
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:GAPDH-TNC (FusionGDB2 ID:32420) |
Fusion Gene Summary for GAPDH-TNC |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: GAPDH-TNC | Fusion gene ID: 32420 | Hgene | Tgene | Gene symbol | GAPDH | TNC | Gene ID | 2597 | 7134 |
| Gene name | glyceraldehyde-3-phosphate dehydrogenase | troponin C1, slow skeletal and cardiac type | |
| Synonyms | G3PD|GAPD|HEL-S-162eP | CMD1Z|CMH13|TN-C|TNC|TNNC | |
| Cytomap | 12p13.31 | 3p21.1 | |
| Type of gene | protein-coding | protein-coding | |
| Description | glyceraldehyde-3-phosphate dehydrogenaseOCAS, p38 componentOct1 coactivator in S phase, 38 Kd componentaging-associated gene 9 proteinepididymis secretory sperm binding protein Li 162ePpeptidyl-cysteine S-nitrosylase GAPDH | troponin C, slow skeletal and cardiac musclescardiac troponin Cslow twitch skeletal/cardiac muscle troponin Ctroponin C type 1 (slow) | |
| Modification date | 20200327 | 20200313 | |
| UniProtAcc | P04406 | . | |
| Ensembl transtripts involved in fusion gene | ENST00000229239, ENST00000396856, ENST00000396858, ENST00000396859, ENST00000396861, | ENST00000481475, ENST00000340094, ENST00000341037, ENST00000345230, ENST00000346706, ENST00000423613, ENST00000535648, ENST00000537320, ENST00000542877, ENST00000350763, | |
| Fusion gene scores | * DoF score | 45 X 34 X 13=19890 | 8 X 8 X 4=256 |
| # samples | 51 | 11 | |
| ** MAII score | log2(51/19890*10)=-5.28540221886225 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(11/256*10)=-1.21864028647534 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: GAPDH [Title/Abstract] AND TNC [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | GAPDH(6646176)-TNC(117783546), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | GAPDH-TNC seems lost the major protein functional domain in Hgene partner, which is a cell metabolism gene due to the frame-shifted ORF. GAPDH-TNC seems lost the major protein functional domain in Hgene partner, which is a essential gene due to the frame-shifted ORF. GAPDH-TNC seems lost the major protein functional domain in Tgene partner, which is a CGC due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | GAPDH | GO:0010951 | negative regulation of endopeptidase activity | 22832495 |
| Hgene | GAPDH | GO:0017148 | negative regulation of translation | 23071094 |
| Hgene | GAPDH | GO:0031640 | killing of cells of other organism | 22832495 |
| Hgene | GAPDH | GO:0050715 | positive regulation of cytokine secretion | 22832495 |
| Hgene | GAPDH | GO:0050832 | defense response to fungus | 22832495 |
| Hgene | GAPDH | GO:0051873 | killing by host of symbiont cells | 22832495 |
| Hgene | GAPDH | GO:0052501 | positive regulation by organism of apoptotic process in other organism involved in symbiotic interaction | 22832495 |
| Hgene | GAPDH | GO:0061844 | antimicrobial humoral immune response mediated by antimicrobial peptide | 22832495 |
| Hgene | GAPDH | GO:0071346 | cellular response to interferon-gamma | 15479637 |
| Tgene | TNC | GO:0006937 | regulation of muscle contraction | 18092822 |
| Tgene | TNC | GO:0060048 | cardiac muscle contraction | 25771144 |
 Fusion gene breakpoints across GAPDH (5'-gene) Fusion gene breakpoints across GAPDH (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
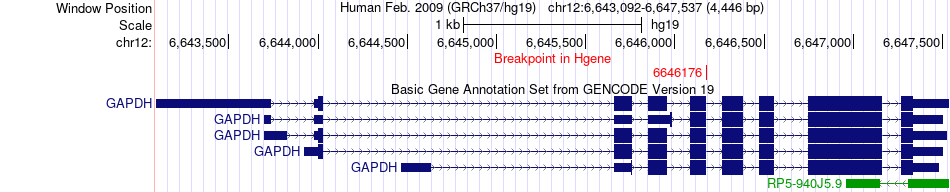 |
 Fusion gene breakpoints across TNC (3'-gene) Fusion gene breakpoints across TNC (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
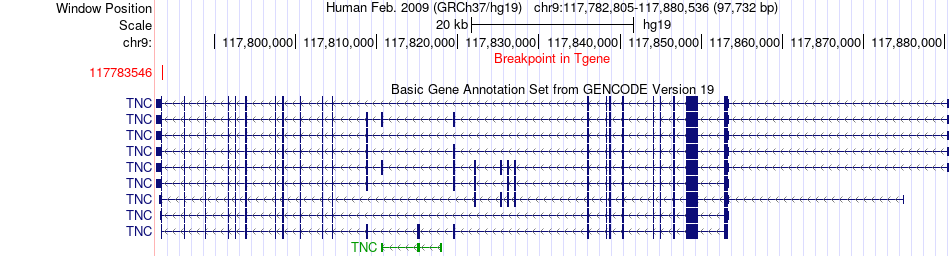 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | HNSC | TCGA-CN-6024-01A | GAPDH | chr12 | 6646176 | + | TNC | chr9 | 117783546 | - |
Top |
Fusion Gene ORF analysis for GAPDH-TNC |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-intron | ENST00000229239 | ENST00000481475 | GAPDH | chr12 | 6646176 | + | TNC | chr9 | 117783546 | - |
| 5CDS-intron | ENST00000396856 | ENST00000481475 | GAPDH | chr12 | 6646176 | + | TNC | chr9 | 117783546 | - |
| 5CDS-intron | ENST00000396858 | ENST00000481475 | GAPDH | chr12 | 6646176 | + | TNC | chr9 | 117783546 | - |
| 5CDS-intron | ENST00000396859 | ENST00000481475 | GAPDH | chr12 | 6646176 | + | TNC | chr9 | 117783546 | - |
| 5CDS-intron | ENST00000396861 | ENST00000481475 | GAPDH | chr12 | 6646176 | + | TNC | chr9 | 117783546 | - |
| Frame-shift | ENST00000229239 | ENST00000340094 | GAPDH | chr12 | 6646176 | + | TNC | chr9 | 117783546 | - |
| Frame-shift | ENST00000229239 | ENST00000341037 | GAPDH | chr12 | 6646176 | + | TNC | chr9 | 117783546 | - |
| Frame-shift | ENST00000229239 | ENST00000345230 | GAPDH | chr12 | 6646176 | + | TNC | chr9 | 117783546 | - |
| Frame-shift | ENST00000229239 | ENST00000346706 | GAPDH | chr12 | 6646176 | + | TNC | chr9 | 117783546 | - |
| Frame-shift | ENST00000229239 | ENST00000423613 | GAPDH | chr12 | 6646176 | + | TNC | chr9 | 117783546 | - |
| Frame-shift | ENST00000229239 | ENST00000535648 | GAPDH | chr12 | 6646176 | + | TNC | chr9 | 117783546 | - |
| Frame-shift | ENST00000229239 | ENST00000537320 | GAPDH | chr12 | 6646176 | + | TNC | chr9 | 117783546 | - |
| Frame-shift | ENST00000229239 | ENST00000542877 | GAPDH | chr12 | 6646176 | + | TNC | chr9 | 117783546 | - |
| Frame-shift | ENST00000396856 | ENST00000340094 | GAPDH | chr12 | 6646176 | + | TNC | chr9 | 117783546 | - |
| Frame-shift | ENST00000396856 | ENST00000341037 | GAPDH | chr12 | 6646176 | + | TNC | chr9 | 117783546 | - |
| Frame-shift | ENST00000396856 | ENST00000345230 | GAPDH | chr12 | 6646176 | + | TNC | chr9 | 117783546 | - |
| Frame-shift | ENST00000396856 | ENST00000346706 | GAPDH | chr12 | 6646176 | + | TNC | chr9 | 117783546 | - |
| Frame-shift | ENST00000396856 | ENST00000423613 | GAPDH | chr12 | 6646176 | + | TNC | chr9 | 117783546 | - |
| Frame-shift | ENST00000396856 | ENST00000535648 | GAPDH | chr12 | 6646176 | + | TNC | chr9 | 117783546 | - |
| Frame-shift | ENST00000396856 | ENST00000537320 | GAPDH | chr12 | 6646176 | + | TNC | chr9 | 117783546 | - |
| Frame-shift | ENST00000396856 | ENST00000542877 | GAPDH | chr12 | 6646176 | + | TNC | chr9 | 117783546 | - |
| Frame-shift | ENST00000396858 | ENST00000340094 | GAPDH | chr12 | 6646176 | + | TNC | chr9 | 117783546 | - |
| Frame-shift | ENST00000396858 | ENST00000341037 | GAPDH | chr12 | 6646176 | + | TNC | chr9 | 117783546 | - |
| Frame-shift | ENST00000396858 | ENST00000345230 | GAPDH | chr12 | 6646176 | + | TNC | chr9 | 117783546 | - |
| Frame-shift | ENST00000396858 | ENST00000346706 | GAPDH | chr12 | 6646176 | + | TNC | chr9 | 117783546 | - |
| Frame-shift | ENST00000396858 | ENST00000423613 | GAPDH | chr12 | 6646176 | + | TNC | chr9 | 117783546 | - |
| Frame-shift | ENST00000396858 | ENST00000535648 | GAPDH | chr12 | 6646176 | + | TNC | chr9 | 117783546 | - |
| Frame-shift | ENST00000396858 | ENST00000537320 | GAPDH | chr12 | 6646176 | + | TNC | chr9 | 117783546 | - |
| Frame-shift | ENST00000396858 | ENST00000542877 | GAPDH | chr12 | 6646176 | + | TNC | chr9 | 117783546 | - |
| Frame-shift | ENST00000396859 | ENST00000340094 | GAPDH | chr12 | 6646176 | + | TNC | chr9 | 117783546 | - |
| Frame-shift | ENST00000396859 | ENST00000341037 | GAPDH | chr12 | 6646176 | + | TNC | chr9 | 117783546 | - |
| Frame-shift | ENST00000396859 | ENST00000345230 | GAPDH | chr12 | 6646176 | + | TNC | chr9 | 117783546 | - |
| Frame-shift | ENST00000396859 | ENST00000346706 | GAPDH | chr12 | 6646176 | + | TNC | chr9 | 117783546 | - |
| Frame-shift | ENST00000396859 | ENST00000423613 | GAPDH | chr12 | 6646176 | + | TNC | chr9 | 117783546 | - |
| Frame-shift | ENST00000396859 | ENST00000535648 | GAPDH | chr12 | 6646176 | + | TNC | chr9 | 117783546 | - |
| Frame-shift | ENST00000396859 | ENST00000537320 | GAPDH | chr12 | 6646176 | + | TNC | chr9 | 117783546 | - |
| Frame-shift | ENST00000396859 | ENST00000542877 | GAPDH | chr12 | 6646176 | + | TNC | chr9 | 117783546 | - |
| Frame-shift | ENST00000396861 | ENST00000340094 | GAPDH | chr12 | 6646176 | + | TNC | chr9 | 117783546 | - |
| Frame-shift | ENST00000396861 | ENST00000341037 | GAPDH | chr12 | 6646176 | + | TNC | chr9 | 117783546 | - |
| Frame-shift | ENST00000396861 | ENST00000345230 | GAPDH | chr12 | 6646176 | + | TNC | chr9 | 117783546 | - |
| Frame-shift | ENST00000396861 | ENST00000346706 | GAPDH | chr12 | 6646176 | + | TNC | chr9 | 117783546 | - |
| Frame-shift | ENST00000396861 | ENST00000423613 | GAPDH | chr12 | 6646176 | + | TNC | chr9 | 117783546 | - |
| Frame-shift | ENST00000396861 | ENST00000535648 | GAPDH | chr12 | 6646176 | + | TNC | chr9 | 117783546 | - |
| Frame-shift | ENST00000396861 | ENST00000537320 | GAPDH | chr12 | 6646176 | + | TNC | chr9 | 117783546 | - |
| Frame-shift | ENST00000396861 | ENST00000542877 | GAPDH | chr12 | 6646176 | + | TNC | chr9 | 117783546 | - |
| In-frame | ENST00000229239 | ENST00000350763 | GAPDH | chr12 | 6646176 | + | TNC | chr9 | 117783546 | - |
| In-frame | ENST00000396856 | ENST00000350763 | GAPDH | chr12 | 6646176 | + | TNC | chr9 | 117783546 | - |
| In-frame | ENST00000396858 | ENST00000350763 | GAPDH | chr12 | 6646176 | + | TNC | chr9 | 117783546 | - |
| In-frame | ENST00000396859 | ENST00000350763 | GAPDH | chr12 | 6646176 | + | TNC | chr9 | 117783546 | - |
| In-frame | ENST00000396861 | ENST00000350763 | GAPDH | chr12 | 6646176 | + | TNC | chr9 | 117783546 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000229239 | GAPDH | chr12 | 6646176 | + | ENST00000350763 | TNC | chr9 | 117783546 | - | 1727 | 993 | 489 | 1103 | 204 |
| ENST00000396856 | GAPDH | chr12 | 6646176 | + | ENST00000350763 | TNC | chr9 | 117783546 | - | 1149 | 415 | 92 | 472 | 126 |
| ENST00000396861 | GAPDH | chr12 | 6646176 | + | ENST00000350763 | TNC | chr9 | 117783546 | - | 1212 | 478 | 121 | 588 | 155 |
| ENST00000396859 | GAPDH | chr12 | 6646176 | + | ENST00000350763 | TNC | chr9 | 117783546 | - | 1140 | 406 | 79 | 516 | 145 |
| ENST00000396858 | GAPDH | chr12 | 6646176 | + | ENST00000350763 | TNC | chr9 | 117783546 | - | 1200 | 466 | 41 | 523 | 160 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000229239 | ENST00000350763 | GAPDH | chr12 | 6646176 | + | TNC | chr9 | 117783546 | - | 0.007140598 | 0.9928595 |
| ENST00000396856 | ENST00000350763 | GAPDH | chr12 | 6646176 | + | TNC | chr9 | 117783546 | - | 0.5729945 | 0.42700556 |
| ENST00000396861 | ENST00000350763 | GAPDH | chr12 | 6646176 | + | TNC | chr9 | 117783546 | - | 0.005875189 | 0.9941248 |
| ENST00000396859 | ENST00000350763 | GAPDH | chr12 | 6646176 | + | TNC | chr9 | 117783546 | - | 0.004173072 | 0.9958269 |
| ENST00000396858 | ENST00000350763 | GAPDH | chr12 | 6646176 | + | TNC | chr9 | 117783546 | - | 0.0484138 | 0.9515862 |
Top |
Fusion Genomic Features for GAPDH-TNC |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
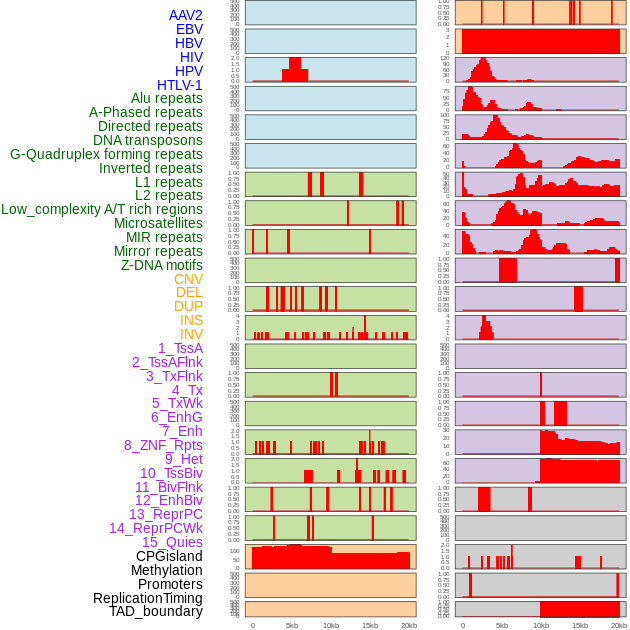 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for GAPDH-TNC |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr12:6646176/chr9:117783546) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| GAPDH | . |
| FUNCTION: Has both glyceraldehyde-3-phosphate dehydrogenase and nitrosylase activities, thereby playing a role in glycolysis and nuclear functions, respectively. Participates in nuclear events including transcription, RNA transport, DNA replication and apoptosis. Nuclear functions are probably due to the nitrosylase activity that mediates cysteine S-nitrosylation of nuclear target proteins such as SIRT1, HDAC2 and PRKDC. Modulates the organization and assembly of the cytoskeleton. Facilitates the CHP1-dependent microtubule and membrane associations through its ability to stimulate the binding of CHP1 to microtubules (By similarity). Glyceraldehyde-3-phosphate dehydrogenase is a key enzyme in glycolysis that catalyzes the first step of the pathway by converting D-glyceraldehyde 3-phosphate (G3P) into 3-phospho-D-glyceroyl phosphate. Component of the GAIT (gamma interferon-activated inhibitor of translation) complex which mediates interferon-gamma-induced transcript-selective translation inhibition in inflammation processes. Upon interferon-gamma treatment assembles into the GAIT complex which binds to stem loop-containing GAIT elements in the 3'-UTR of diverse inflammatory mRNAs (such as ceruplasmin) and suppresses their translation. {ECO:0000250, ECO:0000269|PubMed:11724794, ECO:0000269|PubMed:23071094, ECO:0000269|PubMed:3170585}. | FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | GAPDH | chr12:6646176 | chr9:117783546 | ENST00000229239 | + | 5 | 9 | 13_14 | 109 | 336.0 | Nucleotide binding | NAD |
| Hgene | GAPDH | chr12:6646176 | chr9:117783546 | ENST00000396858 | + | 4 | 8 | 13_14 | 67 | 294.0 | Nucleotide binding | NAD |
| Hgene | GAPDH | chr12:6646176 | chr9:117783546 | ENST00000396859 | + | 4 | 8 | 13_14 | 109 | 336.0 | Nucleotide binding | NAD |
| Hgene | GAPDH | chr12:6646176 | chr9:117783546 | ENST00000396861 | + | 5 | 9 | 13_14 | 109 | 336.0 | Nucleotide binding | NAD |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000340094 | 22 | 24 | 1802_1888 | 1801 | 1838.0 | Domain | Fibronectin type-III 14 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000340094 | 22 | 24 | 1889_1977 | 1801 | 1838.0 | Domain | Fibronectin type-III 15 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000340094 | 22 | 24 | 1975_2190 | 1801 | 1838.0 | Domain | Fibrinogen C-terminal | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000345230 | 19 | 21 | 1533_1621 | 1528 | 1565.0 | Domain | Fibronectin type-III 11 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000345230 | 19 | 21 | 1622_1711 | 1528 | 1565.0 | Domain | Fibronectin type-III 12 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000345230 | 19 | 21 | 1712_1801 | 1528 | 1565.0 | Domain | Fibronectin type-III 13 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000345230 | 19 | 21 | 1802_1888 | 1528 | 1565.0 | Domain | Fibronectin type-III 14 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000345230 | 19 | 21 | 1889_1977 | 1528 | 1565.0 | Domain | Fibronectin type-III 15 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000345230 | 19 | 21 | 1975_2190 | 1528 | 1565.0 | Domain | Fibrinogen C-terminal | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000346706 | 20 | 22 | 1622_1711 | 1619 | 1656.0 | Domain | Fibronectin type-III 12 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000346706 | 20 | 22 | 1712_1801 | 1619 | 1656.0 | Domain | Fibronectin type-III 13 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000346706 | 20 | 22 | 1802_1888 | 1619 | 1656.0 | Domain | Fibronectin type-III 14 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000346706 | 20 | 22 | 1889_1977 | 1619 | 1656.0 | Domain | Fibronectin type-III 15 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000346706 | 20 | 22 | 1975_2190 | 1619 | 1656.0 | Domain | Fibrinogen C-terminal | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000535648 | 21 | 23 | 1712_1801 | 1710 | 1747.0 | Domain | Fibronectin type-III 13 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000535648 | 21 | 23 | 1802_1888 | 1710 | 1747.0 | Domain | Fibronectin type-III 14 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000535648 | 21 | 23 | 1889_1977 | 1710 | 1747.0 | Domain | Fibronectin type-III 15 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000535648 | 21 | 23 | 1975_2190 | 1710 | 1747.0 | Domain | Fibrinogen C-terminal | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000537320 | 18 | 20 | 1533_1621 | 1528 | 1565.0 | Domain | Fibronectin type-III 11 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000537320 | 18 | 20 | 1622_1711 | 1528 | 1565.0 | Domain | Fibronectin type-III 12 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000537320 | 18 | 20 | 1712_1801 | 1528 | 1565.0 | Domain | Fibronectin type-III 13 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000537320 | 18 | 20 | 1802_1888 | 1528 | 1565.0 | Domain | Fibronectin type-III 14 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000537320 | 18 | 20 | 1889_1977 | 1528 | 1565.0 | Domain | Fibronectin type-III 15 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000537320 | 18 | 20 | 1975_2190 | 1528 | 1565.0 | Domain | Fibrinogen C-terminal |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | GAPDH | chr12:6646176 | chr9:117783546 | ENST00000229239 | + | 5 | 9 | 245_250 | 109 | 336.0 | Motif | [IL]-x-C-x-x-[DE] motif |
| Hgene | GAPDH | chr12:6646176 | chr9:117783546 | ENST00000396858 | + | 4 | 8 | 245_250 | 67 | 294.0 | Motif | [IL]-x-C-x-x-[DE] motif |
| Hgene | GAPDH | chr12:6646176 | chr9:117783546 | ENST00000396859 | + | 4 | 8 | 245_250 | 109 | 336.0 | Motif | [IL]-x-C-x-x-[DE] motif |
| Hgene | GAPDH | chr12:6646176 | chr9:117783546 | ENST00000396861 | + | 5 | 9 | 245_250 | 109 | 336.0 | Motif | [IL]-x-C-x-x-[DE] motif |
| Hgene | GAPDH | chr12:6646176 | chr9:117783546 | ENST00000229239 | + | 5 | 9 | 151_153 | 109 | 336.0 | Region | Glyceraldehyde 3-phosphate binding |
| Hgene | GAPDH | chr12:6646176 | chr9:117783546 | ENST00000229239 | + | 5 | 9 | 211_212 | 109 | 336.0 | Region | Glyceraldehyde 3-phosphate binding |
| Hgene | GAPDH | chr12:6646176 | chr9:117783546 | ENST00000396858 | + | 4 | 8 | 151_153 | 67 | 294.0 | Region | Glyceraldehyde 3-phosphate binding |
| Hgene | GAPDH | chr12:6646176 | chr9:117783546 | ENST00000396858 | + | 4 | 8 | 211_212 | 67 | 294.0 | Region | Glyceraldehyde 3-phosphate binding |
| Hgene | GAPDH | chr12:6646176 | chr9:117783546 | ENST00000396859 | + | 4 | 8 | 151_153 | 109 | 336.0 | Region | Glyceraldehyde 3-phosphate binding |
| Hgene | GAPDH | chr12:6646176 | chr9:117783546 | ENST00000396859 | + | 4 | 8 | 211_212 | 109 | 336.0 | Region | Glyceraldehyde 3-phosphate binding |
| Hgene | GAPDH | chr12:6646176 | chr9:117783546 | ENST00000396861 | + | 5 | 9 | 151_153 | 109 | 336.0 | Region | Glyceraldehyde 3-phosphate binding |
| Hgene | GAPDH | chr12:6646176 | chr9:117783546 | ENST00000396861 | + | 5 | 9 | 211_212 | 109 | 336.0 | Region | Glyceraldehyde 3-phosphate binding |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000340094 | 22 | 24 | 118_145 | 1801 | 1838.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000345230 | 19 | 21 | 118_145 | 1528 | 1565.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000346706 | 20 | 22 | 118_145 | 1619 | 1656.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000350763 | 26 | 28 | 118_145 | 2165 | 2202.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000535648 | 21 | 23 | 118_145 | 1710 | 1747.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000537320 | 18 | 20 | 118_145 | 1528 | 1565.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000340094 | 22 | 24 | 1076_1165 | 1801 | 1838.0 | Domain | Fibronectin type-III 6 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000340094 | 22 | 24 | 1167_1256 | 1801 | 1838.0 | Domain | Fibronectin type-III 7 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000340094 | 22 | 24 | 1258_1350 | 1801 | 1838.0 | Domain | Fibronectin type-III 8 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000340094 | 22 | 24 | 1351_1439 | 1801 | 1838.0 | Domain | Fibronectin type-III 9 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000340094 | 22 | 24 | 1440_1531 | 1801 | 1838.0 | Domain | Fibronectin type-III 10 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000340094 | 22 | 24 | 1533_1621 | 1801 | 1838.0 | Domain | Fibronectin type-III 11 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000340094 | 22 | 24 | 1622_1711 | 1801 | 1838.0 | Domain | Fibronectin type-III 12 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000340094 | 22 | 24 | 1712_1801 | 1801 | 1838.0 | Domain | Fibronectin type-III 13 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000340094 | 22 | 24 | 174_186 | 1801 | 1838.0 | Domain | EGF-like 1%3B incomplete | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000340094 | 22 | 24 | 186_217 | 1801 | 1838.0 | Domain | EGF-like 2 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000340094 | 22 | 24 | 217_248 | 1801 | 1838.0 | Domain | EGF-like 3 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000340094 | 22 | 24 | 248_280 | 1801 | 1838.0 | Domain | EGF-like 4 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000340094 | 22 | 24 | 280_311 | 1801 | 1838.0 | Domain | EGF-like 5 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000340094 | 22 | 24 | 311_342 | 1801 | 1838.0 | Domain | EGF-like 6 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000340094 | 22 | 24 | 342_373 | 1801 | 1838.0 | Domain | EGF-like 7 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000340094 | 22 | 24 | 373_404 | 1801 | 1838.0 | Domain | EGF-like 8 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000340094 | 22 | 24 | 404_435 | 1801 | 1838.0 | Domain | EGF-like 9 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000340094 | 22 | 24 | 435_466 | 1801 | 1838.0 | Domain | EGF-like 10 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000340094 | 22 | 24 | 466_497 | 1801 | 1838.0 | Domain | EGF-like 11 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000340094 | 22 | 24 | 497_528 | 1801 | 1838.0 | Domain | EGF-like 12 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000340094 | 22 | 24 | 528_559 | 1801 | 1838.0 | Domain | EGF-like 13 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000340094 | 22 | 24 | 559_590 | 1801 | 1838.0 | Domain | EGF-like 14 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000340094 | 22 | 24 | 590_621 | 1801 | 1838.0 | Domain | EGF-like 15 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000340094 | 22 | 24 | 625_715 | 1801 | 1838.0 | Domain | Fibronectin type-III 1 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000340094 | 22 | 24 | 716_804 | 1801 | 1838.0 | Domain | Fibronectin type-III 2 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000340094 | 22 | 24 | 805_894 | 1801 | 1838.0 | Domain | Fibronectin type-III 3 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000340094 | 22 | 24 | 895_990 | 1801 | 1838.0 | Domain | Fibronectin type-III 4 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000340094 | 22 | 24 | 991_1075 | 1801 | 1838.0 | Domain | Fibronectin type-III 5 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000345230 | 19 | 21 | 1076_1165 | 1528 | 1565.0 | Domain | Fibronectin type-III 6 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000345230 | 19 | 21 | 1167_1256 | 1528 | 1565.0 | Domain | Fibronectin type-III 7 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000345230 | 19 | 21 | 1258_1350 | 1528 | 1565.0 | Domain | Fibronectin type-III 8 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000345230 | 19 | 21 | 1351_1439 | 1528 | 1565.0 | Domain | Fibronectin type-III 9 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000345230 | 19 | 21 | 1440_1531 | 1528 | 1565.0 | Domain | Fibronectin type-III 10 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000345230 | 19 | 21 | 174_186 | 1528 | 1565.0 | Domain | EGF-like 1%3B incomplete | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000345230 | 19 | 21 | 186_217 | 1528 | 1565.0 | Domain | EGF-like 2 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000345230 | 19 | 21 | 217_248 | 1528 | 1565.0 | Domain | EGF-like 3 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000345230 | 19 | 21 | 248_280 | 1528 | 1565.0 | Domain | EGF-like 4 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000345230 | 19 | 21 | 280_311 | 1528 | 1565.0 | Domain | EGF-like 5 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000345230 | 19 | 21 | 311_342 | 1528 | 1565.0 | Domain | EGF-like 6 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000345230 | 19 | 21 | 342_373 | 1528 | 1565.0 | Domain | EGF-like 7 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000345230 | 19 | 21 | 373_404 | 1528 | 1565.0 | Domain | EGF-like 8 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000345230 | 19 | 21 | 404_435 | 1528 | 1565.0 | Domain | EGF-like 9 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000345230 | 19 | 21 | 435_466 | 1528 | 1565.0 | Domain | EGF-like 10 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000345230 | 19 | 21 | 466_497 | 1528 | 1565.0 | Domain | EGF-like 11 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000345230 | 19 | 21 | 497_528 | 1528 | 1565.0 | Domain | EGF-like 12 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000345230 | 19 | 21 | 528_559 | 1528 | 1565.0 | Domain | EGF-like 13 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000345230 | 19 | 21 | 559_590 | 1528 | 1565.0 | Domain | EGF-like 14 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000345230 | 19 | 21 | 590_621 | 1528 | 1565.0 | Domain | EGF-like 15 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000345230 | 19 | 21 | 625_715 | 1528 | 1565.0 | Domain | Fibronectin type-III 1 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000345230 | 19 | 21 | 716_804 | 1528 | 1565.0 | Domain | Fibronectin type-III 2 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000345230 | 19 | 21 | 805_894 | 1528 | 1565.0 | Domain | Fibronectin type-III 3 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000345230 | 19 | 21 | 895_990 | 1528 | 1565.0 | Domain | Fibronectin type-III 4 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000345230 | 19 | 21 | 991_1075 | 1528 | 1565.0 | Domain | Fibronectin type-III 5 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000346706 | 20 | 22 | 1076_1165 | 1619 | 1656.0 | Domain | Fibronectin type-III 6 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000346706 | 20 | 22 | 1167_1256 | 1619 | 1656.0 | Domain | Fibronectin type-III 7 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000346706 | 20 | 22 | 1258_1350 | 1619 | 1656.0 | Domain | Fibronectin type-III 8 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000346706 | 20 | 22 | 1351_1439 | 1619 | 1656.0 | Domain | Fibronectin type-III 9 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000346706 | 20 | 22 | 1440_1531 | 1619 | 1656.0 | Domain | Fibronectin type-III 10 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000346706 | 20 | 22 | 1533_1621 | 1619 | 1656.0 | Domain | Fibronectin type-III 11 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000346706 | 20 | 22 | 174_186 | 1619 | 1656.0 | Domain | EGF-like 1%3B incomplete | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000346706 | 20 | 22 | 186_217 | 1619 | 1656.0 | Domain | EGF-like 2 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000346706 | 20 | 22 | 217_248 | 1619 | 1656.0 | Domain | EGF-like 3 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000346706 | 20 | 22 | 248_280 | 1619 | 1656.0 | Domain | EGF-like 4 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000346706 | 20 | 22 | 280_311 | 1619 | 1656.0 | Domain | EGF-like 5 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000346706 | 20 | 22 | 311_342 | 1619 | 1656.0 | Domain | EGF-like 6 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000346706 | 20 | 22 | 342_373 | 1619 | 1656.0 | Domain | EGF-like 7 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000346706 | 20 | 22 | 373_404 | 1619 | 1656.0 | Domain | EGF-like 8 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000346706 | 20 | 22 | 404_435 | 1619 | 1656.0 | Domain | EGF-like 9 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000346706 | 20 | 22 | 435_466 | 1619 | 1656.0 | Domain | EGF-like 10 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000346706 | 20 | 22 | 466_497 | 1619 | 1656.0 | Domain | EGF-like 11 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000346706 | 20 | 22 | 497_528 | 1619 | 1656.0 | Domain | EGF-like 12 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000346706 | 20 | 22 | 528_559 | 1619 | 1656.0 | Domain | EGF-like 13 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000346706 | 20 | 22 | 559_590 | 1619 | 1656.0 | Domain | EGF-like 14 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000346706 | 20 | 22 | 590_621 | 1619 | 1656.0 | Domain | EGF-like 15 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000346706 | 20 | 22 | 625_715 | 1619 | 1656.0 | Domain | Fibronectin type-III 1 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000346706 | 20 | 22 | 716_804 | 1619 | 1656.0 | Domain | Fibronectin type-III 2 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000346706 | 20 | 22 | 805_894 | 1619 | 1656.0 | Domain | Fibronectin type-III 3 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000346706 | 20 | 22 | 895_990 | 1619 | 1656.0 | Domain | Fibronectin type-III 4 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000346706 | 20 | 22 | 991_1075 | 1619 | 1656.0 | Domain | Fibronectin type-III 5 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000350763 | 26 | 28 | 1076_1165 | 2165 | 2202.0 | Domain | Fibronectin type-III 6 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000350763 | 26 | 28 | 1167_1256 | 2165 | 2202.0 | Domain | Fibronectin type-III 7 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000350763 | 26 | 28 | 1258_1350 | 2165 | 2202.0 | Domain | Fibronectin type-III 8 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000350763 | 26 | 28 | 1351_1439 | 2165 | 2202.0 | Domain | Fibronectin type-III 9 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000350763 | 26 | 28 | 1440_1531 | 2165 | 2202.0 | Domain | Fibronectin type-III 10 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000350763 | 26 | 28 | 1533_1621 | 2165 | 2202.0 | Domain | Fibronectin type-III 11 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000350763 | 26 | 28 | 1622_1711 | 2165 | 2202.0 | Domain | Fibronectin type-III 12 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000350763 | 26 | 28 | 1712_1801 | 2165 | 2202.0 | Domain | Fibronectin type-III 13 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000350763 | 26 | 28 | 174_186 | 2165 | 2202.0 | Domain | EGF-like 1%3B incomplete | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000350763 | 26 | 28 | 1802_1888 | 2165 | 2202.0 | Domain | Fibronectin type-III 14 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000350763 | 26 | 28 | 186_217 | 2165 | 2202.0 | Domain | EGF-like 2 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000350763 | 26 | 28 | 1889_1977 | 2165 | 2202.0 | Domain | Fibronectin type-III 15 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000350763 | 26 | 28 | 1975_2190 | 2165 | 2202.0 | Domain | Fibrinogen C-terminal | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000350763 | 26 | 28 | 217_248 | 2165 | 2202.0 | Domain | EGF-like 3 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000350763 | 26 | 28 | 248_280 | 2165 | 2202.0 | Domain | EGF-like 4 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000350763 | 26 | 28 | 280_311 | 2165 | 2202.0 | Domain | EGF-like 5 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000350763 | 26 | 28 | 311_342 | 2165 | 2202.0 | Domain | EGF-like 6 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000350763 | 26 | 28 | 342_373 | 2165 | 2202.0 | Domain | EGF-like 7 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000350763 | 26 | 28 | 373_404 | 2165 | 2202.0 | Domain | EGF-like 8 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000350763 | 26 | 28 | 404_435 | 2165 | 2202.0 | Domain | EGF-like 9 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000350763 | 26 | 28 | 435_466 | 2165 | 2202.0 | Domain | EGF-like 10 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000350763 | 26 | 28 | 466_497 | 2165 | 2202.0 | Domain | EGF-like 11 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000350763 | 26 | 28 | 497_528 | 2165 | 2202.0 | Domain | EGF-like 12 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000350763 | 26 | 28 | 528_559 | 2165 | 2202.0 | Domain | EGF-like 13 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000350763 | 26 | 28 | 559_590 | 2165 | 2202.0 | Domain | EGF-like 14 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000350763 | 26 | 28 | 590_621 | 2165 | 2202.0 | Domain | EGF-like 15 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000350763 | 26 | 28 | 625_715 | 2165 | 2202.0 | Domain | Fibronectin type-III 1 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000350763 | 26 | 28 | 716_804 | 2165 | 2202.0 | Domain | Fibronectin type-III 2 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000350763 | 26 | 28 | 805_894 | 2165 | 2202.0 | Domain | Fibronectin type-III 3 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000350763 | 26 | 28 | 895_990 | 2165 | 2202.0 | Domain | Fibronectin type-III 4 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000350763 | 26 | 28 | 991_1075 | 2165 | 2202.0 | Domain | Fibronectin type-III 5 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000535648 | 21 | 23 | 1076_1165 | 1710 | 1747.0 | Domain | Fibronectin type-III 6 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000535648 | 21 | 23 | 1167_1256 | 1710 | 1747.0 | Domain | Fibronectin type-III 7 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000535648 | 21 | 23 | 1258_1350 | 1710 | 1747.0 | Domain | Fibronectin type-III 8 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000535648 | 21 | 23 | 1351_1439 | 1710 | 1747.0 | Domain | Fibronectin type-III 9 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000535648 | 21 | 23 | 1440_1531 | 1710 | 1747.0 | Domain | Fibronectin type-III 10 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000535648 | 21 | 23 | 1533_1621 | 1710 | 1747.0 | Domain | Fibronectin type-III 11 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000535648 | 21 | 23 | 1622_1711 | 1710 | 1747.0 | Domain | Fibronectin type-III 12 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000535648 | 21 | 23 | 174_186 | 1710 | 1747.0 | Domain | EGF-like 1%3B incomplete | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000535648 | 21 | 23 | 186_217 | 1710 | 1747.0 | Domain | EGF-like 2 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000535648 | 21 | 23 | 217_248 | 1710 | 1747.0 | Domain | EGF-like 3 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000535648 | 21 | 23 | 248_280 | 1710 | 1747.0 | Domain | EGF-like 4 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000535648 | 21 | 23 | 280_311 | 1710 | 1747.0 | Domain | EGF-like 5 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000535648 | 21 | 23 | 311_342 | 1710 | 1747.0 | Domain | EGF-like 6 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000535648 | 21 | 23 | 342_373 | 1710 | 1747.0 | Domain | EGF-like 7 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000535648 | 21 | 23 | 373_404 | 1710 | 1747.0 | Domain | EGF-like 8 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000535648 | 21 | 23 | 404_435 | 1710 | 1747.0 | Domain | EGF-like 9 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000535648 | 21 | 23 | 435_466 | 1710 | 1747.0 | Domain | EGF-like 10 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000535648 | 21 | 23 | 466_497 | 1710 | 1747.0 | Domain | EGF-like 11 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000535648 | 21 | 23 | 497_528 | 1710 | 1747.0 | Domain | EGF-like 12 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000535648 | 21 | 23 | 528_559 | 1710 | 1747.0 | Domain | EGF-like 13 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000535648 | 21 | 23 | 559_590 | 1710 | 1747.0 | Domain | EGF-like 14 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000535648 | 21 | 23 | 590_621 | 1710 | 1747.0 | Domain | EGF-like 15 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000535648 | 21 | 23 | 625_715 | 1710 | 1747.0 | Domain | Fibronectin type-III 1 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000535648 | 21 | 23 | 716_804 | 1710 | 1747.0 | Domain | Fibronectin type-III 2 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000535648 | 21 | 23 | 805_894 | 1710 | 1747.0 | Domain | Fibronectin type-III 3 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000535648 | 21 | 23 | 895_990 | 1710 | 1747.0 | Domain | Fibronectin type-III 4 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000535648 | 21 | 23 | 991_1075 | 1710 | 1747.0 | Domain | Fibronectin type-III 5 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000537320 | 18 | 20 | 1076_1165 | 1528 | 1565.0 | Domain | Fibronectin type-III 6 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000537320 | 18 | 20 | 1167_1256 | 1528 | 1565.0 | Domain | Fibronectin type-III 7 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000537320 | 18 | 20 | 1258_1350 | 1528 | 1565.0 | Domain | Fibronectin type-III 8 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000537320 | 18 | 20 | 1351_1439 | 1528 | 1565.0 | Domain | Fibronectin type-III 9 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000537320 | 18 | 20 | 1440_1531 | 1528 | 1565.0 | Domain | Fibronectin type-III 10 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000537320 | 18 | 20 | 174_186 | 1528 | 1565.0 | Domain | EGF-like 1%3B incomplete | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000537320 | 18 | 20 | 186_217 | 1528 | 1565.0 | Domain | EGF-like 2 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000537320 | 18 | 20 | 217_248 | 1528 | 1565.0 | Domain | EGF-like 3 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000537320 | 18 | 20 | 248_280 | 1528 | 1565.0 | Domain | EGF-like 4 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000537320 | 18 | 20 | 280_311 | 1528 | 1565.0 | Domain | EGF-like 5 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000537320 | 18 | 20 | 311_342 | 1528 | 1565.0 | Domain | EGF-like 6 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000537320 | 18 | 20 | 342_373 | 1528 | 1565.0 | Domain | EGF-like 7 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000537320 | 18 | 20 | 373_404 | 1528 | 1565.0 | Domain | EGF-like 8 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000537320 | 18 | 20 | 404_435 | 1528 | 1565.0 | Domain | EGF-like 9 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000537320 | 18 | 20 | 435_466 | 1528 | 1565.0 | Domain | EGF-like 10 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000537320 | 18 | 20 | 466_497 | 1528 | 1565.0 | Domain | EGF-like 11 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000537320 | 18 | 20 | 497_528 | 1528 | 1565.0 | Domain | EGF-like 12 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000537320 | 18 | 20 | 528_559 | 1528 | 1565.0 | Domain | EGF-like 13 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000537320 | 18 | 20 | 559_590 | 1528 | 1565.0 | Domain | EGF-like 14 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000537320 | 18 | 20 | 590_621 | 1528 | 1565.0 | Domain | EGF-like 15 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000537320 | 18 | 20 | 625_715 | 1528 | 1565.0 | Domain | Fibronectin type-III 1 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000537320 | 18 | 20 | 716_804 | 1528 | 1565.0 | Domain | Fibronectin type-III 2 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000537320 | 18 | 20 | 805_894 | 1528 | 1565.0 | Domain | Fibronectin type-III 3 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000537320 | 18 | 20 | 895_990 | 1528 | 1565.0 | Domain | Fibronectin type-III 4 | |
| Tgene | TNC | chr12:6646176 | chr9:117783546 | ENST00000537320 | 18 | 20 | 991_1075 | 1528 | 1565.0 | Domain | Fibronectin type-III 5 |
Top |
Fusion Gene Sequence for GAPDH-TNC |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >32420_32420_1_GAPDH-TNC_GAPDH_chr12_6646176_ENST00000229239_TNC_chr9_117783546_ENST00000350763_length(transcript)=1727nt_BP=993nt GGGGGAAGTGGGGGCTGGGAAGGAACCACGGGCCCCCGCCCGAGGCCCATGGGCCCCTCCTAGGCCTTTGCCTGAGCAGTCCGGTGTCAC TACCGCAGAGCCTCGAGGAGAAGTTCCCCAACTTTCCCGCCTCTCAGCCTTTGAAAGAAAGAAAGGGGAGGGGGCAGGCCGCGTGCAGCC GCGAGCGGTGCTGGGCTCCGGCTCCAATTCCCCATCTCAGTCGTTCCCAAAGTCCTCCTGTTTCATCCAAGCGTGTAAGGGTCCCCGTCC TTGACTCCCTAGTGTCCTGCTGCCCACAGTCCAGTCCTGGGAACCAGCACCGATCACCTCCCATCGGGCCAATCTCAGTCCCTTCCCCCC TACGTCGGGGCCCACACGCTCGGTGCGTGCCCAGTTGAACCAGGCGGCTGCGGAAAAAAAAAAGCGGGGAGAAAGTAGGGCCCGGCTACT AGCGGTTTTACGGGCGCACGTAGCTCAGGCCTCAAGACCTTGGGCTGGGACTGGCTGAGCCTGGCGGGAGGCGGGGTCCGAGTCACCGCC TGCCGCCGCGCCCCCGGTTTCTATAAATTGAGCCCGCAGCCTCCCGCTTCGCTCTCTGCTCCTCCTGTTCGACAGTCAGCCGCATCTTCT TTTGCGTCGCCAGCCGAGCCACATCGCTCAGACACCATGGGGAAGGTGAAGGTCGGAGTCAACGGATTTGGTCGTATTGGGCGCCTGGTC ACCAGGGCTGCTTTTAACTCTGGTAAAGTGGATATTGTTGCCATCAATGACCCCTTCATTGACCTCAACTACATGGTTTACATGTTCCAA TATGATTCCACCCATGGCAAATTCCATGGCACCGTCAAGGCTGAGAACGGGAAGCTTGTCATCAATGGAAATCCCATCACCATCTTCCAG GAGCGAGATCCCTCCAAAATCAAGTGGGGCGATGCTGGCGCTGAGTACGTCGTGGAGTCCACTGGCGTCTTCACCACCATGGAGAAGGCT GGGGGCGTTAACTGGTTCCACTGGAAGGGCCACGAACACTCAATCCAGTTTGCTGAGATGAAGCTGAGACCAAGCAACTTCAGAAATCTT GAAGGCAGGCGCAAACGGGCATAAATTCCAGGGACCACTGGGTGAGAGAGGAATAAGGCCCAGAGCGAGGAAAGGATTTTACCAAAGCAT CAATACAACCAGCCCAACCATCGGTCCACACCTGGGCATTTGGTGAGAGTCAAAGCTGACCATGGATCCCTGGGGCCAACGGCAACAGCA TGGGCCTCACCTCCTCTGTGATTTCTTTCTTTGCACCAAAGACATCAGTCTCCAACATGTTTCTGTTTTGTTGTTTGATTCAGCAAAAAT CTCCCAGTGACAACATCGCAATAGTTTTTTACTTCTCTTAGGTGGCTCTGGGAATGGGAGAGGGGTAGGATGTACAGGGGTAGTTTGTTT TAGAACCAGCCGTATTTTACATGAAGCTGTATAATTAATTGTCATTATTTTTGTTAGCAAAGATTAAATGTGTCATTGGAAGCCATCCCT TTTTTTACATTTCATACAACAGAAACCAGAAAAGCAATACTGTTTCCATTTTAAGGATATGATTAATATTATTAATATAATAATGATGAT GATGATGATGAAAACTAAGGATTTTTCAAGAGATCTTTCTTTCCAAAACATTTCTGGACAGTACCTGATTGTATTTTTTTTTTAAATAAA >32420_32420_1_GAPDH-TNC_GAPDH_chr12_6646176_ENST00000229239_TNC_chr9_117783546_ENST00000350763_length(amino acids)=204AA_BP=168 MGWDWLSLAGGGVRVTACRRAPGFYKLSPQPPASLSAPPVRQSAASSFASPAEPHRSDTMGKVKVGVNGFGRIGRLVTRAAFNSGKVDIV AINDPFIDLNYMVYMFQYDSTHGKFHGTVKAENGKLVINGNPITIFQERDPSKIKWGDAGAEYVVESTGVFTTMEKAGGVNWFHWKGHEH -------------------------------------------------------------- >32420_32420_2_GAPDH-TNC_GAPDH_chr12_6646176_ENST00000396856_TNC_chr9_117783546_ENST00000350763_length(transcript)=1149nt_BP=415nt TGTTCGACAGTCAGCCGCATCTTCTTTTGCGTCGCCAGCCGAGCCACATCGCTCAGACACCATGGGGAAGGTGAAGGTCGGAGTCAACGG ATTTGGTCGTATTGGGCGCCTGGTCACCAGGGCTGCTTTTAACTCTGGTAAAGTGGATATTGTTGCCATCAATGACCCCTTCATTGACCT CAACTACATGGTTTACATGTTCCAATATGATTCCACCCATGGCAAATTCCATGGCACCGTCAAGGCTGAGAACGGGAAGCTTGTCATCAA TGGAAATCCCATCACCATCTTCCAGGAGTGAGTGGAAGACAGAATGGAAGAAATGCGAGATCCCTCCAAAATCAAGTGGGGCGATGCTGG CGCTGAGTACGTCGTGGAGTCCACTGGCGTCTTCACCACCATGGAGAAGGCTGGGGGCGTTAACTGGTTCCACTGGAAGGGCCACGAACA CTCAATCCAGTTTGCTGAGATGAAGCTGAGACCAAGCAACTTCAGAAATCTTGAAGGCAGGCGCAAACGGGCATAAATTCCAGGGACCAC TGGGTGAGAGAGGAATAAGGCCCAGAGCGAGGAAAGGATTTTACCAAAGCATCAATACAACCAGCCCAACCATCGGTCCACACCTGGGCA TTTGGTGAGAGTCAAAGCTGACCATGGATCCCTGGGGCCAACGGCAACAGCATGGGCCTCACCTCCTCTGTGATTTCTTTCTTTGCACCA AAGACATCAGTCTCCAACATGTTTCTGTTTTGTTGTTTGATTCAGCAAAAATCTCCCAGTGACAACATCGCAATAGTTTTTTACTTCTCT TAGGTGGCTCTGGGAATGGGAGAGGGGTAGGATGTACAGGGGTAGTTTGTTTTAGAACCAGCCGTATTTTACATGAAGCTGTATAATTAA TTGTCATTATTTTTGTTAGCAAAGATTAAATGTGTCATTGGAAGCCATCCCTTTTTTTACATTTCATACAACAGAAACCAGAAAAGCAAT ACTGTTTCCATTTTAAGGATATGATTAATATTATTAATATAATAATGATGATGATGATGATGAAAACTAAGGATTTTTCAAGAGATCTTT >32420_32420_2_GAPDH-TNC_GAPDH_chr12_6646176_ENST00000396856_TNC_chr9_117783546_ENST00000350763_length(amino acids)=126AA_BP=108 MVVLGAWSPGLLLTLVKWILLPSMTPSLTSTTWFTCSNMIPPMANSMAPSRLRTGSLSSMEIPSPSSRSEWKTEWKKCEIPPKSSGAMLA -------------------------------------------------------------- >32420_32420_3_GAPDH-TNC_GAPDH_chr12_6646176_ENST00000396858_TNC_chr9_117783546_ENST00000350763_length(transcript)=1200nt_BP=466nt TGCAACCGGGAAGGAAATGAATGGGCAGCCGTTAGGAAAGCCTGCCGGTGACTAACCCTGCGCTCCTGCCTCGATGGGTGGAGTCGCGTG TGGCGGGGAAGTCAGGTGGAGCGAGGCTAGCTGGCCCGATTTCTCCTCCGGGTGATGCTTTTCCTAGATTATTCTCTGATTTGGTCGTAT TGGGCGCCTGGTCACCAGGGCTGCTTTTAACTCTGGTAAAGTGGATATTGTTGCCATCAATGACCCCTTCATTGACCTCAACTACATGGT TTACATGTTCCAATATGATTCCACCCATGGCAAATTCCATGGCACCGTCAAGGCTGAGAACGGGAAGCTTGTCATCAATGGAAATCCCAT CACCATCTTCCAGGAGCGAGATCCCTCCAAAATCAAGTGGGGCGATGCTGGCGCTGAGTACGTCGTGGAGTCCACTGGCGTCTTCACCAC CATGGAGAAGGCTGGGGGCGTTAACTGGTTCCACTGGAAGGGCCACGAACACTCAATCCAGTTTGCTGAGATGAAGCTGAGACCAAGCAA CTTCAGAAATCTTGAAGGCAGGCGCAAACGGGCATAAATTCCAGGGACCACTGGGTGAGAGAGGAATAAGGCCCAGAGCGAGGAAAGGAT TTTACCAAAGCATCAATACAACCAGCCCAACCATCGGTCCACACCTGGGCATTTGGTGAGAGTCAAAGCTGACCATGGATCCCTGGGGCC AACGGCAACAGCATGGGCCTCACCTCCTCTGTGATTTCTTTCTTTGCACCAAAGACATCAGTCTCCAACATGTTTCTGTTTTGTTGTTTG ATTCAGCAAAAATCTCCCAGTGACAACATCGCAATAGTTTTTTACTTCTCTTAGGTGGCTCTGGGAATGGGAGAGGGGTAGGATGTACAG GGGTAGTTTGTTTTAGAACCAGCCGTATTTTACATGAAGCTGTATAATTAATTGTCATTATTTTTGTTAGCAAAGATTAAATGTGTCATT GGAAGCCATCCCTTTTTTTACATTTCATACAACAGAAACCAGAAAAGCAATACTGTTTCCATTTTAAGGATATGATTAATATTATTAATA TAATAATGATGATGATGATGATGAAAACTAAGGATTTTTCAAGAGATCTTTCTTTCCAAAACATTTCTGGACAGTACCTGATTGTATTTT >32420_32420_3_GAPDH-TNC_GAPDH_chr12_6646176_ENST00000396858_TNC_chr9_117783546_ENST00000350763_length(amino acids)=160AA_BP=142 MPVTNPALLPRWVESRVAGKSGGARLAGPISPPGDAFPRLFSDLVVLGAWSPGLLLTLVKWILLPSMTPSLTSTTWFTCSNMIPPMANSM -------------------------------------------------------------- >32420_32420_4_GAPDH-TNC_GAPDH_chr12_6646176_ENST00000396859_TNC_chr9_117783546_ENST00000350763_length(transcript)=1140nt_BP=406nt AGGCGTGTGTGTCGGCCGGGGCCACTAGGCGCTCACTGTTCTCTCCCTCCGCGCAGCCGAGCCACATCGCTCAGACACCATGGGGAAGGT GAAGGTCGGAGTCAACGGATTTGGTCGTATTGGGCGCCTGGTCACCAGGGCTGCTTTTAACTCTGGTAAAGTGGATATTGTTGCCATCAA TGACCCCTTCATTGACCTCAACTACATGGTTTACATGTTCCAATATGATTCCACCCATGGCAAATTCCATGGCACCGTCAAGGCTGAGAA CGGGAAGCTTGTCATCAATGGAAATCCCATCACCATCTTCCAGGAGCGAGATCCCTCCAAAATCAAGTGGGGCGATGCTGGCGCTGAGTA CGTCGTGGAGTCCACTGGCGTCTTCACCACCATGGAGAAGGCTGGGGGCGTTAACTGGTTCCACTGGAAGGGCCACGAACACTCAATCCA GTTTGCTGAGATGAAGCTGAGACCAAGCAACTTCAGAAATCTTGAAGGCAGGCGCAAACGGGCATAAATTCCAGGGACCACTGGGTGAGA GAGGAATAAGGCCCAGAGCGAGGAAAGGATTTTACCAAAGCATCAATACAACCAGCCCAACCATCGGTCCACACCTGGGCATTTGGTGAG AGTCAAAGCTGACCATGGATCCCTGGGGCCAACGGCAACAGCATGGGCCTCACCTCCTCTGTGATTTCTTTCTTTGCACCAAAGACATCA GTCTCCAACATGTTTCTGTTTTGTTGTTTGATTCAGCAAAAATCTCCCAGTGACAACATCGCAATAGTTTTTTACTTCTCTTAGGTGGCT CTGGGAATGGGAGAGGGGTAGGATGTACAGGGGTAGTTTGTTTTAGAACCAGCCGTATTTTACATGAAGCTGTATAATTAATTGTCATTA TTTTTGTTAGCAAAGATTAAATGTGTCATTGGAAGCCATCCCTTTTTTTACATTTCATACAACAGAAACCAGAAAAGCAATACTGTTTCC ATTTTAAGGATATGATTAATATTATTAATATAATAATGATGATGATGATGATGAAAACTAAGGATTTTTCAAGAGATCTTTCTTTCCAAA >32420_32420_4_GAPDH-TNC_GAPDH_chr12_6646176_ENST00000396859_TNC_chr9_117783546_ENST00000350763_length(amino acids)=145AA_BP=109 MGKVKVGVNGFGRIGRLVTRAAFNSGKVDIVAINDPFIDLNYMVYMFQYDSTHGKFHGTVKAENGKLVINGNPITIFQERDPSKIKWGDA -------------------------------------------------------------- >32420_32420_5_GAPDH-TNC_GAPDH_chr12_6646176_ENST00000396861_TNC_chr9_117783546_ENST00000350763_length(transcript)=1212nt_BP=478nt TTCGACAGTCAGCCGCATCTTCTTTTGCGTCGCCAGGTGAAGACGGGCGGAGAGAAACCCGGGAGGCTAGGGACGGCCTGAAGGCGGCAG GGGCGGGCGCAGGCCGGATGTGTTCGCGCCGCTGCGGGCCGAGCCACATCGCTCAGACACCATGGGGAAGGTGAAGGTCGGAGTCAACGG ATTTGGTCGTATTGGGCGCCTGGTCACCAGGGCTGCTTTTAACTCTGGTAAAGTGGATATTGTTGCCATCAATGACCCCTTCATTGACCT CAACTACATGGTTTACATGTTCCAATATGATTCCACCCATGGCAAATTCCATGGCACCGTCAAGGCTGAGAACGGGAAGCTTGTCATCAA TGGAAATCCCATCACCATCTTCCAGGAGCGAGATCCCTCCAAAATCAAGTGGGGCGATGCTGGCGCTGAGTACGTCGTGGAGTCCACTGG CGTCTTCACCACCATGGAGAAGGCTGGGGGCGTTAACTGGTTCCACTGGAAGGGCCACGAACACTCAATCCAGTTTGCTGAGATGAAGCT GAGACCAAGCAACTTCAGAAATCTTGAAGGCAGGCGCAAACGGGCATAAATTCCAGGGACCACTGGGTGAGAGAGGAATAAGGCCCAGAG CGAGGAAAGGATTTTACCAAAGCATCAATACAACCAGCCCAACCATCGGTCCACACCTGGGCATTTGGTGAGAGTCAAAGCTGACCATGG ATCCCTGGGGCCAACGGCAACAGCATGGGCCTCACCTCCTCTGTGATTTCTTTCTTTGCACCAAAGACATCAGTCTCCAACATGTTTCTG TTTTGTTGTTTGATTCAGCAAAAATCTCCCAGTGACAACATCGCAATAGTTTTTTACTTCTCTTAGGTGGCTCTGGGAATGGGAGAGGGG TAGGATGTACAGGGGTAGTTTGTTTTAGAACCAGCCGTATTTTACATGAAGCTGTATAATTAATTGTCATTATTTTTGTTAGCAAAGATT AAATGTGTCATTGGAAGCCATCCCTTTTTTTACATTTCATACAACAGAAACCAGAAAAGCAATACTGTTTCCATTTTAAGGATATGATTA ATATTATTAATATAATAATGATGATGATGATGATGAAAACTAAGGATTTTTCAAGAGATCTTTCTTTCCAAAACATTTCTGGACAGTACC >32420_32420_5_GAPDH-TNC_GAPDH_chr12_6646176_ENST00000396861_TNC_chr9_117783546_ENST00000350763_length(amino acids)=155AA_BP=119 MRAEPHRSDTMGKVKVGVNGFGRIGRLVTRAAFNSGKVDIVAINDPFIDLNYMVYMFQYDSTHGKFHGTVKAENGKLVINGNPITIFQER -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for GAPDH-TNC |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
| Hgene | GAPDH | chr12:6646176 | chr9:117783546 | ENST00000229239 | + | 5 | 9 | 2_148 | 109.0 | 336.0 | WARS1 |
| Hgene | GAPDH | chr12:6646176 | chr9:117783546 | ENST00000396858 | + | 4 | 8 | 2_148 | 67.0 | 294.0 | WARS1 |
| Hgene | GAPDH | chr12:6646176 | chr9:117783546 | ENST00000396859 | + | 4 | 8 | 2_148 | 109.0 | 336.0 | WARS1 |
| Hgene | GAPDH | chr12:6646176 | chr9:117783546 | ENST00000396861 | + | 5 | 9 | 2_148 | 109.0 | 336.0 | WARS1 |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for GAPDH-TNC |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for GAPDH-TNC |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
