
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:FUS-KCTD5 (FusionGDB2 ID:31807) |
Fusion Gene Summary for FUS-KCTD5 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: FUS-KCTD5 | Fusion gene ID: 31807 | Hgene | Tgene | Gene symbol | FUS | KCTD5 | Gene ID | 2521 | 54442 |
| Gene name | FUS RNA binding protein | potassium channel tetramerization domain containing 5 | |
| Synonyms | ALS6|ETM4|FUS1|HNRNPP2|POMP75|TLS | - | |
| Cytomap | 16p11.2 | 16p13.3 | |
| Type of gene | protein-coding | protein-coding | |
| Description | RNA-binding protein FUS75 kDa DNA-pairing proteinfus-like proteinfused in sarcomafusion gene in myxoid liposarcomaheterogeneous nuclear ribonucleoprotein P2oncogene FUSoncogene TLStranslocated in liposarcoma protein | BTB/POZ domain-containing protein KCTD5potassium channel tetramerisation domain containing 5 | |
| Modification date | 20200329 | 20200313 | |
| UniProtAcc | P35637 | Q9NXV2 | |
| Ensembl transtripts involved in fusion gene | ENST00000254108, ENST00000380244, ENST00000568685, ENST00000474990, | ENST00000569689, ENST00000564195, ENST00000301738, | |
| Fusion gene scores | * DoF score | 37 X 37 X 16=21904 | 9 X 8 X 8=576 |
| # samples | 44 | 12 | |
| ** MAII score | log2(44/21904*10)=-5.63754701773324 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(12/576*10)=-2.26303440583379 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: FUS [Title/Abstract] AND KCTD5 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | FUS(31191548)-KCTD5(2757299), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | FUS-KCTD5 seems lost the major protein functional domain in Hgene partner, which is a CGC due to the frame-shifted ORF. FUS-KCTD5 seems lost the major protein functional domain in Hgene partner, which is a essential gene due to the frame-shifted ORF. FUS-KCTD5 seems lost the major protein functional domain in Hgene partner, which is a tumor suppressor due to the frame-shifted ORF. FUS-KCTD5 seems lost the major protein functional domain in Tgene partner, which is a essential gene due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | FUS | GO:0006355 | regulation of transcription, DNA-templated | 26124092 |
| Hgene | FUS | GO:0006357 | regulation of transcription by RNA polymerase II | 25453086 |
| Hgene | FUS | GO:0008380 | RNA splicing | 26124092 |
| Hgene | FUS | GO:0043484 | regulation of RNA splicing | 25453086|27731383 |
| Hgene | FUS | GO:0048255 | mRNA stabilization | 27378374 |
| Hgene | FUS | GO:0051260 | protein homooligomerization | 25453086 |
| Hgene | FUS | GO:1905168 | positive regulation of double-strand break repair via homologous recombination | 10567410 |
 Fusion gene breakpoints across FUS (5'-gene) Fusion gene breakpoints across FUS (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
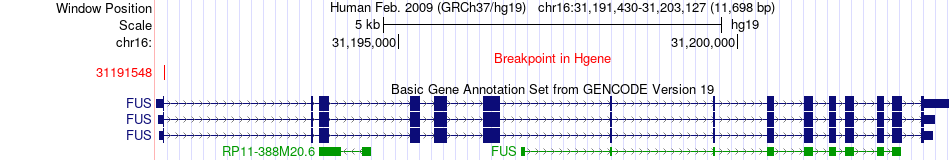 |
 Fusion gene breakpoints across KCTD5 (3'-gene) Fusion gene breakpoints across KCTD5 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
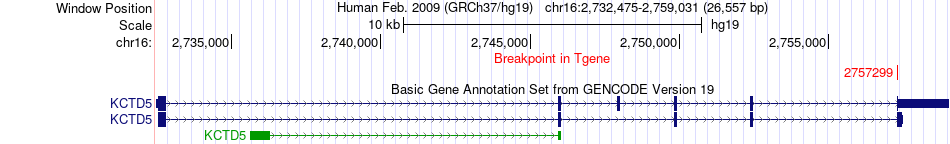 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | Non-Cancer | ERR315469 | FUS | chr16 | 31191548 | + | KCTD5 | chr16 | 2757299 | + |
Top |
Fusion Gene ORF analysis for FUS-KCTD5 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-intron | ENST00000254108 | ENST00000569689 | FUS | chr16 | 31191548 | + | KCTD5 | chr16 | 2757299 | + |
| 5CDS-intron | ENST00000380244 | ENST00000569689 | FUS | chr16 | 31191548 | + | KCTD5 | chr16 | 2757299 | + |
| 5CDS-intron | ENST00000568685 | ENST00000569689 | FUS | chr16 | 31191548 | + | KCTD5 | chr16 | 2757299 | + |
| Frame-shift | ENST00000254108 | ENST00000564195 | FUS | chr16 | 31191548 | + | KCTD5 | chr16 | 2757299 | + |
| Frame-shift | ENST00000380244 | ENST00000564195 | FUS | chr16 | 31191548 | + | KCTD5 | chr16 | 2757299 | + |
| Frame-shift | ENST00000568685 | ENST00000564195 | FUS | chr16 | 31191548 | + | KCTD5 | chr16 | 2757299 | + |
| In-frame | ENST00000254108 | ENST00000301738 | FUS | chr16 | 31191548 | + | KCTD5 | chr16 | 2757299 | + |
| In-frame | ENST00000380244 | ENST00000301738 | FUS | chr16 | 31191548 | + | KCTD5 | chr16 | 2757299 | + |
| In-frame | ENST00000568685 | ENST00000301738 | FUS | chr16 | 31191548 | + | KCTD5 | chr16 | 2757299 | + |
| intron-3CDS | ENST00000474990 | ENST00000301738 | FUS | chr16 | 31191548 | + | KCTD5 | chr16 | 2757299 | + |
| intron-3CDS | ENST00000474990 | ENST00000564195 | FUS | chr16 | 31191548 | + | KCTD5 | chr16 | 2757299 | + |
| intron-intron | ENST00000474990 | ENST00000569689 | FUS | chr16 | 31191548 | + | KCTD5 | chr16 | 2757299 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000254108 | FUS | chr16 | 31191548 | + | ENST00000301738 | KCTD5 | chr16 | 2757299 | + | 1851 | 118 | 319 | 663 | 114 |
| ENST00000380244 | FUS | chr16 | 31191548 | + | ENST00000301738 | KCTD5 | chr16 | 2757299 | + | 1822 | 89 | 290 | 634 | 114 |
| ENST00000568685 | FUS | chr16 | 31191548 | + | ENST00000301738 | KCTD5 | chr16 | 2757299 | + | 1810 | 77 | 278 | 622 | 114 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000254108 | ENST00000301738 | FUS | chr16 | 31191548 | + | KCTD5 | chr16 | 2757299 | + | 0.8202753 | 0.17972462 |
| ENST00000380244 | ENST00000301738 | FUS | chr16 | 31191548 | + | KCTD5 | chr16 | 2757299 | + | 0.9173347 | 0.08266538 |
| ENST00000568685 | ENST00000301738 | FUS | chr16 | 31191548 | + | KCTD5 | chr16 | 2757299 | + | 0.9020645 | 0.09793549 |
Top |
Fusion Genomic Features for FUS-KCTD5 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| FUS | chr16 | 31191548 | + | KCTD5 | chr16 | 2757298 | + | 0.9394157 | 0.060584385 |
| FUS | chr16 | 31191548 | + | KCTD5 | chr16 | 2757298 | + | 0.9394157 | 0.060584385 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
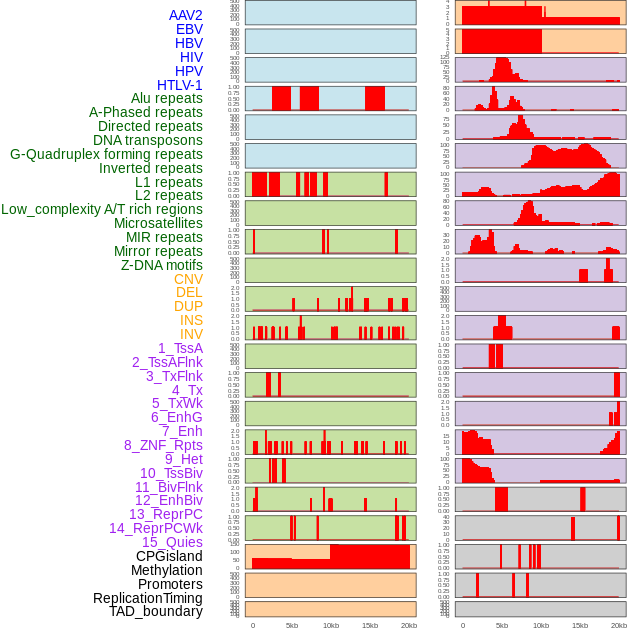 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
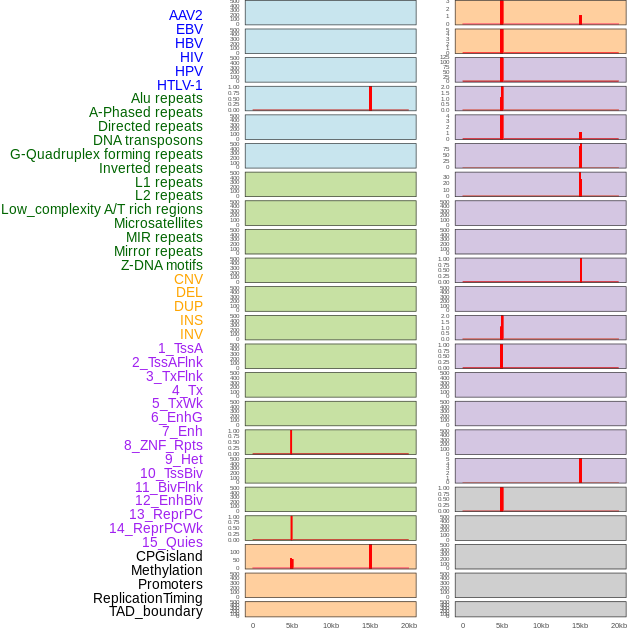 |
Top |
Fusion Protein Features for FUS-KCTD5 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr16:31191548/chr16:2757299) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| FUS | KCTD5 |
| FUNCTION: DNA/RNA-binding protein that plays a role in various cellular processes such as transcription regulation, RNA splicing, RNA transport, DNA repair and damage response (PubMed:27731383). Binds to nascent pre-mRNAs and acts as a molecular mediator between RNA polymerase II and U1 small nuclear ribonucleoprotein thereby coupling transcription and splicing (PubMed:26124092). Binds also its own pre-mRNA and autoregulates its expression; this autoregulation mechanism is mediated by non-sense-mediated decay (PubMed:24204307). Plays a role in DNA repair mechanisms by promoting D-loop formation and homologous recombination during DNA double-strand break repair (PubMed:10567410). In neuronal cells, plays crucial roles in dendritic spine formation and stability, RNA transport, mRNA stability and synaptic homeostasis (By similarity). {ECO:0000250|UniProtKB:P56959, ECO:0000269|PubMed:10567410, ECO:0000269|PubMed:24204307, ECO:0000269|PubMed:26124092, ECO:0000269|PubMed:27731383}. | FUNCTION: Its interaction with CUL3 suggests that it may act as a substrate adapter in some E3 ligase complex (PubMed:18573101). Does not affect the function of Kv channel Kv2.1/KCNB1, Kv1.2/KCNA2, Kv4.2/KCND2 and Kv3.4/KCNC4 (PubMed:19361449). {ECO:0000269|PubMed:18573101, ECO:0000269|PubMed:19361449}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | FUS | chr16:31191548 | chr16:2757299 | ENST00000254108 | + | 1 | 15 | 166_267 | 4 | 527.0 | Compositional bias | Note=Gly-rich |
| Hgene | FUS | chr16:31191548 | chr16:2757299 | ENST00000254108 | + | 1 | 15 | 1_165 | 4 | 527.0 | Compositional bias | Note=Gln/Gly/Ser/Tyr-rich |
| Hgene | FUS | chr16:31191548 | chr16:2757299 | ENST00000254108 | + | 1 | 15 | 371_526 | 4 | 527.0 | Compositional bias | Note=Arg/Gly-rich |
| Hgene | FUS | chr16:31191548 | chr16:2757299 | ENST00000380244 | + | 1 | 15 | 166_267 | 4 | 526.0 | Compositional bias | Note=Gly-rich |
| Hgene | FUS | chr16:31191548 | chr16:2757299 | ENST00000380244 | + | 1 | 15 | 1_165 | 4 | 526.0 | Compositional bias | Note=Gln/Gly/Ser/Tyr-rich |
| Hgene | FUS | chr16:31191548 | chr16:2757299 | ENST00000380244 | + | 1 | 15 | 371_526 | 4 | 526.0 | Compositional bias | Note=Arg/Gly-rich |
| Hgene | FUS | chr16:31191548 | chr16:2757299 | ENST00000254108 | + | 1 | 15 | 285_371 | 4 | 527.0 | Domain | RRM |
| Hgene | FUS | chr16:31191548 | chr16:2757299 | ENST00000380244 | + | 1 | 15 | 285_371 | 4 | 526.0 | Domain | RRM |
| Hgene | FUS | chr16:31191548 | chr16:2757299 | ENST00000254108 | + | 1 | 15 | 422_453 | 4 | 527.0 | Zinc finger | RanBP2-type |
| Hgene | FUS | chr16:31191548 | chr16:2757299 | ENST00000380244 | + | 1 | 15 | 422_453 | 4 | 526.0 | Zinc finger | RanBP2-type |
| Tgene | KCTD5 | chr16:31191548 | chr16:2757299 | ENST00000301738 | 4 | 6 | 44_146 | 225 | 235.0 | Domain | Note=BTB |
Top |
Fusion Gene Sequence for FUS-KCTD5 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >31807_31807_1_FUS-KCTD5_FUS_chr16_31191548_ENST00000254108_KCTD5_chr16_2757299_ENST00000301738_length(transcript)=1851nt_BP=118nt ACTTAAGCTTCGACGCAGGAGGCGGGGCTGCTCAGTCCTCCAGGCGTCGGTACTCAGCGGTGTTGGAACTTCGTTGCTTGCTTGCCTGTG CGCGCGTGCGCGGACATGGCCTCAAACGATTTTGCAAGAACGAGGCTCAAGGATGTGAGGGACACAGTATTGACAGCTGAAGAAATGATT TACGTTTTCCCGAGATGTAATGAACTGCCATGTCCAGGAAGCTTGGCTGTGAGAAGAAACCTGCTTTTGATCATTTTTCTAGAGATCTGG GTGTGAATCCTTTTTTGCCTCTGAGGTGGGTGGTGAGAGACGGGCCCAGCTGTCCAAGGCCAGACGTCCCCAAGTTGGGGGAGCACGGCG GCCGGGTGGGCGCTGCCTCTTGGGGGGGCCTCGCTCTGTTTTTTCCAAGTGCCACGTGGGACTGAGGCAGACACTCCCAGTCAGCCCGCT CGATCCTGAAGATCGTGTGAAGGAAGCGTTCTTGGTGCTACACACAGTCTGGAAAAGCAGCCTGGGCTTCACTGGCAGGAAGCGGCCGCA GCCGCGTCAGTGTCCAGCACGTCGTGGCCTCTGGTCCGACCACCAGGCCCTAGTCTCGGTCAGGGAGTCTGCTCTGCCTCCCAGGCGGGA TCGCTGGTTTCTCTCGACCTCGCAGAGCTCCTGACAAAGGCGGCTTCTGCGTCGTCACTGCTTCCCGGCGCCATTCCGAGGCCGGGCCTT CTTCTGACACGGGCTCCAACCCCACCTGACCAAGCCCGGGACAGTCAAGGCTTCTCAATTTGTATTTTCCAGGCAAGAGAACTTTGTACC CCTTCTCTGTGTGGATAGACTCCCCAGCGTTTTTCCTCTGGAAATGCCCACAGGGCTTGCCGCGTGGAGACTGATCTGTTCCATCCGTTA GCGCGAGGTAGCAGTGTCGCCTCGCCCCTCCCACTGCGGGCTCACGGGGAGCTGGCGTCTGTCAGTGCCTTGTCACGCCTGGCATAGAGG TTGGGCTGGAGGCCTGTCCACTGGCTTCTGGAGCTGAAGGTCTGCGTCACGGTGAAAATTCGGGATGTTTACAGAGCATCACAAATGCCT CTCTCTCGCCGCTCTCTCATTTTCTTTGTATAACTATGCAGCCTTCCCTCTCTTTCTGGGATGTTTGGGACTTCACTCGACCTGCACGGG CTCTGCCCGACATGCCGTGGGAGCTGCTGGGATTCCTTTAGAAACCGCTGCCCGCATGCTTTGAAAACAGACCTTTCTCAGCTGGCTGTG GGGACCTGTCAGAGTCTGGGGACTCGGCGTGCAGGGCGGGCTCCAAGCGCTTTGCTTCCGGAACTCCGGCTTCCCAAGGGGTACTGTGCA GACTGACCACCGGCCTCCCGCCTGCAGGTCAGAGGCTCTGACACTGTCTGGTTTCCAATGCTTCTGGAGACTTCCTGCCTAGGCCTCATC CTCCTCTTTGCCAGTCACCTGATTAACCAATTCTCCAGCATTAGGACTTACCTCCTTCTTTTTGTAAGGTCTCTTGTATGTTGTTAAATG TTTGGCTTAAACAATTTATAAAAGCCTTTCTAGAAGGCAGACTTAGCCCCAGCAGAGCCTGGGAGGAAGGCGGCCGCTGGGGCTCCTGGG CATCCTCTCTGGGGAGCTGCTGGCCGCTTAGCGTTGTTTGATTTTTGACCTTGACATAGTAGATAGGCTTGTGGTTTTTTTAATTTAAAG ATATTAAGACTTAATTCACTAAAATTTATTCTAAGGGGATATTTATACTTTTATACTTTTTTAGGTATCGTATTTTATCAGCTTACAGTT >31807_31807_1_FUS-KCTD5_FUS_chr16_31191548_ENST00000254108_KCTD5_chr16_2757299_ENST00000301738_length(amino acids)=114AA_BP= MSKARRPQVGGARRPGGRCLLGGPRSVFSKCHVGLRQTLPVSPLDPEDRVKEAFLVLHTVWKSSLGFTGRKRPQPRQCPARRGLWSDHQA -------------------------------------------------------------- >31807_31807_2_FUS-KCTD5_FUS_chr16_31191548_ENST00000380244_KCTD5_chr16_2757299_ENST00000301738_length(transcript)=1822nt_BP=89nt GCTCAGTCCTCCAGGCGTCGGTACTCAGCGGTGTTGGAACTTCGTTGCTTGCTTGCCTGTGCGCGCGTGCGCGGACATGGCCTCAAACGA TTTTGCAAGAACGAGGCTCAAGGATGTGAGGGACACAGTATTGACAGCTGAAGAAATGATTTACGTTTTCCCGAGATGTAATGAACTGCC ATGTCCAGGAAGCTTGGCTGTGAGAAGAAACCTGCTTTTGATCATTTTTCTAGAGATCTGGGTGTGAATCCTTTTTTGCCTCTGAGGTGG GTGGTGAGAGACGGGCCCAGCTGTCCAAGGCCAGACGTCCCCAAGTTGGGGGAGCACGGCGGCCGGGTGGGCGCTGCCTCTTGGGGGGGC CTCGCTCTGTTTTTTCCAAGTGCCACGTGGGACTGAGGCAGACACTCCCAGTCAGCCCGCTCGATCCTGAAGATCGTGTGAAGGAAGCGT TCTTGGTGCTACACACAGTCTGGAAAAGCAGCCTGGGCTTCACTGGCAGGAAGCGGCCGCAGCCGCGTCAGTGTCCAGCACGTCGTGGCC TCTGGTCCGACCACCAGGCCCTAGTCTCGGTCAGGGAGTCTGCTCTGCCTCCCAGGCGGGATCGCTGGTTTCTCTCGACCTCGCAGAGCT CCTGACAAAGGCGGCTTCTGCGTCGTCACTGCTTCCCGGCGCCATTCCGAGGCCGGGCCTTCTTCTGACACGGGCTCCAACCCCACCTGA CCAAGCCCGGGACAGTCAAGGCTTCTCAATTTGTATTTTCCAGGCAAGAGAACTTTGTACCCCTTCTCTGTGTGGATAGACTCCCCAGCG TTTTTCCTCTGGAAATGCCCACAGGGCTTGCCGCGTGGAGACTGATCTGTTCCATCCGTTAGCGCGAGGTAGCAGTGTCGCCTCGCCCCT CCCACTGCGGGCTCACGGGGAGCTGGCGTCTGTCAGTGCCTTGTCACGCCTGGCATAGAGGTTGGGCTGGAGGCCTGTCCACTGGCTTCT GGAGCTGAAGGTCTGCGTCACGGTGAAAATTCGGGATGTTTACAGAGCATCACAAATGCCTCTCTCTCGCCGCTCTCTCATTTTCTTTGT ATAACTATGCAGCCTTCCCTCTCTTTCTGGGATGTTTGGGACTTCACTCGACCTGCACGGGCTCTGCCCGACATGCCGTGGGAGCTGCTG GGATTCCTTTAGAAACCGCTGCCCGCATGCTTTGAAAACAGACCTTTCTCAGCTGGCTGTGGGGACCTGTCAGAGTCTGGGGACTCGGCG TGCAGGGCGGGCTCCAAGCGCTTTGCTTCCGGAACTCCGGCTTCCCAAGGGGTACTGTGCAGACTGACCACCGGCCTCCCGCCTGCAGGT CAGAGGCTCTGACACTGTCTGGTTTCCAATGCTTCTGGAGACTTCCTGCCTAGGCCTCATCCTCCTCTTTGCCAGTCACCTGATTAACCA ATTCTCCAGCATTAGGACTTACCTCCTTCTTTTTGTAAGGTCTCTTGTATGTTGTTAAATGTTTGGCTTAAACAATTTATAAAAGCCTTT CTAGAAGGCAGACTTAGCCCCAGCAGAGCCTGGGAGGAAGGCGGCCGCTGGGGCTCCTGGGCATCCTCTCTGGGGAGCTGCTGGCCGCTT AGCGTTGTTTGATTTTTGACCTTGACATAGTAGATAGGCTTGTGGTTTTTTTAATTTAAAGATATTAAGACTTAATTCACTAAAATTTAT TCTAAGGGGATATTTATACTTTTATACTTTTTTAGGTATCGTATTTTATCAGCTTACAGTTTAATGCCTAAGTTTCCCCTGGAAATAGCA >31807_31807_2_FUS-KCTD5_FUS_chr16_31191548_ENST00000380244_KCTD5_chr16_2757299_ENST00000301738_length(amino acids)=114AA_BP= MSKARRPQVGGARRPGGRCLLGGPRSVFSKCHVGLRQTLPVSPLDPEDRVKEAFLVLHTVWKSSLGFTGRKRPQPRQCPARRGLWSDHQA -------------------------------------------------------------- >31807_31807_3_FUS-KCTD5_FUS_chr16_31191548_ENST00000568685_KCTD5_chr16_2757299_ENST00000301738_length(transcript)=1810nt_BP=77nt AGGCGTCGGTACTCAGCGGTGTTGGAACTTCGTTGCTTGCTTGCCTGTGCGCGCGTGCGCGGACATGGCCTCAAACGATTTTGCAAGAAC GAGGCTCAAGGATGTGAGGGACACAGTATTGACAGCTGAAGAAATGATTTACGTTTTCCCGAGATGTAATGAACTGCCATGTCCAGGAAG CTTGGCTGTGAGAAGAAACCTGCTTTTGATCATTTTTCTAGAGATCTGGGTGTGAATCCTTTTTTGCCTCTGAGGTGGGTGGTGAGAGAC GGGCCCAGCTGTCCAAGGCCAGACGTCCCCAAGTTGGGGGAGCACGGCGGCCGGGTGGGCGCTGCCTCTTGGGGGGGCCTCGCTCTGTTT TTTCCAAGTGCCACGTGGGACTGAGGCAGACACTCCCAGTCAGCCCGCTCGATCCTGAAGATCGTGTGAAGGAAGCGTTCTTGGTGCTAC ACACAGTCTGGAAAAGCAGCCTGGGCTTCACTGGCAGGAAGCGGCCGCAGCCGCGTCAGTGTCCAGCACGTCGTGGCCTCTGGTCCGACC ACCAGGCCCTAGTCTCGGTCAGGGAGTCTGCTCTGCCTCCCAGGCGGGATCGCTGGTTTCTCTCGACCTCGCAGAGCTCCTGACAAAGGC GGCTTCTGCGTCGTCACTGCTTCCCGGCGCCATTCCGAGGCCGGGCCTTCTTCTGACACGGGCTCCAACCCCACCTGACCAAGCCCGGGA CAGTCAAGGCTTCTCAATTTGTATTTTCCAGGCAAGAGAACTTTGTACCCCTTCTCTGTGTGGATAGACTCCCCAGCGTTTTTCCTCTGG AAATGCCCACAGGGCTTGCCGCGTGGAGACTGATCTGTTCCATCCGTTAGCGCGAGGTAGCAGTGTCGCCTCGCCCCTCCCACTGCGGGC TCACGGGGAGCTGGCGTCTGTCAGTGCCTTGTCACGCCTGGCATAGAGGTTGGGCTGGAGGCCTGTCCACTGGCTTCTGGAGCTGAAGGT CTGCGTCACGGTGAAAATTCGGGATGTTTACAGAGCATCACAAATGCCTCTCTCTCGCCGCTCTCTCATTTTCTTTGTATAACTATGCAG CCTTCCCTCTCTTTCTGGGATGTTTGGGACTTCACTCGACCTGCACGGGCTCTGCCCGACATGCCGTGGGAGCTGCTGGGATTCCTTTAG AAACCGCTGCCCGCATGCTTTGAAAACAGACCTTTCTCAGCTGGCTGTGGGGACCTGTCAGAGTCTGGGGACTCGGCGTGCAGGGCGGGC TCCAAGCGCTTTGCTTCCGGAACTCCGGCTTCCCAAGGGGTACTGTGCAGACTGACCACCGGCCTCCCGCCTGCAGGTCAGAGGCTCTGA CACTGTCTGGTTTCCAATGCTTCTGGAGACTTCCTGCCTAGGCCTCATCCTCCTCTTTGCCAGTCACCTGATTAACCAATTCTCCAGCAT TAGGACTTACCTCCTTCTTTTTGTAAGGTCTCTTGTATGTTGTTAAATGTTTGGCTTAAACAATTTATAAAAGCCTTTCTAGAAGGCAGA CTTAGCCCCAGCAGAGCCTGGGAGGAAGGCGGCCGCTGGGGCTCCTGGGCATCCTCTCTGGGGAGCTGCTGGCCGCTTAGCGTTGTTTGA TTTTTGACCTTGACATAGTAGATAGGCTTGTGGTTTTTTTAATTTAAAGATATTAAGACTTAATTCACTAAAATTTATTCTAAGGGGATA TTTATACTTTTATACTTTTTTAGGTATCGTATTTTATCAGCTTACAGTTTAATGCCTAAGTTTCCCCTGGAAATAGCAAATAAAATTGTG >31807_31807_3_FUS-KCTD5_FUS_chr16_31191548_ENST00000568685_KCTD5_chr16_2757299_ENST00000301738_length(amino acids)=114AA_BP= MSKARRPQVGGARRPGGRCLLGGPRSVFSKCHVGLRQTLPVSPLDPEDRVKEAFLVLHTVWKSSLGFTGRKRPQPRQCPARRGLWSDHQA -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for FUS-KCTD5 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for FUS-KCTD5 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for FUS-KCTD5 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
