
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:FZR1-CNN1 (FusionGDB2 ID:31969) |
Fusion Gene Summary for FZR1-CNN1 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: FZR1-CNN1 | Fusion gene ID: 31969 | Hgene | Tgene | Gene symbol | FZR1 | CNN1 | Gene ID | 51343 | 1264 |
| Gene name | fizzy and cell division cycle 20 related 1 | calponin 1 | |
| Synonyms | CDC20C|CDH1|FZR|FZR2|HCDH|HCDH1 | HEL-S-14|SMCC|Sm-Calp | |
| Cytomap | 19p13.3 | 19p13.2 | |
| Type of gene | protein-coding | protein-coding | |
| Description | fizzy-related protein homologCDC20 homolog 1CDC20-like 1bCDC20-like protein 1cdh1/Hct1 homologfizzy/cell division cycle 20 related 1 | calponin-1basic calponincalponin 1, basic, smooth musclecalponin H1, smooth musclecalponins, basicepididymis secretory protein Li 14 | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | Q9UM11 | P51911 | |
| Ensembl transtripts involved in fusion gene | ENST00000313639, ENST00000395095, ENST00000441788, | ENST00000544952, ENST00000588468, ENST00000535659, ENST00000592923, ENST00000252456, | |
| Fusion gene scores | * DoF score | 11 X 9 X 10=990 | 4 X 3 X 3=36 |
| # samples | 15 | 5 | |
| ** MAII score | log2(15/990*10)=-2.72246602447109 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(5/36*10)=0.473931188332412 effective Gene in Pan-Cancer Fusion Genes (eGinPCFGs). DoF>8 and MAII>0 | |
| Context | PubMed: FZR1 [Title/Abstract] AND CNN1 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | FZR1(3523056)-CNN1(11651891), # samples:3 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | FZR1 | GO:0031145 | anaphase-promoting complex-dependent catabolic process | 18662541|21596315 |
| Hgene | FZR1 | GO:0072425 | signal transduction involved in G2 DNA damage checkpoint | 18662541 |
| Hgene | FZR1 | GO:1904668 | positive regulation of ubiquitin protein ligase activity | 11459826 |
 Fusion gene breakpoints across FZR1 (5'-gene) Fusion gene breakpoints across FZR1 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
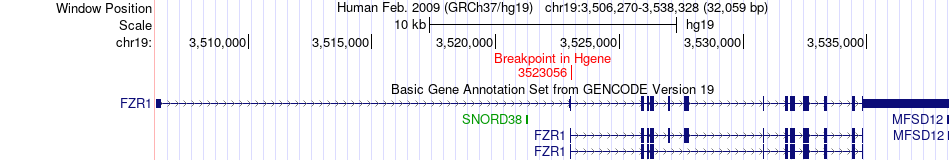 |
 Fusion gene breakpoints across CNN1 (3'-gene) Fusion gene breakpoints across CNN1 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
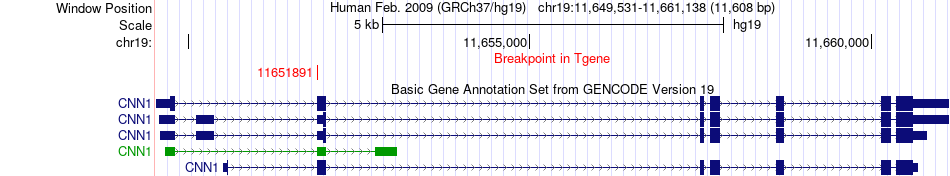 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | LUSC | TCGA-39-5019-01A | FZR1 | chr19 | 3523056 | - | CNN1 | chr19 | 11651891 | + |
| ChimerDB4 | LUSC | TCGA-39-5019-01A | FZR1 | chr19 | 3523056 | + | CNN1 | chr19 | 11651891 | + |
| ChimerDB4 | LUSC | TCGA-39-5019 | FZR1 | chr19 | 3523056 | + | CNN1 | chr19 | 11651891 | + |
Top |
Fusion Gene ORF analysis for FZR1-CNN1 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-3UTR | ENST00000313639 | ENST00000544952 | FZR1 | chr19 | 3523056 | + | CNN1 | chr19 | 11651891 | + |
| 5CDS-3UTR | ENST00000313639 | ENST00000588468 | FZR1 | chr19 | 3523056 | + | CNN1 | chr19 | 11651891 | + |
| 5CDS-3UTR | ENST00000395095 | ENST00000544952 | FZR1 | chr19 | 3523056 | + | CNN1 | chr19 | 11651891 | + |
| 5CDS-3UTR | ENST00000395095 | ENST00000588468 | FZR1 | chr19 | 3523056 | + | CNN1 | chr19 | 11651891 | + |
| 5CDS-3UTR | ENST00000441788 | ENST00000544952 | FZR1 | chr19 | 3523056 | + | CNN1 | chr19 | 11651891 | + |
| 5CDS-3UTR | ENST00000441788 | ENST00000588468 | FZR1 | chr19 | 3523056 | + | CNN1 | chr19 | 11651891 | + |
| 5CDS-5UTR | ENST00000313639 | ENST00000535659 | FZR1 | chr19 | 3523056 | + | CNN1 | chr19 | 11651891 | + |
| 5CDS-5UTR | ENST00000313639 | ENST00000592923 | FZR1 | chr19 | 3523056 | + | CNN1 | chr19 | 11651891 | + |
| 5CDS-5UTR | ENST00000395095 | ENST00000535659 | FZR1 | chr19 | 3523056 | + | CNN1 | chr19 | 11651891 | + |
| 5CDS-5UTR | ENST00000395095 | ENST00000592923 | FZR1 | chr19 | 3523056 | + | CNN1 | chr19 | 11651891 | + |
| 5CDS-5UTR | ENST00000441788 | ENST00000535659 | FZR1 | chr19 | 3523056 | + | CNN1 | chr19 | 11651891 | + |
| 5CDS-5UTR | ENST00000441788 | ENST00000592923 | FZR1 | chr19 | 3523056 | + | CNN1 | chr19 | 11651891 | + |
| In-frame | ENST00000313639 | ENST00000252456 | FZR1 | chr19 | 3523056 | + | CNN1 | chr19 | 11651891 | + |
| In-frame | ENST00000395095 | ENST00000252456 | FZR1 | chr19 | 3523056 | + | CNN1 | chr19 | 11651891 | + |
| In-frame | ENST00000441788 | ENST00000252456 | FZR1 | chr19 | 3523056 | + | CNN1 | chr19 | 11651891 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000441788 | FZR1 | chr19 | 3523056 | + | ENST00000252456 | CNN1 | chr19 | 11651891 | + | 1664 | 305 | 209 | 1135 | 308 |
| ENST00000313639 | FZR1 | chr19 | 3523056 | + | ENST00000252456 | CNN1 | chr19 | 11651891 | + | 1428 | 69 | 0 | 899 | 299 |
| ENST00000395095 | FZR1 | chr19 | 3523056 | + | ENST00000252456 | CNN1 | chr19 | 11651891 | + | 1428 | 69 | 0 | 899 | 299 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000441788 | ENST00000252456 | FZR1 | chr19 | 3523056 | + | CNN1 | chr19 | 11651891 | + | 0.006245925 | 0.9937541 |
| ENST00000313639 | ENST00000252456 | FZR1 | chr19 | 3523056 | + | CNN1 | chr19 | 11651891 | + | 0.005544054 | 0.994456 |
| ENST00000395095 | ENST00000252456 | FZR1 | chr19 | 3523056 | + | CNN1 | chr19 | 11651891 | + | 0.005544054 | 0.994456 |
Top |
Fusion Genomic Features for FZR1-CNN1 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| FZR1 | chr19 | 3523056 | + | CNN1 | chr19 | 11651890 | + | 1.84E-09 | 1 |
| FZR1 | chr19 | 3523056 | + | CNN1 | chr19 | 11651890 | + | 1.84E-09 | 1 |
| FZR1 | chr19 | 3523056 | + | CNN1 | chr19 | 11651890 | + | 1.84E-09 | 1 |
| FZR1 | chr19 | 3523056 | + | CNN1 | chr19 | 11651890 | + | 1.84E-09 | 1 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
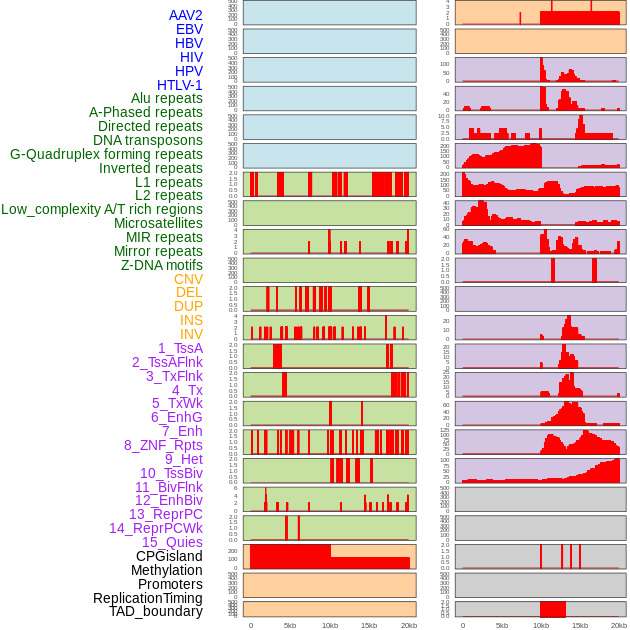 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
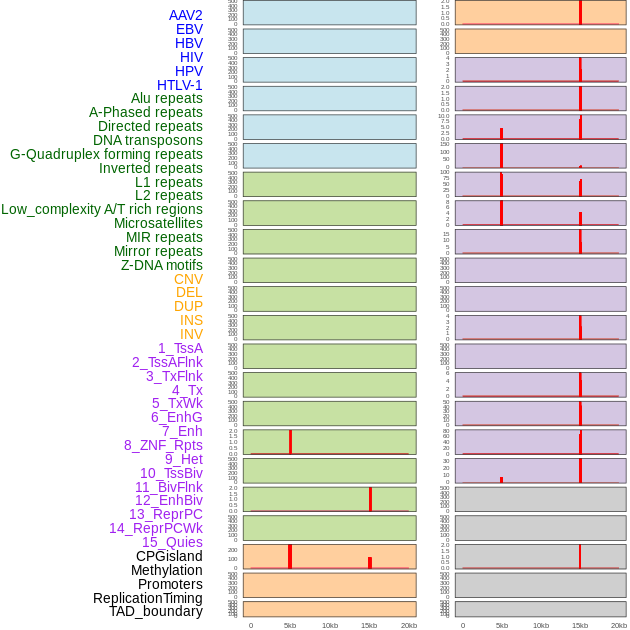 |
Top |
Fusion Protein Features for FZR1-CNN1 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr19:3523056/chr19:11651891) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| FZR1 | CNN1 |
| FUNCTION: Substrate-specific adapter for the anaphase promoting complex/cyclosome (APC/C) E3 ubiquitin-protein ligase complex. Associates with the APC/C in late mitosis, in replacement of CDC20, and activates the APC/C during anaphase and telophase. The APC/C remains active in degrading substrates to ensure that positive regulators of the cell cycle do not accumulate prematurely. At the G1/S transition FZR1 is phosphorylated, leading to its dissociation from the APC/C. Following DNA damage, it is required for the G2 DNA damage checkpoint: its dephosphorylation and reassociation with the APC/C leads to the ubiquitination of PLK1, preventing entry into mitosis. Acts as an adapter for APC/C to target the DNA-end resection factor RBBP8/CtIP for ubiquitination and subsequent proteasomal degradation. Through the regulation of RBBP8/CtIP protein turnover, may play a role in DNA damage response, favoring DNA double-strand repair through error-prone non-homologous end joining (NHEJ) over error-free, RBBP8-mediated homologous recombination (HR) (PubMed:25349192). {ECO:0000269|PubMed:18662541, ECO:0000269|PubMed:21596315, ECO:0000269|PubMed:25349192, ECO:0000269|PubMed:9734353}. | FUNCTION: Thin filament-associated protein that is implicated in the regulation and modulation of smooth muscle contraction. It is capable of binding to actin, calmodulin and tropomyosin. The interaction of calponin with actin inhibits the actomyosin Mg-ATPase activity (By similarity). {ECO:0000250}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | CNN1 | chr19:3523056 | chr19:11651891 | ENST00000252456 | 0 | 7 | 28_131 | 21 | 298.0 | Domain | Calponin-homology (CH) | |
| Tgene | CNN1 | chr19:3523056 | chr19:11651891 | ENST00000252456 | 0 | 7 | 164_189 | 21 | 298.0 | Repeat | Note=Calponin-like 1 | |
| Tgene | CNN1 | chr19:3523056 | chr19:11651891 | ENST00000252456 | 0 | 7 | 204_229 | 21 | 298.0 | Repeat | Note=Calponin-like 2 | |
| Tgene | CNN1 | chr19:3523056 | chr19:11651891 | ENST00000252456 | 0 | 7 | 243_268 | 21 | 298.0 | Repeat | Note=Calponin-like 3 |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | FZR1 | chr19:3523056 | chr19:11651891 | ENST00000313639 | + | 1 | 11 | 47_52 | 23 | 405.0 | Region | Involved in APC/FZR1 E3 ubiquitin-protein ligase complex activity |
| Hgene | FZR1 | chr19:3523056 | chr19:11651891 | ENST00000395095 | + | 1 | 13 | 47_52 | 23 | 497.0 | Region | Involved in APC/FZR1 E3 ubiquitin-protein ligase complex activity |
| Hgene | FZR1 | chr19:3523056 | chr19:11651891 | ENST00000441788 | + | 2 | 14 | 47_52 | 23 | 494.0 | Region | Involved in APC/FZR1 E3 ubiquitin-protein ligase complex activity |
| Hgene | FZR1 | chr19:3523056 | chr19:11651891 | ENST00000313639 | + | 1 | 11 | 182_222 | 23 | 405.0 | Repeat | Note=WD 1 |
| Hgene | FZR1 | chr19:3523056 | chr19:11651891 | ENST00000313639 | + | 1 | 11 | 227_266 | 23 | 405.0 | Repeat | Note=WD 2 |
| Hgene | FZR1 | chr19:3523056 | chr19:11651891 | ENST00000313639 | + | 1 | 11 | 269_306 | 23 | 405.0 | Repeat | Note=WD 3 |
| Hgene | FZR1 | chr19:3523056 | chr19:11651891 | ENST00000313639 | + | 1 | 11 | 311_350 | 23 | 405.0 | Repeat | Note=WD 4 |
| Hgene | FZR1 | chr19:3523056 | chr19:11651891 | ENST00000313639 | + | 1 | 11 | 353_395 | 23 | 405.0 | Repeat | Note=WD 5 |
| Hgene | FZR1 | chr19:3523056 | chr19:11651891 | ENST00000313639 | + | 1 | 11 | 397_438 | 23 | 405.0 | Repeat | Note=WD 6 |
| Hgene | FZR1 | chr19:3523056 | chr19:11651891 | ENST00000313639 | + | 1 | 11 | 441_480 | 23 | 405.0 | Repeat | Note=WD 7 |
| Hgene | FZR1 | chr19:3523056 | chr19:11651891 | ENST00000395095 | + | 1 | 13 | 182_222 | 23 | 497.0 | Repeat | Note=WD 1 |
| Hgene | FZR1 | chr19:3523056 | chr19:11651891 | ENST00000395095 | + | 1 | 13 | 227_266 | 23 | 497.0 | Repeat | Note=WD 2 |
| Hgene | FZR1 | chr19:3523056 | chr19:11651891 | ENST00000395095 | + | 1 | 13 | 269_306 | 23 | 497.0 | Repeat | Note=WD 3 |
| Hgene | FZR1 | chr19:3523056 | chr19:11651891 | ENST00000395095 | + | 1 | 13 | 311_350 | 23 | 497.0 | Repeat | Note=WD 4 |
| Hgene | FZR1 | chr19:3523056 | chr19:11651891 | ENST00000395095 | + | 1 | 13 | 353_395 | 23 | 497.0 | Repeat | Note=WD 5 |
| Hgene | FZR1 | chr19:3523056 | chr19:11651891 | ENST00000395095 | + | 1 | 13 | 397_438 | 23 | 497.0 | Repeat | Note=WD 6 |
| Hgene | FZR1 | chr19:3523056 | chr19:11651891 | ENST00000395095 | + | 1 | 13 | 441_480 | 23 | 497.0 | Repeat | Note=WD 7 |
| Hgene | FZR1 | chr19:3523056 | chr19:11651891 | ENST00000441788 | + | 2 | 14 | 182_222 | 23 | 494.0 | Repeat | Note=WD 1 |
| Hgene | FZR1 | chr19:3523056 | chr19:11651891 | ENST00000441788 | + | 2 | 14 | 227_266 | 23 | 494.0 | Repeat | Note=WD 2 |
| Hgene | FZR1 | chr19:3523056 | chr19:11651891 | ENST00000441788 | + | 2 | 14 | 269_306 | 23 | 494.0 | Repeat | Note=WD 3 |
| Hgene | FZR1 | chr19:3523056 | chr19:11651891 | ENST00000441788 | + | 2 | 14 | 311_350 | 23 | 494.0 | Repeat | Note=WD 4 |
| Hgene | FZR1 | chr19:3523056 | chr19:11651891 | ENST00000441788 | + | 2 | 14 | 353_395 | 23 | 494.0 | Repeat | Note=WD 5 |
| Hgene | FZR1 | chr19:3523056 | chr19:11651891 | ENST00000441788 | + | 2 | 14 | 397_438 | 23 | 494.0 | Repeat | Note=WD 6 |
| Hgene | FZR1 | chr19:3523056 | chr19:11651891 | ENST00000441788 | + | 2 | 14 | 441_480 | 23 | 494.0 | Repeat | Note=WD 7 |
Top |
Fusion Gene Sequence for FZR1-CNN1 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >31969_31969_1_FZR1-CNN1_FZR1_chr19_3523056_ENST00000313639_CNN1_chr19_11651891_ENST00000252456_length(transcript)=1428nt_BP=69nt ATGGACCAGGACTATGAGCGGCGCCTGCTTCGCCAGATCGTCATCCAGAATGAGAACACGATGCCACGCCTGGCCCAGAAGTATGACCAC CAGCGGGAGCAGGAGCTGAGAGAGTGGATCGAGGGGGTGACAGGCCGTCGCATCGGCAACAACTTCATGGACGGCCTCAAAGATGGCATC ATTCTTTGCGAATTCATCAATAAGCTGCAGCCAGGCTCCGTGAAGAAGATCAATGAGTCAACCCAAAATTGGCACCAGCTGGAGAACATC GGCAACTTCATCAAGGCCATCACCAAGTATGGGGTGAAGCCCCACGACATTTTTGAGGCCAACGACCTGTTTGAGAACACCAACCATACA CAGGTGCAGTCCACCCTCCTGGCTTTGGCCAGCATGGCGAAGACGAAAGGAAACAAGGTGAACGTGGGAGTGAAGTACGCAGAGAAGCAG GAGCGGAAATTCGAGCCGGGGAAGCTAAGAGAAGGGCGGAACATCATTGGGCTGCAGATGGGCACCAACAAGTTTGCCAGCCAGCAGGGC ATGACGGCCTATGGCACCCGGCGCCACCTCTACGACCCCAAGCTGGGCACAGACCAGCCTCTGGACCAGGCGACCATCAGCCTGCAGATG GGCACCAACAAAGGAGCCAGCCAGGCTGGCATGACTGCGCCAGGGACCAAGCGGCAGATCTTCGAGCCGGGGCTGGGCATGGAGCACTGC GACACGCTCAATGTCAGCCTGCAGATGGGCAGCAACAAGGGCGCCTCGCAGCGGGGCATGACGGTGTATGGGCTGCCACGCCAGGTCTAC GACCCCAAGTACTGTCTGACTCCCGAGTACCCAGAGCTGGGTGAGCCCGCCCACAACCACCACGCACACAACTACTACAATTCCGCCTAG GGCCACAAGGCCTTCCCTGTTTTCCCCCCAAGGGAGGCTGCTGCTGCTCTTGGCTGGACCCAGCCAGGCCCAGCCGACCCCCTCTCCCTG CATGGCATCCTCCAGCCCCTGTAGAACTCAACCTCTACAGGGTTAGAGTTTGGAGAGAGCAGACTGGCGGGGGGCCCATTGGGGGGAAGG GGACCCTCCGCTCTGTAGTGCTACAGGGTCCAACATAGAGCCGGGTGTCCCCAACAGCGCCCAAAGGACGCACTGAGCAACGCTATTCCA GCTGTCCCCCCACTCCCTCACAAGTGGGTACCCCCAGGACCAGAAGCTCCCCCAGCAAAGCCCCCAGAGCCCAGGCTCGGCCTGCCCCCA CCCCATTCCCGCAGTGGGAGCAAACTGCATGCCCAGAGACCCAGCGGACACACGCGGTTTGGTTTGCAGCGACTGGCATACTATGTGGAT >31969_31969_1_FZR1-CNN1_FZR1_chr19_3523056_ENST00000313639_CNN1_chr19_11651891_ENST00000252456_length(amino acids)=299AA_BP=21 MDQDYERRLLRQIVIQNENTMPRLAQKYDHQREQELREWIEGVTGRRIGNNFMDGLKDGIILCEFINKLQPGSVKKINESTQNWHQLENI GNFIKAITKYGVKPHDIFEANDLFENTNHTQVQSTLLALASMAKTKGNKVNVGVKYAEKQERKFEPGKLREGRNIIGLQMGTNKFASQQG MTAYGTRRHLYDPKLGTDQPLDQATISLQMGTNKGASQAGMTAPGTKRQIFEPGLGMEHCDTLNVSLQMGSNKGASQRGMTVYGLPRQVY -------------------------------------------------------------- >31969_31969_2_FZR1-CNN1_FZR1_chr19_3523056_ENST00000395095_CNN1_chr19_11651891_ENST00000252456_length(transcript)=1428nt_BP=69nt ATGGACCAGGACTATGAGCGGCGCCTGCTTCGCCAGATCGTCATCCAGAATGAGAACACGATGCCACGCCTGGCCCAGAAGTATGACCAC CAGCGGGAGCAGGAGCTGAGAGAGTGGATCGAGGGGGTGACAGGCCGTCGCATCGGCAACAACTTCATGGACGGCCTCAAAGATGGCATC ATTCTTTGCGAATTCATCAATAAGCTGCAGCCAGGCTCCGTGAAGAAGATCAATGAGTCAACCCAAAATTGGCACCAGCTGGAGAACATC GGCAACTTCATCAAGGCCATCACCAAGTATGGGGTGAAGCCCCACGACATTTTTGAGGCCAACGACCTGTTTGAGAACACCAACCATACA CAGGTGCAGTCCACCCTCCTGGCTTTGGCCAGCATGGCGAAGACGAAAGGAAACAAGGTGAACGTGGGAGTGAAGTACGCAGAGAAGCAG GAGCGGAAATTCGAGCCGGGGAAGCTAAGAGAAGGGCGGAACATCATTGGGCTGCAGATGGGCACCAACAAGTTTGCCAGCCAGCAGGGC ATGACGGCCTATGGCACCCGGCGCCACCTCTACGACCCCAAGCTGGGCACAGACCAGCCTCTGGACCAGGCGACCATCAGCCTGCAGATG GGCACCAACAAAGGAGCCAGCCAGGCTGGCATGACTGCGCCAGGGACCAAGCGGCAGATCTTCGAGCCGGGGCTGGGCATGGAGCACTGC GACACGCTCAATGTCAGCCTGCAGATGGGCAGCAACAAGGGCGCCTCGCAGCGGGGCATGACGGTGTATGGGCTGCCACGCCAGGTCTAC GACCCCAAGTACTGTCTGACTCCCGAGTACCCAGAGCTGGGTGAGCCCGCCCACAACCACCACGCACACAACTACTACAATTCCGCCTAG GGCCACAAGGCCTTCCCTGTTTTCCCCCCAAGGGAGGCTGCTGCTGCTCTTGGCTGGACCCAGCCAGGCCCAGCCGACCCCCTCTCCCTG CATGGCATCCTCCAGCCCCTGTAGAACTCAACCTCTACAGGGTTAGAGTTTGGAGAGAGCAGACTGGCGGGGGGCCCATTGGGGGGAAGG GGACCCTCCGCTCTGTAGTGCTACAGGGTCCAACATAGAGCCGGGTGTCCCCAACAGCGCCCAAAGGACGCACTGAGCAACGCTATTCCA GCTGTCCCCCCACTCCCTCACAAGTGGGTACCCCCAGGACCAGAAGCTCCCCCAGCAAAGCCCCCAGAGCCCAGGCTCGGCCTGCCCCCA CCCCATTCCCGCAGTGGGAGCAAACTGCATGCCCAGAGACCCAGCGGACACACGCGGTTTGGTTTGCAGCGACTGGCATACTATGTGGAT >31969_31969_2_FZR1-CNN1_FZR1_chr19_3523056_ENST00000395095_CNN1_chr19_11651891_ENST00000252456_length(amino acids)=299AA_BP=21 MDQDYERRLLRQIVIQNENTMPRLAQKYDHQREQELREWIEGVTGRRIGNNFMDGLKDGIILCEFINKLQPGSVKKINESTQNWHQLENI GNFIKAITKYGVKPHDIFEANDLFENTNHTQVQSTLLALASMAKTKGNKVNVGVKYAEKQERKFEPGKLREGRNIIGLQMGTNKFASQQG MTAYGTRRHLYDPKLGTDQPLDQATISLQMGTNKGASQAGMTAPGTKRQIFEPGLGMEHCDTLNVSLQMGSNKGASQRGMTVYGLPRQVY -------------------------------------------------------------- >31969_31969_3_FZR1-CNN1_FZR1_chr19_3523056_ENST00000441788_CNN1_chr19_11651891_ENST00000252456_length(transcript)=1664nt_BP=305nt GGCCGGGCGCGCGCGGCGGAGGGAGGGAGCCGGGAGGCAGCGGAGGAGCGACCGCCGCCATATTAGGGACCGCGGAGCCCGGGATCCCGT CAGCGGCGGCCGCGGCCGGGCCTGCGGGAGCTGCGGAGGCCGGAGGCGGGCGCTGTGCGGTGCCAGGAGAGGCGGGGTCGGCGGGAGCCA GCGAGCCACGGGAGCGAGCCAGGCTAACCTTGCCGCGGGCCGAGCCCTGCCTCGCCATGGACCAGGACTATGAGCGGCGCCTGCTTCGCC AGATCGTCATCCAGAATGAGAACACGATGCCACGCCTGGCCCAGAAGTATGACCACCAGCGGGAGCAGGAGCTGAGAGAGTGGATCGAGG GGGTGACAGGCCGTCGCATCGGCAACAACTTCATGGACGGCCTCAAAGATGGCATCATTCTTTGCGAATTCATCAATAAGCTGCAGCCAG GCTCCGTGAAGAAGATCAATGAGTCAACCCAAAATTGGCACCAGCTGGAGAACATCGGCAACTTCATCAAGGCCATCACCAAGTATGGGG TGAAGCCCCACGACATTTTTGAGGCCAACGACCTGTTTGAGAACACCAACCATACACAGGTGCAGTCCACCCTCCTGGCTTTGGCCAGCA TGGCGAAGACGAAAGGAAACAAGGTGAACGTGGGAGTGAAGTACGCAGAGAAGCAGGAGCGGAAATTCGAGCCGGGGAAGCTAAGAGAAG GGCGGAACATCATTGGGCTGCAGATGGGCACCAACAAGTTTGCCAGCCAGCAGGGCATGACGGCCTATGGCACCCGGCGCCACCTCTACG ACCCCAAGCTGGGCACAGACCAGCCTCTGGACCAGGCGACCATCAGCCTGCAGATGGGCACCAACAAAGGAGCCAGCCAGGCTGGCATGA CTGCGCCAGGGACCAAGCGGCAGATCTTCGAGCCGGGGCTGGGCATGGAGCACTGCGACACGCTCAATGTCAGCCTGCAGATGGGCAGCA ACAAGGGCGCCTCGCAGCGGGGCATGACGGTGTATGGGCTGCCACGCCAGGTCTACGACCCCAAGTACTGTCTGACTCCCGAGTACCCAG AGCTGGGTGAGCCCGCCCACAACCACCACGCACACAACTACTACAATTCCGCCTAGGGCCACAAGGCCTTCCCTGTTTTCCCCCCAAGGG AGGCTGCTGCTGCTCTTGGCTGGACCCAGCCAGGCCCAGCCGACCCCCTCTCCCTGCATGGCATCCTCCAGCCCCTGTAGAACTCAACCT CTACAGGGTTAGAGTTTGGAGAGAGCAGACTGGCGGGGGGCCCATTGGGGGGAAGGGGACCCTCCGCTCTGTAGTGCTACAGGGTCCAAC ATAGAGCCGGGTGTCCCCAACAGCGCCCAAAGGACGCACTGAGCAACGCTATTCCAGCTGTCCCCCCACTCCCTCACAAGTGGGTACCCC CAGGACCAGAAGCTCCCCCAGCAAAGCCCCCAGAGCCCAGGCTCGGCCTGCCCCCACCCCATTCCCGCAGTGGGAGCAAACTGCATGCCC AGAGACCCAGCGGACACACGCGGTTTGGTTTGCAGCGACTGGCATACTATGTGGATGTGACAGTGGCGTTTGTAATGAGAGCACTTTCTT >31969_31969_3_FZR1-CNN1_FZR1_chr19_3523056_ENST00000441788_CNN1_chr19_11651891_ENST00000252456_length(amino acids)=308AA_BP=30 MPRAEPCLAMDQDYERRLLRQIVIQNENTMPRLAQKYDHQREQELREWIEGVTGRRIGNNFMDGLKDGIILCEFINKLQPGSVKKINEST QNWHQLENIGNFIKAITKYGVKPHDIFEANDLFENTNHTQVQSTLLALASMAKTKGNKVNVGVKYAEKQERKFEPGKLREGRNIIGLQMG TNKFASQQGMTAYGTRRHLYDPKLGTDQPLDQATISLQMGTNKGASQAGMTAPGTKRQIFEPGLGMEHCDTLNVSLQMGSNKGASQRGMT -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for FZR1-CNN1 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for FZR1-CNN1 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for FZR1-CNN1 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
