
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:FUS-ATF1 (FusionGDB2 ID:31790) |
Fusion Gene Summary for FUS-ATF1 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: FUS-ATF1 | Fusion gene ID: 31790 | Hgene | Tgene | Gene symbol | FUS | ATF1 | Gene ID | 2521 | 2668 |
| Gene name | FUS RNA binding protein | glial cell derived neurotrophic factor | |
| Synonyms | ALS6|ETM4|FUS1|HNRNPP2|POMP75|TLS | ATF|ATF1|ATF2|HFB1-GDNF|HSCR3 | |
| Cytomap | 16p11.2 | 5p13.2 | |
| Type of gene | protein-coding | protein-coding | |
| Description | RNA-binding protein FUS75 kDa DNA-pairing proteinfus-like proteinfused in sarcomafusion gene in myxoid liposarcomaheterogeneous nuclear ribonucleoprotein P2oncogene FUSoncogene TLStranslocated in liposarcoma protein | glial cell line-derived neurotrophic factorastrocyte-derived trophic factor | |
| Modification date | 20200329 | 20200314 | |
| UniProtAcc | P35637 | P18846 | |
| Ensembl transtripts involved in fusion gene | ENST00000254108, ENST00000380244, ENST00000568685, ENST00000474990, | ENST00000539132, ENST00000262053, | |
| Fusion gene scores | * DoF score | 37 X 37 X 16=21904 | 10 X 16 X 7=1120 |
| # samples | 44 | 16 | |
| ** MAII score | log2(44/21904*10)=-5.63754701773324 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(16/1120*10)=-2.8073549220576 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: FUS [Title/Abstract] AND ATF1 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | FUS(31195717)-ATF1(51207791), # samples:2 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | FUS | GO:0006355 | regulation of transcription, DNA-templated | 26124092 |
| Hgene | FUS | GO:0006357 | regulation of transcription by RNA polymerase II | 25453086 |
| Hgene | FUS | GO:0008380 | RNA splicing | 26124092 |
| Hgene | FUS | GO:0043484 | regulation of RNA splicing | 25453086|27731383 |
| Hgene | FUS | GO:0048255 | mRNA stabilization | 27378374 |
| Hgene | FUS | GO:0051260 | protein homooligomerization | 25453086 |
| Hgene | FUS | GO:1905168 | positive regulation of double-strand break repair via homologous recombination | 10567410 |
| Tgene | ATF1 | GO:0001755 | neural crest cell migration | 15242795 |
| Tgene | ATF1 | GO:0008284 | positive regulation of cell proliferation | 22897442 |
| Tgene | ATF1 | GO:0031175 | neuron projection development | 15242795 |
| Tgene | ATF1 | GO:0032770 | positive regulation of monooxygenase activity | 12358785 |
| Tgene | ATF1 | GO:0043524 | negative regulation of neuron apoptotic process | 8493557 |
| Tgene | ATF1 | GO:0045944 | positive regulation of transcription by RNA polymerase II | 12358785 |
| Tgene | ATF1 | GO:0048255 | mRNA stabilization | 12358785 |
| Tgene | ATF1 | GO:0051584 | regulation of dopamine uptake involved in synaptic transmission | 8493557 |
| Tgene | ATF1 | GO:0072107 | positive regulation of ureteric bud formation | 8657307 |
| Tgene | ATF1 | GO:0090190 | positive regulation of branching involved in ureteric bud morphogenesis | 8657307|17229286 |
| Tgene | ATF1 | GO:2001240 | negative regulation of extrinsic apoptotic signaling pathway in absence of ligand | 10921886 |
 Fusion gene breakpoints across FUS (5'-gene) Fusion gene breakpoints across FUS (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
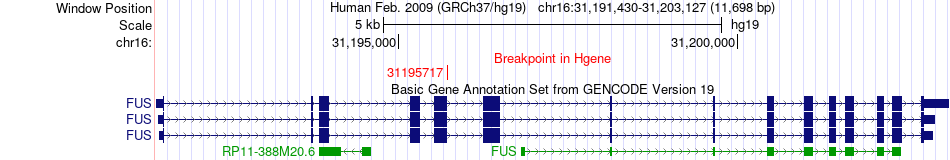 |
 Fusion gene breakpoints across ATF1 (3'-gene) Fusion gene breakpoints across ATF1 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
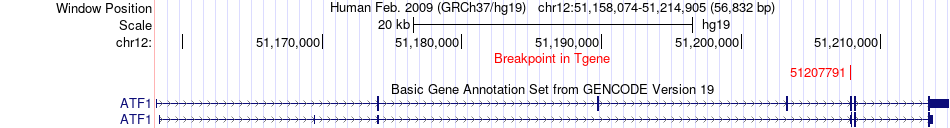 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | refractory anemia;angiomatoid malignant fibrous histiocytoma | AJ295163 | FUS | chr16 | 31195717 | ATF1 | chr12 | 51207791 | ||
| ChimerKB3 | . | . | FUS | chr16 | 31195323 | + | ATF1 | chr12 | 51157788 | + |
| ChimerKB3 | . | . | FUS | chr16 | 31195705 | + | ATF1 | chr12 | 51207792 | + |
| ChimerKB3 | . | . | FUS | chr16 | 31195705 | + | ATF1 | chr5 | 37835929 | - |
| ChimerKB3 | . | . | FUS | chr16 | 31195717 | + | ATF1 | chr12 | 51207791 | + |
| ChiTaRS5.0 | N/A | AJ295163 | FUS | chr16 | 31195717 | + | ATF1 | chr12 | 51207791 | + |
Top |
Fusion Gene ORF analysis for FUS-ATF1 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-5UTR | ENST00000254108 | ENST00000539132 | FUS | chr16 | 31195717 | ATF1 | chr12 | 51207791 | ||
| 5CDS-5UTR | ENST00000254108 | ENST00000539132 | FUS | chr16 | 31195705 | + | ATF1 | chr12 | 51207792 | + |
| 5CDS-5UTR | ENST00000254108 | ENST00000539132 | FUS | chr16 | 31195717 | + | ATF1 | chr12 | 51207791 | + |
| 5CDS-5UTR | ENST00000380244 | ENST00000539132 | FUS | chr16 | 31195717 | ATF1 | chr12 | 51207791 | ||
| 5CDS-5UTR | ENST00000380244 | ENST00000539132 | FUS | chr16 | 31195705 | + | ATF1 | chr12 | 51207792 | + |
| 5CDS-5UTR | ENST00000380244 | ENST00000539132 | FUS | chr16 | 31195717 | + | ATF1 | chr12 | 51207791 | + |
| 5CDS-5UTR | ENST00000568685 | ENST00000539132 | FUS | chr16 | 31195717 | ATF1 | chr12 | 51207791 | ||
| 5CDS-5UTR | ENST00000568685 | ENST00000539132 | FUS | chr16 | 31195705 | + | ATF1 | chr12 | 51207792 | + |
| 5CDS-5UTR | ENST00000568685 | ENST00000539132 | FUS | chr16 | 31195717 | + | ATF1 | chr12 | 51207791 | + |
| 5CDS-intron | ENST00000254108 | ENST00000262053 | FUS | chr16 | 31195323 | + | ATF1 | chr12 | 51157788 | + |
| 5CDS-intron | ENST00000254108 | ENST00000262053 | FUS | chr16 | 31195705 | + | ATF1 | chr5 | 37835929 | - |
| 5CDS-intron | ENST00000254108 | ENST00000539132 | FUS | chr16 | 31195323 | + | ATF1 | chr12 | 51157788 | + |
| 5CDS-intron | ENST00000254108 | ENST00000539132 | FUS | chr16 | 31195705 | + | ATF1 | chr5 | 37835929 | - |
| 5CDS-intron | ENST00000380244 | ENST00000262053 | FUS | chr16 | 31195323 | + | ATF1 | chr12 | 51157788 | + |
| 5CDS-intron | ENST00000380244 | ENST00000262053 | FUS | chr16 | 31195705 | + | ATF1 | chr5 | 37835929 | - |
| 5CDS-intron | ENST00000380244 | ENST00000539132 | FUS | chr16 | 31195323 | + | ATF1 | chr12 | 51157788 | + |
| 5CDS-intron | ENST00000380244 | ENST00000539132 | FUS | chr16 | 31195705 | + | ATF1 | chr5 | 37835929 | - |
| 5CDS-intron | ENST00000568685 | ENST00000262053 | FUS | chr16 | 31195323 | + | ATF1 | chr12 | 51157788 | + |
| 5CDS-intron | ENST00000568685 | ENST00000262053 | FUS | chr16 | 31195705 | + | ATF1 | chr5 | 37835929 | - |
| 5CDS-intron | ENST00000568685 | ENST00000539132 | FUS | chr16 | 31195323 | + | ATF1 | chr12 | 51157788 | + |
| 5CDS-intron | ENST00000568685 | ENST00000539132 | FUS | chr16 | 31195705 | + | ATF1 | chr5 | 37835929 | - |
| In-frame | ENST00000254108 | ENST00000262053 | FUS | chr16 | 31195717 | ATF1 | chr12 | 51207791 | ||
| In-frame | ENST00000254108 | ENST00000262053 | FUS | chr16 | 31195705 | + | ATF1 | chr12 | 51207792 | + |
| In-frame | ENST00000254108 | ENST00000262053 | FUS | chr16 | 31195717 | + | ATF1 | chr12 | 51207791 | + |
| In-frame | ENST00000380244 | ENST00000262053 | FUS | chr16 | 31195717 | ATF1 | chr12 | 51207791 | ||
| In-frame | ENST00000380244 | ENST00000262053 | FUS | chr16 | 31195705 | + | ATF1 | chr12 | 51207792 | + |
| In-frame | ENST00000380244 | ENST00000262053 | FUS | chr16 | 31195717 | + | ATF1 | chr12 | 51207791 | + |
| In-frame | ENST00000568685 | ENST00000262053 | FUS | chr16 | 31195717 | ATF1 | chr12 | 51207791 | ||
| In-frame | ENST00000568685 | ENST00000262053 | FUS | chr16 | 31195705 | + | ATF1 | chr12 | 51207792 | + |
| In-frame | ENST00000568685 | ENST00000262053 | FUS | chr16 | 31195717 | + | ATF1 | chr12 | 51207791 | + |
| intron-3CDS | ENST00000474990 | ENST00000262053 | FUS | chr16 | 31195717 | ATF1 | chr12 | 51207791 | ||
| intron-3CDS | ENST00000474990 | ENST00000262053 | FUS | chr16 | 31195705 | + | ATF1 | chr12 | 51207792 | + |
| intron-3CDS | ENST00000474990 | ENST00000262053 | FUS | chr16 | 31195717 | + | ATF1 | chr12 | 51207791 | + |
| intron-5UTR | ENST00000474990 | ENST00000539132 | FUS | chr16 | 31195717 | ATF1 | chr12 | 51207791 | ||
| intron-5UTR | ENST00000474990 | ENST00000539132 | FUS | chr16 | 31195705 | + | ATF1 | chr12 | 51207792 | + |
| intron-5UTR | ENST00000474990 | ENST00000539132 | FUS | chr16 | 31195717 | + | ATF1 | chr12 | 51207791 | + |
| intron-intron | ENST00000474990 | ENST00000262053 | FUS | chr16 | 31195323 | + | ATF1 | chr12 | 51157788 | + |
| intron-intron | ENST00000474990 | ENST00000262053 | FUS | chr16 | 31195705 | + | ATF1 | chr5 | 37835929 | - |
| intron-intron | ENST00000474990 | ENST00000539132 | FUS | chr16 | 31195323 | + | ATF1 | chr12 | 51157788 | + |
| intron-intron | ENST00000474990 | ENST00000539132 | FUS | chr16 | 31195705 | + | ATF1 | chr5 | 37835929 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000254108 | FUS | chr16 | 31195705 | + | ENST00000262053 | ATF1 | chr12 | 51207792 | + | 2283 | 452 | 73 | 939 | 288 |
| ENST00000380244 | FUS | chr16 | 31195705 | + | ENST00000262053 | ATF1 | chr12 | 51207792 | + | 2251 | 420 | 44 | 907 | 287 |
| ENST00000568685 | FUS | chr16 | 31195705 | + | ENST00000262053 | ATF1 | chr12 | 51207792 | + | 2242 | 411 | 32 | 898 | 288 |
| ENST00000254108 | FUS | chr16 | 31195717 | + | ENST00000262053 | ATF1 | chr12 | 51207791 | + | 2459 | 628 | 27 | 1115 | 362 |
| ENST00000380244 | FUS | chr16 | 31195717 | + | ENST00000262053 | ATF1 | chr12 | 51207791 | + | 2427 | 596 | 52 | 1083 | 343 |
| ENST00000568685 | FUS | chr16 | 31195717 | + | ENST00000262053 | ATF1 | chr12 | 51207791 | + | 2418 | 587 | 40 | 1074 | 344 |
| ENST00000254108 | FUS | chr16 | 31195717 | ENST00000262053 | ATF1 | chr12 | 51207791 | 2459 | 628 | 27 | 1115 | 362 | ||
| ENST00000380244 | FUS | chr16 | 31195717 | ENST00000262053 | ATF1 | chr12 | 51207791 | 2427 | 596 | 52 | 1083 | 343 | ||
| ENST00000568685 | FUS | chr16 | 31195717 | ENST00000262053 | ATF1 | chr12 | 51207791 | 2418 | 587 | 40 | 1074 | 344 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000254108 | ENST00000262053 | FUS | chr16 | 31195717 | ATF1 | chr12 | 51207791 | 0.000567612 | 0.9994324 | ||
| ENST00000380244 | ENST00000262053 | FUS | chr16 | 31195717 | ATF1 | chr12 | 51207791 | 0.000121627 | 0.9998784 | ||
| ENST00000568685 | ENST00000262053 | FUS | chr16 | 31195717 | ATF1 | chr12 | 51207791 | 0.00054754 | 0.9994524 |
Top |
Fusion Genomic Features for FUS-ATF1 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| FUS | chr16 | 31195717 | + | ATF1 | chr12 | 51207792 | + | 0.014104997 | 0.98589504 |
| FUS | chr16 | 31195717 | + | ATF1 | chr12 | 51207792 | + | 0.014104997 | 0.98589504 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
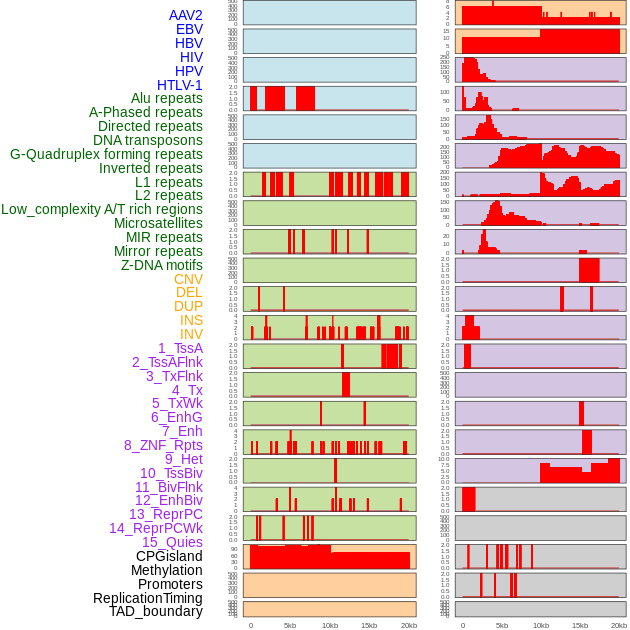 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
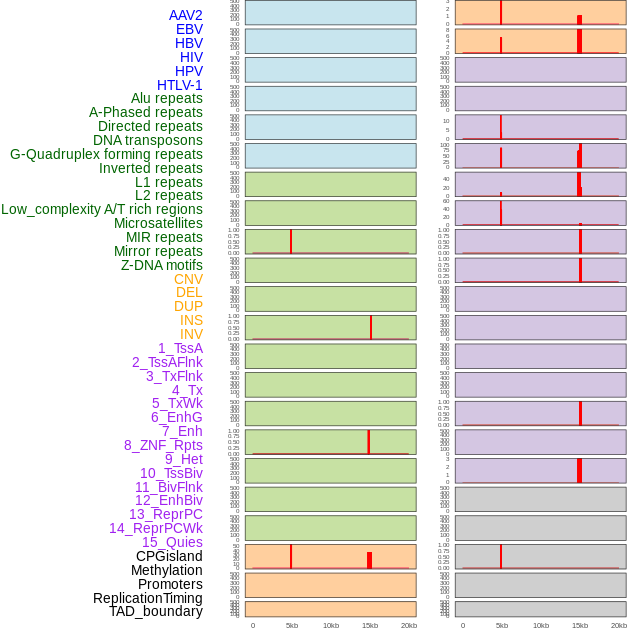 |
Top |
Fusion Protein Features for FUS-ATF1 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr16:31195717/chr12:51207791) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| FUS | ATF1 |
| FUNCTION: DNA/RNA-binding protein that plays a role in various cellular processes such as transcription regulation, RNA splicing, RNA transport, DNA repair and damage response (PubMed:27731383). Binds to nascent pre-mRNAs and acts as a molecular mediator between RNA polymerase II and U1 small nuclear ribonucleoprotein thereby coupling transcription and splicing (PubMed:26124092). Binds also its own pre-mRNA and autoregulates its expression; this autoregulation mechanism is mediated by non-sense-mediated decay (PubMed:24204307). Plays a role in DNA repair mechanisms by promoting D-loop formation and homologous recombination during DNA double-strand break repair (PubMed:10567410). In neuronal cells, plays crucial roles in dendritic spine formation and stability, RNA transport, mRNA stability and synaptic homeostasis (By similarity). {ECO:0000250|UniProtKB:P56959, ECO:0000269|PubMed:10567410, ECO:0000269|PubMed:24204307, ECO:0000269|PubMed:26124092, ECO:0000269|PubMed:27731383}. | FUNCTION: This protein binds the cAMP response element (CRE) (consensus: 5'-GTGACGT[AC][AG]-3'), a sequence present in many viral and cellular promoters. Binds to the Tax-responsive element (TRE) of HTLV-I. Mediates PKA-induced stimulation of CRE-reporter genes. Represses the expression of FTH1 and other antioxidant detoxification genes. Triggers cell proliferation and transformation. {ECO:0000269|PubMed:18794154, ECO:0000269|PubMed:20980392}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | FUS | chr16:31195717 | chr12:51207791 | ENST00000254108 | + | 5 | 15 | 1_165 | 174 | 527.0 | Compositional bias | Note=Gln/Gly/Ser/Tyr-rich |
| Hgene | FUS | chr16:31195717 | chr12:51207791 | ENST00000380244 | + | 5 | 15 | 1_165 | 173 | 526.0 | Compositional bias | Note=Gln/Gly/Ser/Tyr-rich |
| Tgene | ATF1 | chr16:31195717 | chr12:51207791 | ENST00000262053 | 3 | 7 | 213_271 | 109 | 272.0 | Domain | bZIP | |
| Tgene | ATF1 | chr16:31195717 | chr12:51207791 | ENST00000262053 | 3 | 7 | 215_239 | 109 | 272.0 | Region | Basic motif | |
| Tgene | ATF1 | chr16:31195717 | chr12:51207791 | ENST00000262053 | 3 | 7 | 241_262 | 109 | 272.0 | Region | Leucine-zipper |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | FUS | chr16:31195717 | chr12:51207791 | ENST00000254108 | + | 5 | 15 | 166_267 | 174 | 527.0 | Compositional bias | Note=Gly-rich |
| Hgene | FUS | chr16:31195717 | chr12:51207791 | ENST00000254108 | + | 5 | 15 | 371_526 | 174 | 527.0 | Compositional bias | Note=Arg/Gly-rich |
| Hgene | FUS | chr16:31195717 | chr12:51207791 | ENST00000380244 | + | 5 | 15 | 166_267 | 173 | 526.0 | Compositional bias | Note=Gly-rich |
| Hgene | FUS | chr16:31195717 | chr12:51207791 | ENST00000380244 | + | 5 | 15 | 371_526 | 173 | 526.0 | Compositional bias | Note=Arg/Gly-rich |
| Hgene | FUS | chr16:31195717 | chr12:51207791 | ENST00000254108 | + | 5 | 15 | 285_371 | 174 | 527.0 | Domain | RRM |
| Hgene | FUS | chr16:31195717 | chr12:51207791 | ENST00000380244 | + | 5 | 15 | 285_371 | 173 | 526.0 | Domain | RRM |
| Hgene | FUS | chr16:31195717 | chr12:51207791 | ENST00000254108 | + | 5 | 15 | 422_453 | 174 | 527.0 | Zinc finger | RanBP2-type |
| Hgene | FUS | chr16:31195717 | chr12:51207791 | ENST00000380244 | + | 5 | 15 | 422_453 | 173 | 526.0 | Zinc finger | RanBP2-type |
| Tgene | ATF1 | chr16:31195717 | chr12:51207791 | ENST00000262053 | 3 | 7 | 31_90 | 109 | 272.0 | Domain | KID |
Top |
Fusion Gene Sequence for FUS-ATF1 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >31790_31790_1_FUS-ATF1_FUS_chr16_31195705_ENST00000254108_ATF1_chr12_51207792_ENST00000262053_length(transcript)=2283nt_BP=452nt ACTTAAGCTTCGACGCAGGAGGCGGGGCTGCTCAGTCCTCCAGGCGTCGGTACTCAGCGGTGTTGGAACTTCGTTGCTTGCTTGCCTGTG CGCGCGTGCGCGGACATGGCCTCAAACGATTATACCCAACAAGCAACCCAAAGCTATGGGGCCTACCCCACCCAGCCCGGGCAGGGCTAT TCCCAGCAGAGCAGTCAGCCCTACGGACAGCAGAGTTACAGTGGTTATAGCCAGTCCACGGACACTTCAGGCTATGGCCAGAGCAGCTAT TCTTCTTATGGCCAGAGCCAGAACACAGGCTATGGAACTCAGTCAACTCCCCAGGGATATGGCTCGACTGGCGGCTATGGCAGTAGCCAG AGCTCCCAATCGTCTTACGGGCAGCAGTCCTCCTACCCTGGCTATGGCCAGCAGCCAGCTCCCAGCAGCACCTCGGGAAGGTGGAGGTGG AGTTGCCATTGCCCCAAATGGAGCCTTACAGTTGGCAAGTCCAGGCACAGATGGAGTACAGGGACTTCAGACATTAACCATGACAAATTC AGGCAGTACTCAGCAAGGTACAACTATTCTTCAGTATGCACAGACCTCTGATGGACAGCAGATACTTGTGCCCAGCAATCAGGTGGTCGT ACAAACTGCATCAGGAGATATGCAAACATATCAGATCCGAACTACACCTTCAGCTACTTCTCTGCCACAAACTGTGGTGATGACATCTCC TGTGACTCTCACCTCTCAGACAACTAAGACAGATGACCCCCAATTGAAAAGAGAAATAAGGTTAATGAAAAACAGAGAAGCTGCTCGAGA ATGTCGCAGAAAGAAGAAAGAATATGTGAAATGCCTGGAAAACCGAGTTGCAGTCCTGGAAAATCAAAATAAAACTCTAATAGAAGAGTT AAAAACTTTGAAGGATCTTTATTCCAATAAAAGTGTTTGATTCCTAAGAAAGAAAATATTTTTGTGGACATGCATAAAAATTAAATGGAT TTCCTAGTGGAGTTTTATAAATTAAAAGGTCAAAACTGAAGCTTTTTATTTAGGCTTTTCCAAATCAAGGATAAATATCTTACGCACGAT ATCTAGTGACAGAGGAGAAAGTGGAAAATGACCTCAAGGAAGCTACGGGCACAACTGGAAGCTTTGTAGAAATTAAACATATTCAAGGAG CAAGAAATGAACTTTCAGCAGTCTAAATTTTCTAAATAACCAATAGTTGCCAATCTAAAATGGCAGAGAAGATGAAATTTGATAAACTGA ATTTTTTTTAAAAATCCATTTACCCTACAGGTTTGCATTTGTTTGCTGAAATTTACCTTTTTTTAGTTATATATATGTGTGTGTGTGTGT GTAATTTCTGCCAATAAATTCTAAATTACAAAGGTAAGAGAAAACCTAGTACATTACTAAATATATAAAGTATATGTTCTGATTATGTAT ACTTGTTCTAGTGTCAAGTCTTTTTAAGTGGGTTTTTAAAAGTTTGTTATTGGACTTGAATGGATTTTTGAGACTAGGTTAATTATTTTT GAGGTCTTATCCTAAAAGGCATCTAAGGTACATGAATGGAGTATGGTGATTTTATAACATTTTTTATCAGAATGGAAAAAGAACTGTTTA AAAGTTTGATACTTTTAAATAGTTGGTTTTTTTGCTTACTCTGGTAATGATTTTCTACAAATACATAATAAATTGTTTTTTTGAGTCTAT ATTCTGTATGCAGTTGAATATCCATTACTTATTCTGCTGTGCTTTAATAGAATGGAATGTTTACAGGCCCTTAAAATATTATTTTTAAAA AACCTTCTGAAGATACATACCAAAGTTTTTCCAAGAAGATTTTATAATCAATTTAATAATGTAAGGTTTATCAGATTCTATAATAGTATA GTTATTAAGGCAATTTTATGTTAGAGACTATTTTGTAATGTAGTGAGTGGTACCTTTATAAGAAAAGTGACTGCCAATATATTTTTATAG CTGATCTTTATAAATTCTAATGTTGAGTTTTTAATGATTATTTTAAATGTTTATATAGTTTAGTAAAATTTGCATCTCAAAGTATCATTT TTATATTATGGGACGTTTTCAGATTGGCTAATATTTGCATTGTAAATTTTGTATGCAGTTTATCTAAAATTCAAAAATACTGTCAGTACA CCAGCGTTTAACATCTATATTCCAATTTGTATACAGTTTAAAATTGTACTGCAAAACTATTGTGTGCTCTTACACAGTATGCATCATATT >31790_31790_1_FUS-ATF1_FUS_chr16_31195705_ENST00000254108_ATF1_chr12_51207792_ENST00000262053_length(amino acids)=288AA_BP=126 MLACLCARARTWPQTIIPNKQPKAMGPTPPSPGRAIPSRAVSPTDSRVTVVIASPRTLQAMARAAILLMARARTQAMELSQLPRDMARLA AMAVARAPNRLTGSSPPTLAMASSQLPAAPREGGGGVAIAPNGALQLASPGTDGVQGLQTLTMTNSGSTQQGTTILQYAQTSDGQQILVP SNQVVVQTASGDMQTYQIRTTPSATSLPQTVVMTSPVTLTSQTTKTDDPQLKREIRLMKNREAARECRRKKKEYVKCLENRVAVLENQNK -------------------------------------------------------------- >31790_31790_2_FUS-ATF1_FUS_chr16_31195705_ENST00000380244_ATF1_chr12_51207792_ENST00000262053_length(transcript)=2251nt_BP=420nt GCTCAGTCCTCCAGGCGTCGGTACTCAGCGGTGTTGGAACTTCGTTGCTTGCTTGCCTGTGCGCGCGTGCGCGGACATGGCCTCAAACGA TTATACCCAACAAGCAACCCAAAGCTATGGGGCCTACCCCACCCAGCCCGGGCAGGGCTATTCCCAGCAGAGCAGTCAGCCCTACGGACA GCAGAGTTACAGTGGTTATAGCCAGTCCACGGACACTTCAGGCTATGGCCAGAGCAGCTATTCTTCTTATGGCCAGAGCCAGAACAGCTA TGGAACTCAGTCAACTCCCCAGGGATATGGCTCGACTGGCGGCTATGGCAGTAGCCAGAGCTCCCAATCGTCTTACGGGCAGCAGTCCTC CTACCCTGGCTATGGCCAGCAGCCAGCTCCCAGCAGCACCTCGGGAAGGTGGAGGTGGAGTTGCCATTGCCCCAAATGGAGCCTTACAGT TGGCAAGTCCAGGCACAGATGGAGTACAGGGACTTCAGACATTAACCATGACAAATTCAGGCAGTACTCAGCAAGGTACAACTATTCTTC AGTATGCACAGACCTCTGATGGACAGCAGATACTTGTGCCCAGCAATCAGGTGGTCGTACAAACTGCATCAGGAGATATGCAAACATATC AGATCCGAACTACACCTTCAGCTACTTCTCTGCCACAAACTGTGGTGATGACATCTCCTGTGACTCTCACCTCTCAGACAACTAAGACAG ATGACCCCCAATTGAAAAGAGAAATAAGGTTAATGAAAAACAGAGAAGCTGCTCGAGAATGTCGCAGAAAGAAGAAAGAATATGTGAAAT GCCTGGAAAACCGAGTTGCAGTCCTGGAAAATCAAAATAAAACTCTAATAGAAGAGTTAAAAACTTTGAAGGATCTTTATTCCAATAAAA GTGTTTGATTCCTAAGAAAGAAAATATTTTTGTGGACATGCATAAAAATTAAATGGATTTCCTAGTGGAGTTTTATAAATTAAAAGGTCA AAACTGAAGCTTTTTATTTAGGCTTTTCCAAATCAAGGATAAATATCTTACGCACGATATCTAGTGACAGAGGAGAAAGTGGAAAATGAC CTCAAGGAAGCTACGGGCACAACTGGAAGCTTTGTAGAAATTAAACATATTCAAGGAGCAAGAAATGAACTTTCAGCAGTCTAAATTTTC TAAATAACCAATAGTTGCCAATCTAAAATGGCAGAGAAGATGAAATTTGATAAACTGAATTTTTTTTAAAAATCCATTTACCCTACAGGT TTGCATTTGTTTGCTGAAATTTACCTTTTTTTAGTTATATATATGTGTGTGTGTGTGTGTAATTTCTGCCAATAAATTCTAAATTACAAA GGTAAGAGAAAACCTAGTACATTACTAAATATATAAAGTATATGTTCTGATTATGTATACTTGTTCTAGTGTCAAGTCTTTTTAAGTGGG TTTTTAAAAGTTTGTTATTGGACTTGAATGGATTTTTGAGACTAGGTTAATTATTTTTGAGGTCTTATCCTAAAAGGCATCTAAGGTACA TGAATGGAGTATGGTGATTTTATAACATTTTTTATCAGAATGGAAAAAGAACTGTTTAAAAGTTTGATACTTTTAAATAGTTGGTTTTTT TGCTTACTCTGGTAATGATTTTCTACAAATACATAATAAATTGTTTTTTTGAGTCTATATTCTGTATGCAGTTGAATATCCATTACTTAT TCTGCTGTGCTTTAATAGAATGGAATGTTTACAGGCCCTTAAAATATTATTTTTAAAAAACCTTCTGAAGATACATACCAAAGTTTTTCC AAGAAGATTTTATAATCAATTTAATAATGTAAGGTTTATCAGATTCTATAATAGTATAGTTATTAAGGCAATTTTATGTTAGAGACTATT TTGTAATGTAGTGAGTGGTACCTTTATAAGAAAAGTGACTGCCAATATATTTTTATAGCTGATCTTTATAAATTCTAATGTTGAGTTTTT AATGATTATTTTAAATGTTTATATAGTTTAGTAAAATTTGCATCTCAAAGTATCATTTTTATATTATGGGACGTTTTCAGATTGGCTAAT ATTTGCATTGTAAATTTTGTATGCAGTTTATCTAAAATTCAAAAATACTGTCAGTACACCAGCGTTTAACATCTATATTCCAATTTGTAT ACAGTTTAAAATTGTACTGCAAAACTATTGTGTGCTCTTACACAGTATGCATCATATTGTTGTCTGTGAAATTAAAGGACATTTGATAGT >31790_31790_2_FUS-ATF1_FUS_chr16_31195705_ENST00000380244_ATF1_chr12_51207792_ENST00000262053_length(amino acids)=287AA_BP=125 MLACLCARARTWPQTIIPNKQPKAMGPTPPSPGRAIPSRAVSPTDSRVTVVIASPRTLQAMARAAILLMARARTAMELSQLPRDMARLAA MAVARAPNRLTGSSPPTLAMASSQLPAAPREGGGGVAIAPNGALQLASPGTDGVQGLQTLTMTNSGSTQQGTTILQYAQTSDGQQILVPS NQVVVQTASGDMQTYQIRTTPSATSLPQTVVMTSPVTLTSQTTKTDDPQLKREIRLMKNREAARECRRKKKEYVKCLENRVAVLENQNKT -------------------------------------------------------------- >31790_31790_3_FUS-ATF1_FUS_chr16_31195705_ENST00000568685_ATF1_chr12_51207792_ENST00000262053_length(transcript)=2242nt_BP=411nt AGGCGTCGGTACTCAGCGGTGTTGGAACTTCGTTGCTTGCTTGCCTGTGCGCGCGTGCGCGGACATGGCCTCAAACGATTATACCCAACA AGCAACCCAAAGCTATGGGGCCTACCCCACCCAGCCCGGGCAGGGCTATTCCCAGCAGAGCAGTCAGCCCTACGGACAGCAGAGTTACAG TGGTTATAGCCAGTCCACGGACACTTCAGGCTATGGCCAGAGCAGCTATTCTTCTTATGGCCAGAGCCAGAACACAGGCTATGGAACTCA GTCAACTCCCCAGGGATATGGCTCGACTGGCGGCTATGGCAGTAGCCAGAGCTCCCAATCGTCTTACGGGCAGCAGTCCTCCTACCCTGG CTATGGCCAGCAGCCAGCTCCCAGCAGCACCTCGGGAAGGTGGAGGTGGAGTTGCCATTGCCCCAAATGGAGCCTTACAGTTGGCAAGTC CAGGCACAGATGGAGTACAGGGACTTCAGACATTAACCATGACAAATTCAGGCAGTACTCAGCAAGGTACAACTATTCTTCAGTATGCAC AGACCTCTGATGGACAGCAGATACTTGTGCCCAGCAATCAGGTGGTCGTACAAACTGCATCAGGAGATATGCAAACATATCAGATCCGAA CTACACCTTCAGCTACTTCTCTGCCACAAACTGTGGTGATGACATCTCCTGTGACTCTCACCTCTCAGACAACTAAGACAGATGACCCCC AATTGAAAAGAGAAATAAGGTTAATGAAAAACAGAGAAGCTGCTCGAGAATGTCGCAGAAAGAAGAAAGAATATGTGAAATGCCTGGAAA ACCGAGTTGCAGTCCTGGAAAATCAAAATAAAACTCTAATAGAAGAGTTAAAAACTTTGAAGGATCTTTATTCCAATAAAAGTGTTTGAT TCCTAAGAAAGAAAATATTTTTGTGGACATGCATAAAAATTAAATGGATTTCCTAGTGGAGTTTTATAAATTAAAAGGTCAAAACTGAAG CTTTTTATTTAGGCTTTTCCAAATCAAGGATAAATATCTTACGCACGATATCTAGTGACAGAGGAGAAAGTGGAAAATGACCTCAAGGAA GCTACGGGCACAACTGGAAGCTTTGTAGAAATTAAACATATTCAAGGAGCAAGAAATGAACTTTCAGCAGTCTAAATTTTCTAAATAACC AATAGTTGCCAATCTAAAATGGCAGAGAAGATGAAATTTGATAAACTGAATTTTTTTTAAAAATCCATTTACCCTACAGGTTTGCATTTG TTTGCTGAAATTTACCTTTTTTTAGTTATATATATGTGTGTGTGTGTGTGTAATTTCTGCCAATAAATTCTAAATTACAAAGGTAAGAGA AAACCTAGTACATTACTAAATATATAAAGTATATGTTCTGATTATGTATACTTGTTCTAGTGTCAAGTCTTTTTAAGTGGGTTTTTAAAA GTTTGTTATTGGACTTGAATGGATTTTTGAGACTAGGTTAATTATTTTTGAGGTCTTATCCTAAAAGGCATCTAAGGTACATGAATGGAG TATGGTGATTTTATAACATTTTTTATCAGAATGGAAAAAGAACTGTTTAAAAGTTTGATACTTTTAAATAGTTGGTTTTTTTGCTTACTC TGGTAATGATTTTCTACAAATACATAATAAATTGTTTTTTTGAGTCTATATTCTGTATGCAGTTGAATATCCATTACTTATTCTGCTGTG CTTTAATAGAATGGAATGTTTACAGGCCCTTAAAATATTATTTTTAAAAAACCTTCTGAAGATACATACCAAAGTTTTTCCAAGAAGATT TTATAATCAATTTAATAATGTAAGGTTTATCAGATTCTATAATAGTATAGTTATTAAGGCAATTTTATGTTAGAGACTATTTTGTAATGT AGTGAGTGGTACCTTTATAAGAAAAGTGACTGCCAATATATTTTTATAGCTGATCTTTATAAATTCTAATGTTGAGTTTTTAATGATTAT TTTAAATGTTTATATAGTTTAGTAAAATTTGCATCTCAAAGTATCATTTTTATATTATGGGACGTTTTCAGATTGGCTAATATTTGCATT GTAAATTTTGTATGCAGTTTATCTAAAATTCAAAAATACTGTCAGTACACCAGCGTTTAACATCTATATTCCAATTTGTATACAGTTTAA >31790_31790_3_FUS-ATF1_FUS_chr16_31195705_ENST00000568685_ATF1_chr12_51207792_ENST00000262053_length(amino acids)=288AA_BP=126 MLACLCARARTWPQTIIPNKQPKAMGPTPPSPGRAIPSRAVSPTDSRVTVVIASPRTLQAMARAAILLMARARTQAMELSQLPRDMARLA AMAVARAPNRLTGSSPPTLAMASSQLPAAPREGGGGVAIAPNGALQLASPGTDGVQGLQTLTMTNSGSTQQGTTILQYAQTSDGQQILVP SNQVVVQTASGDMQTYQIRTTPSATSLPQTVVMTSPVTLTSQTTKTDDPQLKREIRLMKNREAARECRRKKKEYVKCLENRVAVLENQNK -------------------------------------------------------------- >31790_31790_4_FUS-ATF1_FUS_chr16_31195717_ENST00000254108_ATF1_chr12_51207791_ENST00000262053_length(transcript)=2459nt_BP=628nt ACTTAAGCTTCGACGCAGGAGGCGGGGCTGCTCAGTCCTCCAGGCGTCGGTACTCAGCGGTGTTGGAACTTCGTTGCTTGCTTGCCTGTG CGCGCGTGCGCGGACATGGCCTCAAACGATTATACCCAACAAGCAACCCAAAGCTATGGGGCCTACCCCACCCAGCCCGGGCAGGGCTAT TCCCAGCAGAGCAGTCAGCCCTACGGACAGCAGAGTTACAGTGGTTATAGCCAGTCCACGGACACTTCAGGCTATGGCCAGAGCAGCTAT TCTTCTTATGGCCAGAGCCAGAACACAGGCTATGGAACTCAGTCAACTCCCCAGGGATATGGCTCGACTGGCGGCTATGGCAGTAGCCAG AGCTCCCAATCGTCTTACGGGCAGCAGTCCTCCTACCCTGGCTATGGCCAGCAGCCAGCTCCCAGCAGCACCTCGGGAAGTTACGGTAGC AGTTCTCAGAGCAGCAGCTATGGGCAGCCCCAGAGTGGGAGCTACAGCCAGCAGCCTAGCTATGGTGGACAGCAGCAAAGCTATGGACAG CAGCAAAGCTATAATCCCCCTCAGGGCTATGGACAGCAGAACCAGTACAACAGCAGCAGTGGTGGTGGAGGTGGAGGTGGAGGTGGAGTT GCCATTGCCCCAAATGGAGCCTTACAGTTGGCAAGTCCAGGCACAGATGGAGTACAGGGACTTCAGACATTAACCATGACAAATTCAGGC AGTACTCAGCAAGGTACAACTATTCTTCAGTATGCACAGACCTCTGATGGACAGCAGATACTTGTGCCCAGCAATCAGGTGGTCGTACAA ACTGCATCAGGAGATATGCAAACATATCAGATCCGAACTACACCTTCAGCTACTTCTCTGCCACAAACTGTGGTGATGACATCTCCTGTG ACTCTCACCTCTCAGACAACTAAGACAGATGACCCCCAATTGAAAAGAGAAATAAGGTTAATGAAAAACAGAGAAGCTGCTCGAGAATGT CGCAGAAAGAAGAAAGAATATGTGAAATGCCTGGAAAACCGAGTTGCAGTCCTGGAAAATCAAAATAAAACTCTAATAGAAGAGTTAAAA ACTTTGAAGGATCTTTATTCCAATAAAAGTGTTTGATTCCTAAGAAAGAAAATATTTTTGTGGACATGCATAAAAATTAAATGGATTTCC TAGTGGAGTTTTATAAATTAAAAGGTCAAAACTGAAGCTTTTTATTTAGGCTTTTCCAAATCAAGGATAAATATCTTACGCACGATATCT AGTGACAGAGGAGAAAGTGGAAAATGACCTCAAGGAAGCTACGGGCACAACTGGAAGCTTTGTAGAAATTAAACATATTCAAGGAGCAAG AAATGAACTTTCAGCAGTCTAAATTTTCTAAATAACCAATAGTTGCCAATCTAAAATGGCAGAGAAGATGAAATTTGATAAACTGAATTT TTTTTAAAAATCCATTTACCCTACAGGTTTGCATTTGTTTGCTGAAATTTACCTTTTTTTAGTTATATATATGTGTGTGTGTGTGTGTAA TTTCTGCCAATAAATTCTAAATTACAAAGGTAAGAGAAAACCTAGTACATTACTAAATATATAAAGTATATGTTCTGATTATGTATACTT GTTCTAGTGTCAAGTCTTTTTAAGTGGGTTTTTAAAAGTTTGTTATTGGACTTGAATGGATTTTTGAGACTAGGTTAATTATTTTTGAGG TCTTATCCTAAAAGGCATCTAAGGTACATGAATGGAGTATGGTGATTTTATAACATTTTTTATCAGAATGGAAAAAGAACTGTTTAAAAG TTTGATACTTTTAAATAGTTGGTTTTTTTGCTTACTCTGGTAATGATTTTCTACAAATACATAATAAATTGTTTTTTTGAGTCTATATTC TGTATGCAGTTGAATATCCATTACTTATTCTGCTGTGCTTTAATAGAATGGAATGTTTACAGGCCCTTAAAATATTATTTTTAAAAAACC TTCTGAAGATACATACCAAAGTTTTTCCAAGAAGATTTTATAATCAATTTAATAATGTAAGGTTTATCAGATTCTATAATAGTATAGTTA TTAAGGCAATTTTATGTTAGAGACTATTTTGTAATGTAGTGAGTGGTACCTTTATAAGAAAAGTGACTGCCAATATATTTTTATAGCTGA TCTTTATAAATTCTAATGTTGAGTTTTTAATGATTATTTTAAATGTTTATATAGTTTAGTAAAATTTGCATCTCAAAGTATCATTTTTAT ATTATGGGACGTTTTCAGATTGGCTAATATTTGCATTGTAAATTTTGTATGCAGTTTATCTAAAATTCAAAAATACTGTCAGTACACCAG CGTTTAACATCTATATTCCAATTTGTATACAGTTTAAAATTGTACTGCAAAACTATTGTGTGCTCTTACACAGTATGCATCATATTGTTG >31790_31790_4_FUS-ATF1_FUS_chr16_31195717_ENST00000254108_ATF1_chr12_51207791_ENST00000262053_length(amino acids)=362AA_BP=200 MLSPPGVGTQRCWNFVACLPVRACADMASNDYTQQATQSYGAYPTQPGQGYSQQSSQPYGQQSYSGYSQSTDTSGYGQSSYSSYGQSQNT GYGTQSTPQGYGSTGGYGSSQSSQSSYGQQSSYPGYGQQPAPSSTSGSYGSSSQSSSYGQPQSGSYSQQPSYGGQQQSYGQQQSYNPPQG YGQQNQYNSSSGGGGGGGGGVAIAPNGALQLASPGTDGVQGLQTLTMTNSGSTQQGTTILQYAQTSDGQQILVPSNQVVVQTASGDMQTY QIRTTPSATSLPQTVVMTSPVTLTSQTTKTDDPQLKREIRLMKNREAARECRRKKKEYVKCLENRVAVLENQNKTLIEELKTLKDLYSNK -------------------------------------------------------------- >31790_31790_5_FUS-ATF1_FUS_chr16_31195717_ENST00000380244_ATF1_chr12_51207791_ENST00000262053_length(transcript)=2427nt_BP=596nt GCTCAGTCCTCCAGGCGTCGGTACTCAGCGGTGTTGGAACTTCGTTGCTTGCTTGCCTGTGCGCGCGTGCGCGGACATGGCCTCAAACGA TTATACCCAACAAGCAACCCAAAGCTATGGGGCCTACCCCACCCAGCCCGGGCAGGGCTATTCCCAGCAGAGCAGTCAGCCCTACGGACA GCAGAGTTACAGTGGTTATAGCCAGTCCACGGACACTTCAGGCTATGGCCAGAGCAGCTATTCTTCTTATGGCCAGAGCCAGAACAGCTA TGGAACTCAGTCAACTCCCCAGGGATATGGCTCGACTGGCGGCTATGGCAGTAGCCAGAGCTCCCAATCGTCTTACGGGCAGCAGTCCTC CTACCCTGGCTATGGCCAGCAGCCAGCTCCCAGCAGCACCTCGGGAAGTTACGGTAGCAGTTCTCAGAGCAGCAGCTATGGGCAGCCCCA GAGTGGGAGCTACAGCCAGCAGCCTAGCTATGGTGGACAGCAGCAAAGCTATGGACAGCAGCAAAGCTATAATCCCCCTCAGGGCTATGG ACAGCAGAACCAGTACAACAGCAGCAGTGGTGGTGGAGGTGGAGGTGGAGGTGGAGTTGCCATTGCCCCAAATGGAGCCTTACAGTTGGC AAGTCCAGGCACAGATGGAGTACAGGGACTTCAGACATTAACCATGACAAATTCAGGCAGTACTCAGCAAGGTACAACTATTCTTCAGTA TGCACAGACCTCTGATGGACAGCAGATACTTGTGCCCAGCAATCAGGTGGTCGTACAAACTGCATCAGGAGATATGCAAACATATCAGAT CCGAACTACACCTTCAGCTACTTCTCTGCCACAAACTGTGGTGATGACATCTCCTGTGACTCTCACCTCTCAGACAACTAAGACAGATGA CCCCCAATTGAAAAGAGAAATAAGGTTAATGAAAAACAGAGAAGCTGCTCGAGAATGTCGCAGAAAGAAGAAAGAATATGTGAAATGCCT GGAAAACCGAGTTGCAGTCCTGGAAAATCAAAATAAAACTCTAATAGAAGAGTTAAAAACTTTGAAGGATCTTTATTCCAATAAAAGTGT TTGATTCCTAAGAAAGAAAATATTTTTGTGGACATGCATAAAAATTAAATGGATTTCCTAGTGGAGTTTTATAAATTAAAAGGTCAAAAC TGAAGCTTTTTATTTAGGCTTTTCCAAATCAAGGATAAATATCTTACGCACGATATCTAGTGACAGAGGAGAAAGTGGAAAATGACCTCA AGGAAGCTACGGGCACAACTGGAAGCTTTGTAGAAATTAAACATATTCAAGGAGCAAGAAATGAACTTTCAGCAGTCTAAATTTTCTAAA TAACCAATAGTTGCCAATCTAAAATGGCAGAGAAGATGAAATTTGATAAACTGAATTTTTTTTAAAAATCCATTTACCCTACAGGTTTGC ATTTGTTTGCTGAAATTTACCTTTTTTTAGTTATATATATGTGTGTGTGTGTGTGTAATTTCTGCCAATAAATTCTAAATTACAAAGGTA AGAGAAAACCTAGTACATTACTAAATATATAAAGTATATGTTCTGATTATGTATACTTGTTCTAGTGTCAAGTCTTTTTAAGTGGGTTTT TAAAAGTTTGTTATTGGACTTGAATGGATTTTTGAGACTAGGTTAATTATTTTTGAGGTCTTATCCTAAAAGGCATCTAAGGTACATGAA TGGAGTATGGTGATTTTATAACATTTTTTATCAGAATGGAAAAAGAACTGTTTAAAAGTTTGATACTTTTAAATAGTTGGTTTTTTTGCT TACTCTGGTAATGATTTTCTACAAATACATAATAAATTGTTTTTTTGAGTCTATATTCTGTATGCAGTTGAATATCCATTACTTATTCTG CTGTGCTTTAATAGAATGGAATGTTTACAGGCCCTTAAAATATTATTTTTAAAAAACCTTCTGAAGATACATACCAAAGTTTTTCCAAGA AGATTTTATAATCAATTTAATAATGTAAGGTTTATCAGATTCTATAATAGTATAGTTATTAAGGCAATTTTATGTTAGAGACTATTTTGT AATGTAGTGAGTGGTACCTTTATAAGAAAAGTGACTGCCAATATATTTTTATAGCTGATCTTTATAAATTCTAATGTTGAGTTTTTAATG ATTATTTTAAATGTTTATATAGTTTAGTAAAATTTGCATCTCAAAGTATCATTTTTATATTATGGGACGTTTTCAGATTGGCTAATATTT GCATTGTAAATTTTGTATGCAGTTTATCTAAAATTCAAAAATACTGTCAGTACACCAGCGTTTAACATCTATATTCCAATTTGTATACAG >31790_31790_5_FUS-ATF1_FUS_chr16_31195717_ENST00000380244_ATF1_chr12_51207791_ENST00000262053_length(amino acids)=343AA_BP=181 MPVRACADMASNDYTQQATQSYGAYPTQPGQGYSQQSSQPYGQQSYSGYSQSTDTSGYGQSSYSSYGQSQNSYGTQSTPQGYGSTGGYGS SQSSQSSYGQQSSYPGYGQQPAPSSTSGSYGSSSQSSSYGQPQSGSYSQQPSYGGQQQSYGQQQSYNPPQGYGQQNQYNSSSGGGGGGGG GVAIAPNGALQLASPGTDGVQGLQTLTMTNSGSTQQGTTILQYAQTSDGQQILVPSNQVVVQTASGDMQTYQIRTTPSATSLPQTVVMTS -------------------------------------------------------------- >31790_31790_6_FUS-ATF1_FUS_chr16_31195717_ENST00000568685_ATF1_chr12_51207791_ENST00000262053_length(transcript)=2418nt_BP=587nt AGGCGTCGGTACTCAGCGGTGTTGGAACTTCGTTGCTTGCTTGCCTGTGCGCGCGTGCGCGGACATGGCCTCAAACGATTATACCCAACA AGCAACCCAAAGCTATGGGGCCTACCCCACCCAGCCCGGGCAGGGCTATTCCCAGCAGAGCAGTCAGCCCTACGGACAGCAGAGTTACAG TGGTTATAGCCAGTCCACGGACACTTCAGGCTATGGCCAGAGCAGCTATTCTTCTTATGGCCAGAGCCAGAACACAGGCTATGGAACTCA GTCAACTCCCCAGGGATATGGCTCGACTGGCGGCTATGGCAGTAGCCAGAGCTCCCAATCGTCTTACGGGCAGCAGTCCTCCTACCCTGG CTATGGCCAGCAGCCAGCTCCCAGCAGCACCTCGGGAAGTTACGGTAGCAGTTCTCAGAGCAGCAGCTATGGGCAGCCCCAGAGTGGGAG CTACAGCCAGCAGCCTAGCTATGGTGGACAGCAGCAAAGCTATGGACAGCAGCAAAGCTATAATCCCCCTCAGGGCTATGGACAGCAGAA CCAGTACAACAGCAGCAGTGGTGGTGGAGGTGGAGGTGGAGGTGGAGTTGCCATTGCCCCAAATGGAGCCTTACAGTTGGCAAGTCCAGG CACAGATGGAGTACAGGGACTTCAGACATTAACCATGACAAATTCAGGCAGTACTCAGCAAGGTACAACTATTCTTCAGTATGCACAGAC CTCTGATGGACAGCAGATACTTGTGCCCAGCAATCAGGTGGTCGTACAAACTGCATCAGGAGATATGCAAACATATCAGATCCGAACTAC ACCTTCAGCTACTTCTCTGCCACAAACTGTGGTGATGACATCTCCTGTGACTCTCACCTCTCAGACAACTAAGACAGATGACCCCCAATT GAAAAGAGAAATAAGGTTAATGAAAAACAGAGAAGCTGCTCGAGAATGTCGCAGAAAGAAGAAAGAATATGTGAAATGCCTGGAAAACCG AGTTGCAGTCCTGGAAAATCAAAATAAAACTCTAATAGAAGAGTTAAAAACTTTGAAGGATCTTTATTCCAATAAAAGTGTTTGATTCCT AAGAAAGAAAATATTTTTGTGGACATGCATAAAAATTAAATGGATTTCCTAGTGGAGTTTTATAAATTAAAAGGTCAAAACTGAAGCTTT TTATTTAGGCTTTTCCAAATCAAGGATAAATATCTTACGCACGATATCTAGTGACAGAGGAGAAAGTGGAAAATGACCTCAAGGAAGCTA CGGGCACAACTGGAAGCTTTGTAGAAATTAAACATATTCAAGGAGCAAGAAATGAACTTTCAGCAGTCTAAATTTTCTAAATAACCAATA GTTGCCAATCTAAAATGGCAGAGAAGATGAAATTTGATAAACTGAATTTTTTTTAAAAATCCATTTACCCTACAGGTTTGCATTTGTTTG CTGAAATTTACCTTTTTTTAGTTATATATATGTGTGTGTGTGTGTGTAATTTCTGCCAATAAATTCTAAATTACAAAGGTAAGAGAAAAC CTAGTACATTACTAAATATATAAAGTATATGTTCTGATTATGTATACTTGTTCTAGTGTCAAGTCTTTTTAAGTGGGTTTTTAAAAGTTT GTTATTGGACTTGAATGGATTTTTGAGACTAGGTTAATTATTTTTGAGGTCTTATCCTAAAAGGCATCTAAGGTACATGAATGGAGTATG GTGATTTTATAACATTTTTTATCAGAATGGAAAAAGAACTGTTTAAAAGTTTGATACTTTTAAATAGTTGGTTTTTTTGCTTACTCTGGT AATGATTTTCTACAAATACATAATAAATTGTTTTTTTGAGTCTATATTCTGTATGCAGTTGAATATCCATTACTTATTCTGCTGTGCTTT AATAGAATGGAATGTTTACAGGCCCTTAAAATATTATTTTTAAAAAACCTTCTGAAGATACATACCAAAGTTTTTCCAAGAAGATTTTAT AATCAATTTAATAATGTAAGGTTTATCAGATTCTATAATAGTATAGTTATTAAGGCAATTTTATGTTAGAGACTATTTTGTAATGTAGTG AGTGGTACCTTTATAAGAAAAGTGACTGCCAATATATTTTTATAGCTGATCTTTATAAATTCTAATGTTGAGTTTTTAATGATTATTTTA AATGTTTATATAGTTTAGTAAAATTTGCATCTCAAAGTATCATTTTTATATTATGGGACGTTTTCAGATTGGCTAATATTTGCATTGTAA ATTTTGTATGCAGTTTATCTAAAATTCAAAAATACTGTCAGTACACCAGCGTTTAACATCTATATTCCAATTTGTATACAGTTTAAAATT >31790_31790_6_FUS-ATF1_FUS_chr16_31195717_ENST00000568685_ATF1_chr12_51207791_ENST00000262053_length(amino acids)=344AA_BP=182 MPVRACADMASNDYTQQATQSYGAYPTQPGQGYSQQSSQPYGQQSYSGYSQSTDTSGYGQSSYSSYGQSQNTGYGTQSTPQGYGSTGGYG SSQSSQSSYGQQSSYPGYGQQPAPSSTSGSYGSSSQSSSYGQPQSGSYSQQPSYGGQQQSYGQQQSYNPPQGYGQQNQYNSSSGGGGGGG GGVAIAPNGALQLASPGTDGVQGLQTLTMTNSGSTQQGTTILQYAQTSDGQQILVPSNQVVVQTASGDMQTYQIRTTPSATSLPQTVVMT -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for FUS-ATF1 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for FUS-ATF1 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for FUS-ATF1 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
