
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:AJUBA-CEP192 (FusionGDB2 ID:3231) |
Fusion Gene Summary for AJUBA-CEP192 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: AJUBA-CEP192 | Fusion gene ID: 3231 | Hgene | Tgene | Gene symbol | AJUBA | CEP192 | Gene ID | 84962 | 55125 |
| Gene name | ajuba LIM protein | centrosomal protein 192 | |
| Synonyms | JUB | PPP1R62 | |
| Cytomap | 14q11.2 | 18p11.21 | |
| Type of gene | protein-coding | protein-coding | |
| Description | LIM domain-containing protein ajubajub, ajuba homolog | centrosomal protein of 192 kDa192 kDa centrosomal proteincentrosomal protein 192kDacep192/SPD-2protein phosphatase 1, regulatory subunit 62 | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | Q96IF1 | Q8TEP8 | |
| Ensembl transtripts involved in fusion gene | ENST00000262713, ENST00000361265, ENST00000397388, | ENST00000540847, ENST00000325971, ENST00000430049, ENST00000506447, | |
| Fusion gene scores | * DoF score | 2 X 2 X 2=8 | 4 X 4 X 4=64 |
| # samples | 2 | 4 | |
| ** MAII score | log2(2/8*10)=1.32192809488736 | log2(4/64*10)=-0.678071905112638 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: AJUBA [Title/Abstract] AND CEP192 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | AJUBA(23445854)-CEP192(13087016), # samples:2 | ||
| Anticipated loss of major functional domain due to fusion event. | AJUBA-CEP192 seems lost the major protein functional domain in Tgene partner, which is a essential gene due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | AJUBA | GO:0001666 | response to hypoxia | 22286099 |
| Hgene | AJUBA | GO:0031334 | positive regulation of protein complex assembly | 15870274 |
| Hgene | AJUBA | GO:0035331 | negative regulation of hippo signaling | 20303269 |
| Hgene | AJUBA | GO:1900037 | regulation of cellular response to hypoxia | 22286099 |
| Tgene | CEP192 | GO:0010923 | negative regulation of phosphatase activity | 19389623 |
 Fusion gene breakpoints across AJUBA (5'-gene) Fusion gene breakpoints across AJUBA (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
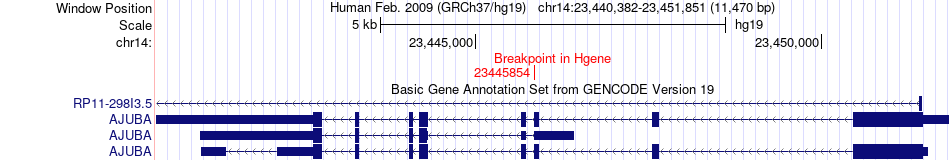 |
 Fusion gene breakpoints across CEP192 (3'-gene) Fusion gene breakpoints across CEP192 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
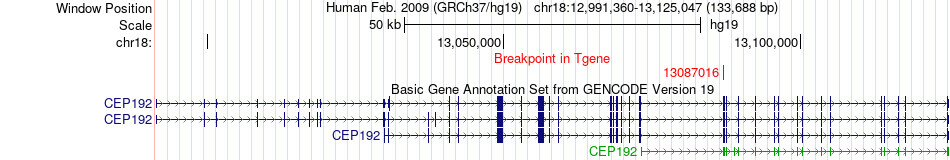 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | CHOL | TCGA-W6-AA0S-01A | AJUBA | chr14 | 23445854 | - | CEP192 | chr18 | 13087016 | + |
Top |
Fusion Gene ORF analysis for AJUBA-CEP192 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-3UTR | ENST00000262713 | ENST00000540847 | AJUBA | chr14 | 23445854 | - | CEP192 | chr18 | 13087016 | + |
| 5CDS-3UTR | ENST00000361265 | ENST00000540847 | AJUBA | chr14 | 23445854 | - | CEP192 | chr18 | 13087016 | + |
| 5UTR-3CDS | ENST00000397388 | ENST00000325971 | AJUBA | chr14 | 23445854 | - | CEP192 | chr18 | 13087016 | + |
| 5UTR-3CDS | ENST00000397388 | ENST00000430049 | AJUBA | chr14 | 23445854 | - | CEP192 | chr18 | 13087016 | + |
| 5UTR-3CDS | ENST00000397388 | ENST00000506447 | AJUBA | chr14 | 23445854 | - | CEP192 | chr18 | 13087016 | + |
| 5UTR-3UTR | ENST00000397388 | ENST00000540847 | AJUBA | chr14 | 23445854 | - | CEP192 | chr18 | 13087016 | + |
| Frame-shift | ENST00000361265 | ENST00000325971 | AJUBA | chr14 | 23445854 | - | CEP192 | chr18 | 13087016 | + |
| Frame-shift | ENST00000361265 | ENST00000430049 | AJUBA | chr14 | 23445854 | - | CEP192 | chr18 | 13087016 | + |
| Frame-shift | ENST00000361265 | ENST00000506447 | AJUBA | chr14 | 23445854 | - | CEP192 | chr18 | 13087016 | + |
| In-frame | ENST00000262713 | ENST00000325971 | AJUBA | chr14 | 23445854 | - | CEP192 | chr18 | 13087016 | + |
| In-frame | ENST00000262713 | ENST00000430049 | AJUBA | chr14 | 23445854 | - | CEP192 | chr18 | 13087016 | + |
| In-frame | ENST00000262713 | ENST00000506447 | AJUBA | chr14 | 23445854 | - | CEP192 | chr18 | 13087016 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000262713 | AJUBA | chr14 | 23445854 | - | ENST00000506447 | CEP192 | chr18 | 13087016 | + | 3816 | 1552 | 277 | 3549 | 1090 |
| ENST00000262713 | AJUBA | chr14 | 23445854 | - | ENST00000325971 | CEP192 | chr18 | 13087016 | + | 3828 | 1552 | 277 | 3549 | 1090 |
| ENST00000262713 | AJUBA | chr14 | 23445854 | - | ENST00000430049 | CEP192 | chr18 | 13087016 | + | 3775 | 1552 | 277 | 3549 | 1090 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000262713 | ENST00000506447 | AJUBA | chr14 | 23445854 | - | CEP192 | chr18 | 13087016 | + | 0.000392333 | 0.99960774 |
| ENST00000262713 | ENST00000325971 | AJUBA | chr14 | 23445854 | - | CEP192 | chr18 | 13087016 | + | 0.000384192 | 0.9996158 |
| ENST00000262713 | ENST00000430049 | AJUBA | chr14 | 23445854 | - | CEP192 | chr18 | 13087016 | + | 0.000429507 | 0.9995704 |
Top |
Fusion Genomic Features for AJUBA-CEP192 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| AJUBA | chr14 | 23445853 | - | CEP192 | chr18 | 13087015 | + | 0.002187998 | 0.99781203 |
| AJUBA | chr14 | 23445853 | - | CEP192 | chr18 | 13087015 | + | 0.002187998 | 0.99781203 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
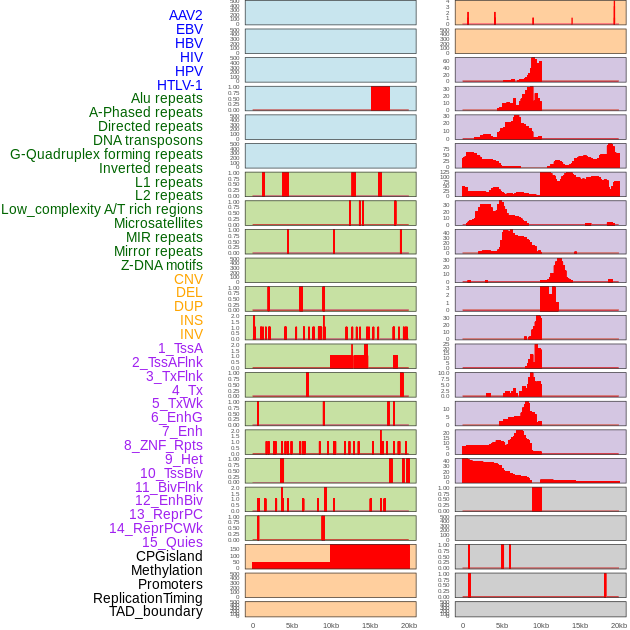 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
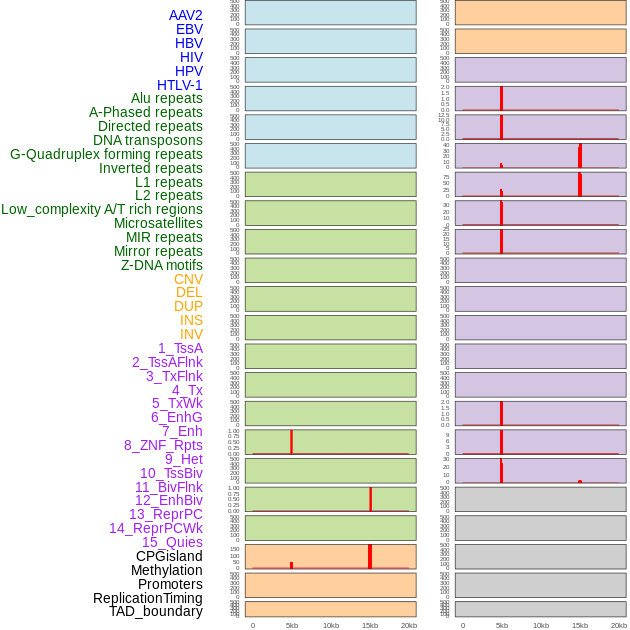 |
Top |
Fusion Protein Features for AJUBA-CEP192 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr14:23445854/chr18:13087016) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| AJUBA | CEP192 |
| FUNCTION: Adapter or scaffold protein which participates in the assembly of numerous protein complexes and is involved in several cellular processes such as cell fate determination, cytoskeletal organization, repression of gene transcription, mitosis, cell-cell adhesion, cell differentiation, proliferation and migration. Contributes to the linking and/or strengthening of epithelia cell-cell junctions in part by linking adhesive receptors to the actin cytoskeleton. May be involved in signal transduction from cell adhesion sites to the nucleus. Plays an important role in regulation of the kinase activity of AURKA for mitotic commitment. Also a component of the IL-1 signaling pathway modulating IL-1-induced NFKB1 activation by influencing the assembly and activity of the PRKCZ-SQSTM1-TRAF6 multiprotein signaling complex. Functions as an HDAC-dependent corepressor for a subset of GFI1 target genes. Acts as a transcriptional corepressor for SNAI1 and SNAI2/SLUG-dependent repression of E-cadherin transcription. Acts as a hypoxic regulator by bridging an association between the prolyl hydroxylases and VHL enabling efficient degradation of HIF1A. Positively regulates microRNA (miRNA)-mediated gene silencing. Negatively regulates the Hippo signaling pathway and antagonizes phosphorylation of YAP1. {ECO:0000269|PubMed:12417594, ECO:0000269|PubMed:13678582, ECO:0000269|PubMed:15870274, ECO:0000269|PubMed:16413547, ECO:0000269|PubMed:17909014, ECO:0000269|PubMed:18805794, ECO:0000269|PubMed:20303269, ECO:0000269|PubMed:20616046, ECO:0000269|PubMed:22286099}. | FUNCTION: Required for mitotic centrosome maturation and bipolar spindle assembly (PubMed:25042804, PubMed:17980596, PubMed:18207742). Appears to be a major regulator of pericentriolar material (PCM) recruitment, centrosome maturation, and centriole duplication (PubMed:25042804, PubMed:17980596, PubMed:18207742). Centrosome-specific activating scaffold for AURKA and PLK1 (PubMed:25042804). {ECO:0000269|PubMed:17980596, ECO:0000269|PubMed:18207742, ECO:0000269|PubMed:25042804}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | AJUBA | chr14:23445854 | chr18:13087016 | ENST00000262713 | - | 3 | 8 | 280_288 | 392 | 539.0 | Motif | Nuclear localization signal |
| Hgene | AJUBA | chr14:23445854 | chr18:13087016 | ENST00000361265 | - | 3 | 9 | 280_288 | 392 | 481.0 | Motif | Nuclear localization signal |
| Hgene | AJUBA | chr14:23445854 | chr18:13087016 | ENST00000262713 | - | 3 | 8 | 1_335 | 392 | 539.0 | Region | Note=PreLIM |
| Hgene | AJUBA | chr14:23445854 | chr18:13087016 | ENST00000361265 | - | 3 | 9 | 1_335 | 392 | 481.0 | Region | Note=PreLIM |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | AJUBA | chr14:23445854 | chr18:13087016 | ENST00000262713 | - | 3 | 8 | 336_397 | 392 | 539.0 | Domain | LIM zinc-binding 1 |
| Hgene | AJUBA | chr14:23445854 | chr18:13087016 | ENST00000262713 | - | 3 | 8 | 401_461 | 392 | 539.0 | Domain | LIM zinc-binding 2 |
| Hgene | AJUBA | chr14:23445854 | chr18:13087016 | ENST00000262713 | - | 3 | 8 | 462_530 | 392 | 539.0 | Domain | LIM zinc-binding 3 |
| Hgene | AJUBA | chr14:23445854 | chr18:13087016 | ENST00000361265 | - | 3 | 9 | 336_397 | 392 | 481.0 | Domain | LIM zinc-binding 1 |
| Hgene | AJUBA | chr14:23445854 | chr18:13087016 | ENST00000361265 | - | 3 | 9 | 401_461 | 392 | 481.0 | Domain | LIM zinc-binding 2 |
| Hgene | AJUBA | chr14:23445854 | chr18:13087016 | ENST00000361265 | - | 3 | 9 | 462_530 | 392 | 481.0 | Domain | LIM zinc-binding 3 |
| Hgene | AJUBA | chr14:23445854 | chr18:13087016 | ENST00000397388 | - | 1 | 6 | 336_397 | 0 | 122.0 | Domain | LIM zinc-binding 1 |
| Hgene | AJUBA | chr14:23445854 | chr18:13087016 | ENST00000397388 | - | 1 | 6 | 401_461 | 0 | 122.0 | Domain | LIM zinc-binding 2 |
| Hgene | AJUBA | chr14:23445854 | chr18:13087016 | ENST00000397388 | - | 1 | 6 | 462_530 | 0 | 122.0 | Domain | LIM zinc-binding 3 |
| Hgene | AJUBA | chr14:23445854 | chr18:13087016 | ENST00000397388 | - | 1 | 6 | 280_288 | 0 | 122.0 | Motif | Nuclear localization signal |
| Hgene | AJUBA | chr14:23445854 | chr18:13087016 | ENST00000397388 | - | 1 | 6 | 1_335 | 0 | 122.0 | Region | Note=PreLIM |
Top |
Fusion Gene Sequence for AJUBA-CEP192 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >3231_3231_1_AJUBA-CEP192_AJUBA_chr14_23445854_ENST00000262713_CEP192_chr18_13087016_ENST00000325971_length(transcript)=3828nt_BP=1552nt AGATCCGGCCAGGCAGACAAAAGTTTGGGAGAGAAGTTCGGTCTGTGGCTAGCGTGAGAGCTAGGGGGGCGGGGGCCGGGGAGAGTGGGG CAGTCGGGCGAGTCGACGCGTGAACAGATAGACCTGCGGACTGGACAGCCGCGGCCAGAGACCCTGCTAGCCCCGCTCAGCCCCAGATGC GCGGGGGGACGCAGCCCCTCCCGCTGGGGGATGCTGTGGGATTCCTGGCGCAGGGCATCCAGGCCGCCCGCTAAGCCCCTGTGCCTCCCC TGTGCCCCTGGGGAACCAGAGTCCGGCTGCAGGGAAAGAGAACCGGCCGCCGAGACGCCGCAGGGTGCCAGGCGGGGAGGGGGCGAGAGG CCCCAGGCCCGAGGGCATGGAGCGGTTAGGAGAGAAAGCCAGTCGCCTGCTGGAGAAGTTCGGCCGCAGAAAGGGTGAATCTAGCCGGTC TGGGTCTGACGGGACCCCCGGGCCGGGCAAGGGGCGCCTAAGTGGGTTGGGGGGACCTAGGAAGTCAGGGCCCCGAGGAGCTACTGGGGG ACCTGGGGATGAGCCGTTGGAGCCGGCCCGGGAGCAAGGTTCCCTGGACGCTGAGCGAAATCAGCGCGGCTCCTTTGAGGCGCCGCGCTA CGAAGGCTCTTTTCCCGCGGGGCCGCCGCCCACCCGGGCCTTGCCTCTACCTCAGTCGTTGCCCCCCGATTTTCGGCTGGAGCCCACGGC CCCGGCCCTCAGCCCCCGCTCTAGCTTCGCCAGTAGCTCGGCCAGCGACGCGAGCAAGCCGTCCAGCCCCCGGGGCAGCCTGCTGCTGGA CGGGGCGGGGGCTGGCGGAGCTGGAGGTAGCCGGCCCTGCAGCAATCGCACCAGCGGCATCAGCATGGGCTACGACCAGCGCCACGGGAG CCCCTTGCCAGCGGGGCCGTGCCTGTTTGGCCCACCCCTGGCCGGAGCACCGGCAGGCTATTCTCCCGGAGGGGTCCCGTCCGCCTACCC GGAGCTCCACGCCGCCCTGGACCGATTGTACGCTCAGCGGCCCGCGGGGTTCGGCTGCCAGGAAAGCCGCCACTCGTATCCCCCGGCCCT GGGCAGCCCTGGAGCTCTAGCCGGGGCCGGAGTGGGAGCGGCGGGGCCCTTGGAGAGACGGGGGGCGCAACCCGGACGACACTCTGTGAC CGGCTACGGGGACTGCGCCGTGGGCGCCCGGTACCAGGACGAGCTAACAGCTTTGCTTCGCCTGACGGTGGGCACCGGTGGGCGAGAAGC CGGAGCCCGCGGAGAACCCTCGGGGATTGAGCCGTCGGGTCTGGAGGAGCCACCAGGTCCTTTCGTTCCGGAGGCCGCCCGGGCCCGGAT GCGGGAGCCAGAGGCCAGGGAGGACTACTTCGGCACCTGTATCAAGTGCAACAAAGGCATCTATGGGCAGAGCAATGCCTGCCAGGCCCT GGACAGCCTCTACCACACCCAGTGCTTTGTTTGCTGCTCTTGTGGGCGAACTTTGCGTTGCAAGGCTTTCTACAGTGTCAATGGCTCTGT GTACTGTGAGGAAGATTATCTGATACCTTTGTCTGGATATGGAGGAACAAGCAATCTTATTTTGGAAGGCGTTAAAAAATTATCTGACAG TTACATGGTAACAGTGAATGGCTTAGTACCTGGCAAAGAAAGTAAAATTGTTTTTTCTGTCCGCAACACTGGCTCCCGAGCAGCTTTTGT TAAAGCAGTAGGTTTTAAGGATTCTCAGAAAAAAGTTTTGCTGGATCCTAAAGTATTGAGGATTTTTCCAGATAAATTTGTACTCAAGGA AAGAACACAAGAAAATGTTACTTTAATATATAATCCATCAGACAGAGGAATCAATAATAAAACTGCAACAGAACTATCAACTGTATACTT ATTTGGTGGAGATGAAATTTCAAGACAGCAGTATCGCAGGGCCCTGTTACATAAACCAGAGATGATAAAACAGATACTTCCAGAACATAG TGTGCTTCAAAACATTAATTTTGTTGAAGCATTTCAAGATGAGCTATTAGTAACTGAAGTATATGATCTTCCCCAACGACCTAATGATGT TCAGCTCTTTTATGGAAGCATGTGTAAAATTATACTTTCAGTAATTGGAGAATTCAGAGATTGCATTTCTAGCAGAGAATTCCTTCAGCC TTCTTCCAAAGCTAGCTTGGAATCTACAAGCGACTTGGGAGCTTCTGGGAAACATGGTGGCAACGTCTCTTTGGATGTTTTACCAGTCAA AGGTCCTCAGGGTTCTCCTCTTCTCTCACGGGCGGCTCGCCCGCCTCTGGATCAGCTGGCCTCCGAAGAGCCGTGGACTGTCCTACCCGA GCACTTGATTCTGGTAGCTCCTTCTCCTTGTGACATGGCAAAAACTGGACGTTTCCAGATTGTGAATAACTCTGTGAGGTTACTGAGATT TGAGCTGTGCTGGCCAGCGCATTGCCTCACAGTCACGCCGCAGCATGGATGTGTCGCGCCAGAGAGTAAACTACAAATTCTTGTGAGTCC TAATTCCTCCTTATCCACAAAACAGTCAATGTTCCCGTGGAGTGGTTTGATCTATATACACTGTGACGATGGACAGAAGAAAATTGTGAA AGTTCAAATTCGAGAAGATTTAACTCAAGTGGAACTTTTAACTCGTTTGACCTCCAAACCATTTGGAATTCTTTCCCCAGTATCTGAGCC TTCAGTTAGTCATTTGGTCAAACCAATGACAAAACCGCCTTCCACAAAAGTTGAAATAAGAAACAAGAGTATTACTTTTCCTACAACAGA ACCTGGTGAAACTTCAGAGAGCTGTCTAGAACTCGAGAATCATGGCACCACAGACGTGAAATGGCATCTGTCATCTTTAGCGCCACCTTA TGTCAAGGGAGTTGATGAAAGTGGAGATGTTTTTAGAGCTACCTATGCAGCATTCAGATGTTCTCCTATTTCTGGTCTGCTGGAAAGCCA TGGGATCCAAAAAGTCTCCATCACATTTTTGCCCAGAGGTAGGGGGGATTATGCCCAGTTTTGGGATGTTGAATGTCACCCTCTTAAGGA GCCTCACATGAAACACACGTTGAGATTCCAACTCTCTGGACAAAGCATCGAAGCAGAAAATGAGCCTGAAAACGCATGCCTTTCCACGGA TTCCCTCATTAAAATAGATCATTTAGTTAAGCCCCGAAGACAAGCTGTGTCAGAGGCTTCTGCTCGCATACCTGAGCAGCTTGATGTGAC TGCTCGTGGAGTTTATGCCCCAGAGGATGTGTACAGGTTCCGGCCGACTAGTGTGGGGGAATCACGGACACTTAAAGTCAATCTGCGAAA TAATTCTTTTATTACACACTCACTGAAGTTTTTGAGTCCCAGAGAGCCATTCTATGTCAAACATTCCAAGTACTCTTTGAGAGCCCAGCA TTACATCAACATGCCCGTGCAGTTCAAACCGAAGTCCGCAGGCAAATTTGAAGCTTTGCTTGTCATTCAAACAGATGAAGGCAAGAGTAT TGCTATTCGACTAATTGGTGAAGCTCTTGGAAAAAATTAACTAGAATACATTTTTGTGTAAAGTAAATTACATAAGTTGTATTTTGTTAA CTTTATCTTTCTACACTACAATTATGCTTTTGTATATATATTTTGTATGATGGATATCTATAATTGTAGATTTTGTTTTTACAAGCTAAT ACTGAAGACTCGACTGAAATATTATGTATCTAGCCCATAGTATTGTACTTAACTTTTACAGGTGAGAAGAGAGTTCTGTGTTTGCATTGA >3231_3231_1_AJUBA-CEP192_AJUBA_chr14_23445854_ENST00000262713_CEP192_chr18_13087016_ENST00000325971_length(amino acids)=1090AA_BP=425 MGNQSPAAGKENRPPRRRRVPGGEGARGPRPEGMERLGEKASRLLEKFGRRKGESSRSGSDGTPGPGKGRLSGLGGPRKSGPRGATGGPG DEPLEPAREQGSLDAERNQRGSFEAPRYEGSFPAGPPPTRALPLPQSLPPDFRLEPTAPALSPRSSFASSSASDASKPSSPRGSLLLDGA GAGGAGGSRPCSNRTSGISMGYDQRHGSPLPAGPCLFGPPLAGAPAGYSPGGVPSAYPELHAALDRLYAQRPAGFGCQESRHSYPPALGS PGALAGAGVGAAGPLERRGAQPGRHSVTGYGDCAVGARYQDELTALLRLTVGTGGREAGARGEPSGIEPSGLEEPPGPFVPEAARARMRE PEAREDYFGTCIKCNKGIYGQSNACQALDSLYHTQCFVCCSCGRTLRCKAFYSVNGSVYCEEDYLIPLSGYGGTSNLILEGVKKLSDSYM VTVNGLVPGKESKIVFSVRNTGSRAAFVKAVGFKDSQKKVLLDPKVLRIFPDKFVLKERTQENVTLIYNPSDRGINNKTATELSTVYLFG GDEISRQQYRRALLHKPEMIKQILPEHSVLQNINFVEAFQDELLVTEVYDLPQRPNDVQLFYGSMCKIILSVIGEFRDCISSREFLQPSS KASLESTSDLGASGKHGGNVSLDVLPVKGPQGSPLLSRAARPPLDQLASEEPWTVLPEHLILVAPSPCDMAKTGRFQIVNNSVRLLRFEL CWPAHCLTVTPQHGCVAPESKLQILVSPNSSLSTKQSMFPWSGLIYIHCDDGQKKIVKVQIREDLTQVELLTRLTSKPFGILSPVSEPSV SHLVKPMTKPPSTKVEIRNKSITFPTTEPGETSESCLELENHGTTDVKWHLSSLAPPYVKGVDESGDVFRATYAAFRCSPISGLLESHGI QKVSITFLPRGRGDYAQFWDVECHPLKEPHMKHTLRFQLSGQSIEAENEPENACLSTDSLIKIDHLVKPRRQAVSEASARIPEQLDVTAR GVYAPEDVYRFRPTSVGESRTLKVNLRNNSFITHSLKFLSPREPFYVKHSKYSLRAQHYINMPVQFKPKSAGKFEALLVIQTDEGKSIAI -------------------------------------------------------------- >3231_3231_2_AJUBA-CEP192_AJUBA_chr14_23445854_ENST00000262713_CEP192_chr18_13087016_ENST00000430049_length(transcript)=3775nt_BP=1552nt AGATCCGGCCAGGCAGACAAAAGTTTGGGAGAGAAGTTCGGTCTGTGGCTAGCGTGAGAGCTAGGGGGGCGGGGGCCGGGGAGAGTGGGG CAGTCGGGCGAGTCGACGCGTGAACAGATAGACCTGCGGACTGGACAGCCGCGGCCAGAGACCCTGCTAGCCCCGCTCAGCCCCAGATGC GCGGGGGGACGCAGCCCCTCCCGCTGGGGGATGCTGTGGGATTCCTGGCGCAGGGCATCCAGGCCGCCCGCTAAGCCCCTGTGCCTCCCC TGTGCCCCTGGGGAACCAGAGTCCGGCTGCAGGGAAAGAGAACCGGCCGCCGAGACGCCGCAGGGTGCCAGGCGGGGAGGGGGCGAGAGG CCCCAGGCCCGAGGGCATGGAGCGGTTAGGAGAGAAAGCCAGTCGCCTGCTGGAGAAGTTCGGCCGCAGAAAGGGTGAATCTAGCCGGTC TGGGTCTGACGGGACCCCCGGGCCGGGCAAGGGGCGCCTAAGTGGGTTGGGGGGACCTAGGAAGTCAGGGCCCCGAGGAGCTACTGGGGG ACCTGGGGATGAGCCGTTGGAGCCGGCCCGGGAGCAAGGTTCCCTGGACGCTGAGCGAAATCAGCGCGGCTCCTTTGAGGCGCCGCGCTA CGAAGGCTCTTTTCCCGCGGGGCCGCCGCCCACCCGGGCCTTGCCTCTACCTCAGTCGTTGCCCCCCGATTTTCGGCTGGAGCCCACGGC CCCGGCCCTCAGCCCCCGCTCTAGCTTCGCCAGTAGCTCGGCCAGCGACGCGAGCAAGCCGTCCAGCCCCCGGGGCAGCCTGCTGCTGGA CGGGGCGGGGGCTGGCGGAGCTGGAGGTAGCCGGCCCTGCAGCAATCGCACCAGCGGCATCAGCATGGGCTACGACCAGCGCCACGGGAG CCCCTTGCCAGCGGGGCCGTGCCTGTTTGGCCCACCCCTGGCCGGAGCACCGGCAGGCTATTCTCCCGGAGGGGTCCCGTCCGCCTACCC GGAGCTCCACGCCGCCCTGGACCGATTGTACGCTCAGCGGCCCGCGGGGTTCGGCTGCCAGGAAAGCCGCCACTCGTATCCCCCGGCCCT GGGCAGCCCTGGAGCTCTAGCCGGGGCCGGAGTGGGAGCGGCGGGGCCCTTGGAGAGACGGGGGGCGCAACCCGGACGACACTCTGTGAC CGGCTACGGGGACTGCGCCGTGGGCGCCCGGTACCAGGACGAGCTAACAGCTTTGCTTCGCCTGACGGTGGGCACCGGTGGGCGAGAAGC CGGAGCCCGCGGAGAACCCTCGGGGATTGAGCCGTCGGGTCTGGAGGAGCCACCAGGTCCTTTCGTTCCGGAGGCCGCCCGGGCCCGGAT GCGGGAGCCAGAGGCCAGGGAGGACTACTTCGGCACCTGTATCAAGTGCAACAAAGGCATCTATGGGCAGAGCAATGCCTGCCAGGCCCT GGACAGCCTCTACCACACCCAGTGCTTTGTTTGCTGCTCTTGTGGGCGAACTTTGCGTTGCAAGGCTTTCTACAGTGTCAATGGCTCTGT GTACTGTGAGGAAGATTATCTGATACCTTTGTCTGGATATGGAGGAACAAGCAATCTTATTTTGGAAGGCGTTAAAAAATTATCTGACAG TTACATGGTAACAGTGAATGGCTTAGTACCTGGCAAAGAAAGTAAAATTGTTTTTTCTGTCCGCAACACTGGCTCCCGAGCAGCTTTTGT TAAAGCAGTAGGTTTTAAGGATTCTCAGAAAAAAGTTTTGCTGGATCCTAAAGTATTGAGGATTTTTCCAGATAAATTTGTACTCAAGGA AAGAACACAAGAAAATGTTACTTTAATATATAATCCATCAGACAGAGGAATCAATAATAAAACTGCAACAGAACTATCAACTGTATACTT ATTTGGTGGAGATGAAATTTCAAGACAGCAGTATCGCAGGGCCCTGTTACATAAACCAGAGATGATAAAACAGATACTTCCAGAACATAG TGTGCTTCAAAACATTAATTTTGTTGAAGCATTTCAAGATGAGCTATTAGTAACTGAAGTATATGATCTTCCCCAACGACCTAATGATGT TCAGCTCTTTTATGGAAGCATGTGTAAAATTATACTTTCAGTAATTGGAGAATTCAGAGATTGCATTTCTAGCAGAGAATTCCTTCAGCC TTCTTCCAAAGCTAGCTTGGAATCTACAAGCGACTTGGGAGCTTCTGGGAAACATGGTGGCAACGTCTCTTTGGATGTTTTACCAGTCAA AGGTCCTCAGGGTTCTCCTCTTCTCTCACGGGCGGCTCGCCCGCCTCTGGATCAGCTGGCCTCCGAAGAGCCGTGGACTGTCCTACCCGA GCACTTGATTCTGGTAGCTCCTTCTCCTTGTGACATGGCAAAAACTGGACGTTTCCAGATTGTGAATAACTCTGTGAGGTTACTGAGATT TGAGCTGTGCTGGCCAGCGCATTGCCTCACAGTCACGCCGCAGCATGGATGTGTCGCGCCAGAGAGTAAACTACAAATTCTTGTGAGTCC TAATTCCTCCTTATCCACAAAACAGTCAATGTTCCCGTGGAGTGGTTTGATCTATATACACTGTGACGATGGACAGAAGAAAATTGTGAA AGTTCAAATTCGAGAAGATTTAACTCAAGTGGAACTTTTAACTCGTTTGACCTCCAAACCATTTGGAATTCTTTCCCCAGTATCTGAGCC TTCAGTTAGTCATTTGGTCAAACCAATGACAAAACCGCCTTCCACAAAAGTTGAAATAAGAAACAAGAGTATTACTTTTCCTACAACAGA ACCTGGTGAAACTTCAGAGAGCTGTCTAGAACTCGAGAATCATGGCACCACAGACGTGAAATGGCATCTGTCATCTTTAGCGCCACCTTA TGTCAAGGGAGTTGATGAAAGTGGAGATGTTTTTAGAGCTACCTATGCAGCATTCAGATGTTCTCCTATTTCTGGTCTGCTGGAAAGCCA TGGGATCCAAAAAGTCTCCATCACATTTTTGCCCAGAGGTAGGGGGGATTATGCCCAGTTTTGGGATGTTGAATGTCACCCTCTTAAGGA GCCTCACATGAAACACACGTTGAGATTCCAACTCTCTGGACAAAGCATCGAAGCAGAAAATGAGCCTGAAAACGCATGCCTTTCCACGGA TTCCCTCATTAAAATAGATCATTTAGTTAAGCCCCGAAGACAAGCTGTGTCAGAGGCTTCTGCTCGCATACCTGAGCAGCTTGATGTGAC TGCTCGTGGAGTTTATGCCCCAGAGGATGTGTACAGGTTCCGGCCGACTAGTGTGGGGGAATCACGGACACTTAAAGTCAATCTGCGAAA TAATTCTTTTATTACACACTCACTGAAGTTTTTGAGTCCCAGAGAGCCATTCTATGTCAAACATTCCAAGTACTCTTTGAGAGCCCAGCA TTACATCAACATGCCCGTGCAGTTCAAACCGAAGTCCGCAGGCAAATTTGAAGCTTTGCTTGTCATTCAAACAGATGAAGGCAAGAGTAT TGCTATTCGACTAATTGGTGAAGCTCTTGGAAAAAATTAACTAGAATACATTTTTGTGTAAAGTAAATTACATAAGTTGTATTTTGTTAA CTTTATCTTTCTACACTACAATTATGCTTTTGTATATATATTTTGTATGATGGATATCTATAATTGTAGATTTTGTTTTTACAAGCTAAT >3231_3231_2_AJUBA-CEP192_AJUBA_chr14_23445854_ENST00000262713_CEP192_chr18_13087016_ENST00000430049_length(amino acids)=1090AA_BP=425 MGNQSPAAGKENRPPRRRRVPGGEGARGPRPEGMERLGEKASRLLEKFGRRKGESSRSGSDGTPGPGKGRLSGLGGPRKSGPRGATGGPG DEPLEPAREQGSLDAERNQRGSFEAPRYEGSFPAGPPPTRALPLPQSLPPDFRLEPTAPALSPRSSFASSSASDASKPSSPRGSLLLDGA GAGGAGGSRPCSNRTSGISMGYDQRHGSPLPAGPCLFGPPLAGAPAGYSPGGVPSAYPELHAALDRLYAQRPAGFGCQESRHSYPPALGS PGALAGAGVGAAGPLERRGAQPGRHSVTGYGDCAVGARYQDELTALLRLTVGTGGREAGARGEPSGIEPSGLEEPPGPFVPEAARARMRE PEAREDYFGTCIKCNKGIYGQSNACQALDSLYHTQCFVCCSCGRTLRCKAFYSVNGSVYCEEDYLIPLSGYGGTSNLILEGVKKLSDSYM VTVNGLVPGKESKIVFSVRNTGSRAAFVKAVGFKDSQKKVLLDPKVLRIFPDKFVLKERTQENVTLIYNPSDRGINNKTATELSTVYLFG GDEISRQQYRRALLHKPEMIKQILPEHSVLQNINFVEAFQDELLVTEVYDLPQRPNDVQLFYGSMCKIILSVIGEFRDCISSREFLQPSS KASLESTSDLGASGKHGGNVSLDVLPVKGPQGSPLLSRAARPPLDQLASEEPWTVLPEHLILVAPSPCDMAKTGRFQIVNNSVRLLRFEL CWPAHCLTVTPQHGCVAPESKLQILVSPNSSLSTKQSMFPWSGLIYIHCDDGQKKIVKVQIREDLTQVELLTRLTSKPFGILSPVSEPSV SHLVKPMTKPPSTKVEIRNKSITFPTTEPGETSESCLELENHGTTDVKWHLSSLAPPYVKGVDESGDVFRATYAAFRCSPISGLLESHGI QKVSITFLPRGRGDYAQFWDVECHPLKEPHMKHTLRFQLSGQSIEAENEPENACLSTDSLIKIDHLVKPRRQAVSEASARIPEQLDVTAR GVYAPEDVYRFRPTSVGESRTLKVNLRNNSFITHSLKFLSPREPFYVKHSKYSLRAQHYINMPVQFKPKSAGKFEALLVIQTDEGKSIAI -------------------------------------------------------------- >3231_3231_3_AJUBA-CEP192_AJUBA_chr14_23445854_ENST00000262713_CEP192_chr18_13087016_ENST00000506447_length(transcript)=3816nt_BP=1552nt AGATCCGGCCAGGCAGACAAAAGTTTGGGAGAGAAGTTCGGTCTGTGGCTAGCGTGAGAGCTAGGGGGGCGGGGGCCGGGGAGAGTGGGG CAGTCGGGCGAGTCGACGCGTGAACAGATAGACCTGCGGACTGGACAGCCGCGGCCAGAGACCCTGCTAGCCCCGCTCAGCCCCAGATGC GCGGGGGGACGCAGCCCCTCCCGCTGGGGGATGCTGTGGGATTCCTGGCGCAGGGCATCCAGGCCGCCCGCTAAGCCCCTGTGCCTCCCC TGTGCCCCTGGGGAACCAGAGTCCGGCTGCAGGGAAAGAGAACCGGCCGCCGAGACGCCGCAGGGTGCCAGGCGGGGAGGGGGCGAGAGG CCCCAGGCCCGAGGGCATGGAGCGGTTAGGAGAGAAAGCCAGTCGCCTGCTGGAGAAGTTCGGCCGCAGAAAGGGTGAATCTAGCCGGTC TGGGTCTGACGGGACCCCCGGGCCGGGCAAGGGGCGCCTAAGTGGGTTGGGGGGACCTAGGAAGTCAGGGCCCCGAGGAGCTACTGGGGG ACCTGGGGATGAGCCGTTGGAGCCGGCCCGGGAGCAAGGTTCCCTGGACGCTGAGCGAAATCAGCGCGGCTCCTTTGAGGCGCCGCGCTA CGAAGGCTCTTTTCCCGCGGGGCCGCCGCCCACCCGGGCCTTGCCTCTACCTCAGTCGTTGCCCCCCGATTTTCGGCTGGAGCCCACGGC CCCGGCCCTCAGCCCCCGCTCTAGCTTCGCCAGTAGCTCGGCCAGCGACGCGAGCAAGCCGTCCAGCCCCCGGGGCAGCCTGCTGCTGGA CGGGGCGGGGGCTGGCGGAGCTGGAGGTAGCCGGCCCTGCAGCAATCGCACCAGCGGCATCAGCATGGGCTACGACCAGCGCCACGGGAG CCCCTTGCCAGCGGGGCCGTGCCTGTTTGGCCCACCCCTGGCCGGAGCACCGGCAGGCTATTCTCCCGGAGGGGTCCCGTCCGCCTACCC GGAGCTCCACGCCGCCCTGGACCGATTGTACGCTCAGCGGCCCGCGGGGTTCGGCTGCCAGGAAAGCCGCCACTCGTATCCCCCGGCCCT GGGCAGCCCTGGAGCTCTAGCCGGGGCCGGAGTGGGAGCGGCGGGGCCCTTGGAGAGACGGGGGGCGCAACCCGGACGACACTCTGTGAC CGGCTACGGGGACTGCGCCGTGGGCGCCCGGTACCAGGACGAGCTAACAGCTTTGCTTCGCCTGACGGTGGGCACCGGTGGGCGAGAAGC CGGAGCCCGCGGAGAACCCTCGGGGATTGAGCCGTCGGGTCTGGAGGAGCCACCAGGTCCTTTCGTTCCGGAGGCCGCCCGGGCCCGGAT GCGGGAGCCAGAGGCCAGGGAGGACTACTTCGGCACCTGTATCAAGTGCAACAAAGGCATCTATGGGCAGAGCAATGCCTGCCAGGCCCT GGACAGCCTCTACCACACCCAGTGCTTTGTTTGCTGCTCTTGTGGGCGAACTTTGCGTTGCAAGGCTTTCTACAGTGTCAATGGCTCTGT GTACTGTGAGGAAGATTATCTGATACCTTTGTCTGGATATGGAGGAACAAGCAATCTTATTTTGGAAGGCGTTAAAAAATTATCTGACAG TTACATGGTAACAGTGAATGGCTTAGTACCTGGCAAAGAAAGTAAAATTGTTTTTTCTGTCCGCAACACTGGCTCCCGAGCAGCTTTTGT TAAAGCAGTAGGTTTTAAGGATTCTCAGAAAAAAGTTTTGCTGGATCCTAAAGTATTGAGGATTTTTCCAGATAAATTTGTACTCAAGGA AAGAACACAAGAAAATGTTACTTTAATATATAATCCATCAGACAGAGGAATCAATAATAAAACTGCAACAGAACTATCAACTGTATACTT ATTTGGTGGAGATGAAATTTCAAGACAGCAGTATCGCAGGGCCCTGTTACATAAACCAGAGATGATAAAACAGATACTTCCAGAACATAG TGTGCTTCAAAACATTAATTTTGTTGAAGCATTTCAAGATGAGCTATTAGTAACTGAAGTATATGATCTTCCCCAACGACCTAATGATGT TCAGCTCTTTTATGGAAGCATGTGTAAAATTATACTTTCAGTAATTGGAGAATTCAGAGATTGCATTTCTAGCAGAGAATTCCTTCAGCC TTCTTCCAAAGCTAGCTTGGAATCTACAAGCGACTTGGGAGCTTCTGGGAAACATGGTGGCAACGTCTCTTTGGATGTTTTACCAGTCAA AGGTCCTCAGGGTTCTCCTCTTCTCTCACGGGCGGCTCGCCCGCCTCTGGATCAGCTGGCCTCCGAAGAGCCGTGGACTGTCCTACCCGA GCACTTGATTCTGGTAGCTCCTTCTCCTTGTGACATGGCAAAAACTGGACGTTTCCAGATTGTGAATAACTCTGTGAGGTTACTGAGATT TGAGCTGTGCTGGCCAGCGCATTGCCTCACAGTCACGCCGCAGCATGGATGTGTCGCGCCAGAGAGTAAACTACAAATTCTTGTGAGTCC TAATTCCTCCTTATCCACAAAACAGTCAATGTTCCCGTGGAGTGGTTTGATCTATATACACTGTGACGATGGACAGAAGAAAATTGTGAA AGTTCAAATTCGAGAAGATTTAACTCAAGTGGAACTTTTAACTCGTTTGACCTCCAAACCATTTGGAATTCTTTCCCCAGTATCTGAGCC TTCAGTTAGTCATTTGGTCAAACCAATGACAAAACCGCCTTCCACAAAAGTTGAAATAAGAAACAAGAGTATTACTTTTCCTACAACAGA ACCTGGTGAAACTTCAGAGAGCTGTCTAGAACTCGAGAATCATGGCACCACAGACGTGAAATGGCATCTGTCATCTTTAGCGCCACCTTA TGTCAAGGGAGTTGATGAAAGTGGAGATGTTTTTAGAGCTACCTATGCAGCATTCAGATGTTCTCCTATTTCTGGTCTGCTGGAAAGCCA TGGGATCCAAAAAGTCTCCATCACATTTTTGCCCAGAGGTAGGGGGGATTATGCCCAGTTTTGGGATGTTGAATGTCACCCTCTTAAGGA GCCTCACATGAAACACACGTTGAGATTCCAACTCTCTGGACAAAGCATCGAAGCAGAAAATGAGCCTGAAAACGCATGCCTTTCCACGGA TTCCCTCATTAAAATAGATCATTTAGTTAAGCCCCGAAGACAAGCTGTGTCAGAGGCTTCTGCTCGCATACCTGAGCAGCTTGATGTGAC TGCTCGTGGAGTTTATGCCCCAGAGGATGTGTACAGGTTCCGGCCGACTAGTGTGGGGGAATCACGGACACTTAAAGTCAATCTGCGAAA TAATTCTTTTATTACACACTCACTGAAGTTTTTGAGTCCCAGAGAGCCATTCTATGTCAAACATTCCAAGTACTCTTTGAGAGCCCAGCA TTACATCAACATGCCCGTGCAGTTCAAACCGAAGTCCGCAGGCAAATTTGAAGCTTTGCTTGTCATTCAAACAGATGAAGGCAAGAGTAT TGCTATTCGACTAATTGGTGAAGCTCTTGGAAAAAATTAACTAGAATACATTTTTGTGTAAAGTAAATTACATAAGTTGTATTTTGTTAA CTTTATCTTTCTACACTACAATTATGCTTTTGTATATATATTTTGTATGATGGATATCTATAATTGTAGATTTTGTTTTTACAAGCTAAT ACTGAAGACTCGACTGAAATATTATGTATCTAGCCCATAGTATTGTACTTAACTTTTACAGGTGAGAAGAGAGTTCTGTGTTTGCATTGA >3231_3231_3_AJUBA-CEP192_AJUBA_chr14_23445854_ENST00000262713_CEP192_chr18_13087016_ENST00000506447_length(amino acids)=1090AA_BP=425 MGNQSPAAGKENRPPRRRRVPGGEGARGPRPEGMERLGEKASRLLEKFGRRKGESSRSGSDGTPGPGKGRLSGLGGPRKSGPRGATGGPG DEPLEPAREQGSLDAERNQRGSFEAPRYEGSFPAGPPPTRALPLPQSLPPDFRLEPTAPALSPRSSFASSSASDASKPSSPRGSLLLDGA GAGGAGGSRPCSNRTSGISMGYDQRHGSPLPAGPCLFGPPLAGAPAGYSPGGVPSAYPELHAALDRLYAQRPAGFGCQESRHSYPPALGS PGALAGAGVGAAGPLERRGAQPGRHSVTGYGDCAVGARYQDELTALLRLTVGTGGREAGARGEPSGIEPSGLEEPPGPFVPEAARARMRE PEAREDYFGTCIKCNKGIYGQSNACQALDSLYHTQCFVCCSCGRTLRCKAFYSVNGSVYCEEDYLIPLSGYGGTSNLILEGVKKLSDSYM VTVNGLVPGKESKIVFSVRNTGSRAAFVKAVGFKDSQKKVLLDPKVLRIFPDKFVLKERTQENVTLIYNPSDRGINNKTATELSTVYLFG GDEISRQQYRRALLHKPEMIKQILPEHSVLQNINFVEAFQDELLVTEVYDLPQRPNDVQLFYGSMCKIILSVIGEFRDCISSREFLQPSS KASLESTSDLGASGKHGGNVSLDVLPVKGPQGSPLLSRAARPPLDQLASEEPWTVLPEHLILVAPSPCDMAKTGRFQIVNNSVRLLRFEL CWPAHCLTVTPQHGCVAPESKLQILVSPNSSLSTKQSMFPWSGLIYIHCDDGQKKIVKVQIREDLTQVELLTRLTSKPFGILSPVSEPSV SHLVKPMTKPPSTKVEIRNKSITFPTTEPGETSESCLELENHGTTDVKWHLSSLAPPYVKGVDESGDVFRATYAAFRCSPISGLLESHGI QKVSITFLPRGRGDYAQFWDVECHPLKEPHMKHTLRFQLSGQSIEAENEPENACLSTDSLIKIDHLVKPRRQAVSEASARIPEQLDVTAR GVYAPEDVYRFRPTSVGESRTLKVNLRNNSFITHSLKFLSPREPFYVKHSKYSLRAQHYINMPVQFKPKSAGKFEALLVIQTDEGKSIAI -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for AJUBA-CEP192 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for AJUBA-CEP192 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for AJUBA-CEP192 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
