATAAGA ATAGGGATAGCTTCCTGAGCCACCACCCAGAGAAGCCAAAGAAAGAGCTGGATATCTTTGGATTGAACCCTGCTGATGAAAGCACACGGA GCTATGTTATTTTATCTTTTGAAAACAATGGTGACTACATGGACATGAAGCAGGCTGATACTACACAGTATGTCCCCATGCTAGAAAGGA AAGAGGTTTCTAAATATTCCGACATCCAGAGATCACTCTATGATCGTCCAGCCTCATATAAGAAGAAATCTATGTTAGACTCAGAAGTCA AAAACCTCCTTTCAGATGATAACTCAGAAGGCCTTACTTTATTGGATTTGTTGAGCTTCACCTATCAAGTTGCCCGAGGAATGGAGTTTT TGGCTTCAAAAAATTGTGTCCACCGTGATCTGGCTGCTCGCAACGTCCTCCTGGCACAAGGAAAAATTGTGAAGATCTGTGACTTTGGCC TGGCCAGAGACATCATGCATGATTCGAACTATGTGTCGAAAGGCAGTACCTTTCTGCCCGTGAAGTGGATGGCTCCTGAGAGCATCTTTG ACAACCTCTACACCACACTGAGTGATGTCTGGTCTTATGGCATTCTGCTCTGGGAGATCTTTTCCCTTGGTGGCACCCCTTACCCCGGCA TGATGGTGGATTCTACTTTCTACAATAAGATCAAGAGTGGGTACCGGATGGCCAAGCCTGACCACGCTACCAGTGAAGTCTACGAGATCA TGGTGAAATGCTGGAACAGTGAGCCGGAGAAGAGACCCTCCTTTTACCACCTGAGTGAGATTGTGGAGAATCTGCTGCCTGGACAATATA AAAAGAGTTATGAAAAAATTCACCTGGACTTCCTGAAGAGTGACCATCCTGCTGTGGCACGCATGCGTGTGGACTCAGACAATGCATACA TTGGTGTCACCTACAAAAACGAGGAAGACAAGCTGAAGGACTGGGAGGGTGGTCTGGATGAGCAGAGACTGAGCGCTGACAGTGGCTACA TCATTCCTCTGCCTGACATTGACCCTGTCCCTGAGGAGGAGGACCTGGGCAAGAGGAACAGACACAGCTCGCAGACCTCTGAAGAGAGTG CCATTGAGACGGGTTCCAGCAGTTCCACCTTCATCAAGAGAGAGGACGAGACCATTGAAGACATCGACATGATGGATGACATCGGCATAG >31765_31765_4_FUCA2-FIP1L1_FUCA2_chr6_143828374_ENST00000438118_FIP1L1_chr4_54265897_ENST00000507166_length(amino acids)=617AA_BP=1 MKVQQGRTGNSEKETALPSTKAEFTSPPSLFKTGLPPSRNSTSSQSQTSTASRKANSSVGKWQDRYGRAESPDLRRLPGAIDVIGQTITI SRVEGRRRANENSNIQLPYDSRWEFPRDGLVLGRVLGSGAFGKVVEGTAYGLSRSQPVMKVAVKMLKPTARSSEKQALMSELKIMTHLGP HLNIVNLLGACTKSGPIYIITEYCFYGDLVNYLHKNRDSFLSHHPEKPKKELDIFGLNPADESTRSYVILSFENNGDYMDMKQADTTQYV PMLERKEVSKYSDIQRSLYDRPASYKKKSMLDSEVKNLLSDDNSEGLTLLDLLSFTYQVARGMEFLASKNCVHRDLAARNVLLAQGKIVK ICDFGLARDIMHDSNYVSKGSTFLPVKWMAPESIFDNLYTTLSDVWSYGILLWEIFSLGGTPYPGMMVDSTFYNKIKSGYRMAKPDHATS EVYEIMVKCWNSEPEKRPSFYHLSEIVENLLPGQYKKSYEKIHLDFLKSDHPAVARMRVDSDNAYIGVTYKNEEDKLKDWEGGLDEQRLS -------------------------------------------------------------- >31765_31765_5_FUCA2-FIP1L1_FUCA2_chr6_143828374_ENST00000438118_FIP1L1_chr4_54265897_ENST00000507922_length(transcript)=947nt_BP=440nt AGAGAGGACAGCCGGCCTGCGCCGGGACATGCGGCCCCAGGAGCTCCCCAGGCTCGCGTTCCCGTTGCTGCTGTTGCTGTTGCTGCTGCT GCCGCCGCCGCCGTGCCCTGCCCACAGCGCCACGCGCTTCGACCCCACCTGGGAGTCCCTGGACGCCCGCCAGCTGCCCGCGTGGTTTGA CCAGGCCAAGTTCGGCATCTTCATCCACTGGGGAGTGTTTTCCGTGCCCAGCTTCGGTAGCGAGTGGTTCTGGTGGTATTGGCAAAAGGA AAAGATACCGAAGTATGTGGAATTTATGAAAGATAATTACCCTCCTAGTTTCAAATATGAAGATTTTGGACCACTATTTACAGCAAAATT TTTTAATGCCAACCAGTGGGCAGATATTTTTCAGGCCTCTGGTGCCAAATACATTGTCTTAACTTCCAAACATCATGAAGGTACAGCAGG GAAGAACTGGAAACTCAGAGAAAGAAACTGCCCTTCCATCTACAAAAGCTGAGTTTACTTCTCCTCCTTCTTTGTTCAAGACTGGGCTTC CACCGAGCAGAAACAGCACTTCTTCTCAGTCTCAGACAAGTACTGCCTCCAGAAAAGCCAATTCAAGCGTTGGGAAGTGGCAGGATCGAT ATGGGAGGGCCGAATCACCTGATCTAAGGAGATTACCTGGGGCAATTGATGTTATCGGTCAGACTATAACTATCAGCCGAGTAGAAGGCA GGCGACGGGCAAATGAGAACAGCAACATACAGGTCCTTTCTGAAAGATCTGCTACTGAAGTAGACAACAATTTTAGCAAACCACCTCCGT TTTTCCCTCCAGGAGCTCCTCCCACTCACCTTCCACCTCCTCCATTTCTTCCACCTCCTCCGACTGTCAGCACTGCTCCACCTCTGATTC >31765_31765_5_FUCA2-FIP1L1_FUCA2_chr6_143828374_ENST00000438118_FIP1L1_chr4_54265897_ENST00000507922_length(amino acids)=161AA_BP=141 MRRDMRPQELPRLAFPLLLLLLLLLPPPPCPAHSATRFDPTWESLDARQLPAWFDQAKFGIFIHWGVFSVPSFGSEWFWWYWQKEKIPKY -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for FUCA2-FIP1L1 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for FUCA2-FIP1L1 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for FUCA2-FIP1L1 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
