
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:GBF1-WBP1L (FusionGDB2 ID:32730) |
Fusion Gene Summary for GBF1-WBP1L |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: GBF1-WBP1L | Fusion gene ID: 32730 | Hgene | Tgene | Gene symbol | GBF1 | WBP1L | Gene ID | 80142 | 54838 |
| Gene name | prostaglandin E synthase 2 | WW domain binding protein 1 like | |
| Synonyms | C9orf15|GBF-1|GBF1|PGES2|mPGES-2 | C10orf26|OPA1L|OPAL1 | |
| Cytomap | 9q34.11 | 10q24.32 | |
| Type of gene | protein-coding | protein-coding | |
| Description | prostaglandin E synthase 2GATE-binding factor 1gamma-interferon-activated transcriptional element-binding factor 1mPGE synthase-2membrane-associated prostaglandin E synthase 2microsomal prostaglandin E synthase-2prostaglandin-H(2) E-isomerase | WW domain binding protein 1-likeoutcome predictor in acute leukemia 1 | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | Q92538 | Q9NX94 | |
| Ensembl transtripts involved in fusion gene | ENST00000369983, ENST00000476019, | ENST00000369889, ENST00000448841, | |
| Fusion gene scores | * DoF score | 15 X 10 X 9=1350 | 7 X 8 X 3=168 |
| # samples | 16 | 9 | |
| ** MAII score | log2(16/1350*10)=-3.07681559705083 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(9/168*10)=-0.900464326449086 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: GBF1 [Title/Abstract] AND WBP1L [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | GBF1(104103939)-WBP1L(104569650), # samples:2 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
 Fusion gene breakpoints across GBF1 (5'-gene) Fusion gene breakpoints across GBF1 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
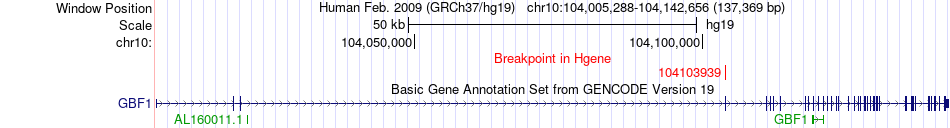 |
 Fusion gene breakpoints across WBP1L (3'-gene) Fusion gene breakpoints across WBP1L (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
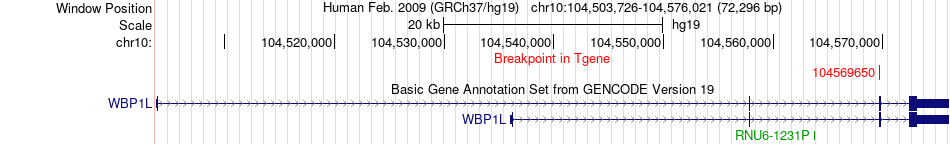 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | PRAD | TCGA-G9-6364-01A | GBF1 | chr10 | 104103939 | + | WBP1L | chr10 | 104569650 | + |
| ChimerDB4 | PRAD | TCGA-G9-6364 | GBF1 | chr10 | 104103939 | + | WBP1L | chr10 | 104569650 | + |
Top |
Fusion Gene ORF analysis for GBF1-WBP1L |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| In-frame | ENST00000369983 | ENST00000369889 | GBF1 | chr10 | 104103939 | + | WBP1L | chr10 | 104569650 | + |
| In-frame | ENST00000369983 | ENST00000448841 | GBF1 | chr10 | 104103939 | + | WBP1L | chr10 | 104569650 | + |
| intron-3CDS | ENST00000476019 | ENST00000369889 | GBF1 | chr10 | 104103939 | + | WBP1L | chr10 | 104569650 | + |
| intron-3CDS | ENST00000476019 | ENST00000448841 | GBF1 | chr10 | 104103939 | + | WBP1L | chr10 | 104569650 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000369983 | GBF1 | chr10 | 104103939 | + | ENST00000448841 | WBP1L | chr10 | 104569650 | + | 4363 | 555 | 260 | 1453 | 397 |
| ENST00000369983 | GBF1 | chr10 | 104103939 | + | ENST00000369889 | WBP1L | chr10 | 104569650 | + | 4387 | 555 | 260 | 1453 | 397 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000369983 | ENST00000448841 | GBF1 | chr10 | 104103939 | + | WBP1L | chr10 | 104569650 | + | 0.005409883 | 0.99459016 |
| ENST00000369983 | ENST00000369889 | GBF1 | chr10 | 104103939 | + | WBP1L | chr10 | 104569650 | + | 0.005066453 | 0.99493355 |
Top |
Fusion Genomic Features for GBF1-WBP1L |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| GBF1 | chr10 | 104103939 | + | WBP1L | chr10 | 104569649 | + | 2.27E-08 | 1 |
| GBF1 | chr10 | 104103939 | + | WBP1L | chr10 | 104569649 | + | 2.27E-08 | 1 |
| GBF1 | chr10 | 104103939 | + | WBP1L | chr10 | 104569649 | + | 2.27E-08 | 1 |
| GBF1 | chr10 | 104103939 | + | WBP1L | chr10 | 104569649 | + | 2.27E-08 | 1 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
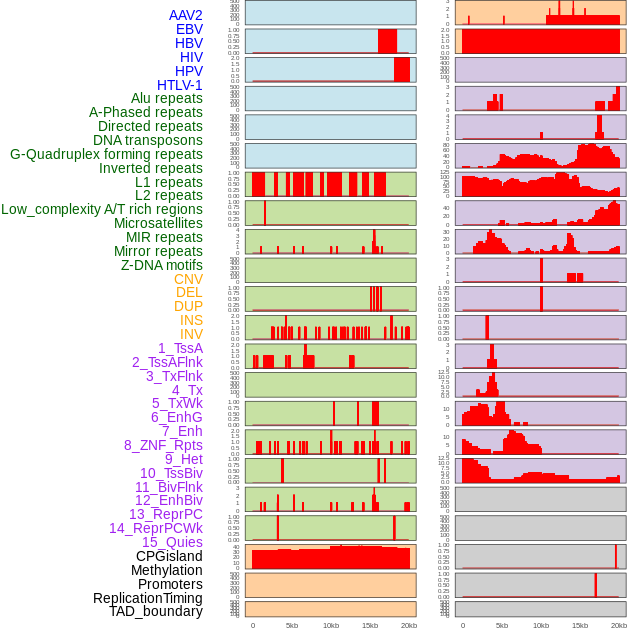 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
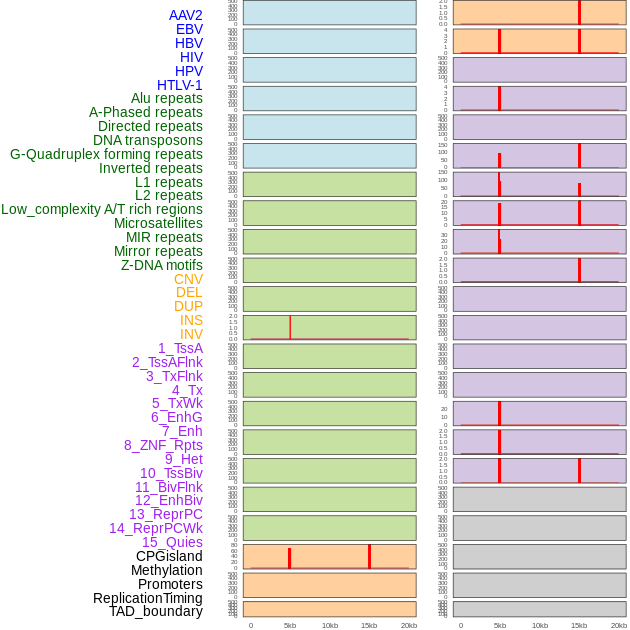 |
Top |
Fusion Protein Features for GBF1-WBP1L |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr10:104103939/chr10:104569650) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| GBF1 | WBP1L |
| FUNCTION: Guanine-nucleotide exchange factor (GEF) for members of the Arf family of small GTPases involved in trafficking in the early secretory pathway; its GEF activity initiates the coating of nascent vesicles via the localized generation of activated ARFs through replacement of GDP with GTP. Recruitment to cis-Golgi membranes requires membrane association of Arf-GDP and can be regulated by ARF1, ARF3, ARF4 and ARF5. Involved in the recruitment of the COPI coat complex to the endoplasmic reticulum exit sites (ERES), and the endoplasmic reticulum-Golgi intermediate (ERGIC) and cis-Golgi compartments which implicates ARF1 activation. Involved in COPI vesicle-dependent retrograde transport from the ERGIC and cis-Golgi compartments to the endoplasmic reticulum (ER) (PubMed:16926190, PubMed:17956946, PubMed:18003980, PubMed:12047556, PubMed:12808027, PubMed:19039328, PubMed:24213530). Involved in the trans-Golgi network recruitment of GGA1, GGA2, GGA3, BIG1, BIG2, and the AP-1 adapter protein complex related to chlathrin-dependent transport; the function requires its GEF activity (probably at least in part on ARF4 and ARF5) (PubMed:23386609). Has GEF activity towards ARF1 (PubMed:15616190). Has in vitro GEF activity towards ARF5 (By similarity). Involved in the processing of PSAP (PubMed:17666033). Required for the assembly of the Golgi apparatus (PubMed:12808027, PubMed:18003980). The AMPK-phosphorylated form is involved in Golgi disassembly during mitotis and under stress conditions (PubMed:18063581, PubMed:23418352). May be involved in the COPI vesicle-dependent recruitment of PNPLA2 to lipid droplets; however, this function is under debate (PubMed:19461073, PubMed:22185782). In neutrophils, involved in G protein-coupled receptor (GPCR)-mediated chemotaxis und superoxide production. Proposed to be recruited by phosphatidylinositol-phosphates generated upon GPCR stimulation to the leading edge where it recruits and activates ARF1, and is involved in recruitment of GIT2 and the NADPH oxidase complex (PubMed:22573891). Plays a role in maintaining mitochondrial morphology (PubMed:25190516). {ECO:0000250|UniProtKB:Q9R1D7, ECO:0000269|PubMed:12047556, ECO:0000269|PubMed:12808027, ECO:0000269|PubMed:15616190, ECO:0000269|PubMed:16926190, ECO:0000269|PubMed:17666033, ECO:0000269|PubMed:17956946, ECO:0000269|PubMed:18003980, ECO:0000269|PubMed:18063581, ECO:0000269|PubMed:19461073, ECO:0000269|PubMed:22185782, ECO:0000269|PubMed:22573891, ECO:0000269|PubMed:23386609, ECO:0000269|PubMed:23418352, ECO:0000269|PubMed:24213530, ECO:0000269|PubMed:25190516, ECO:0000305|PubMed:19039328, ECO:0000305|PubMed:22573891}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | WBP1L | chr10:104103939 | chr10:104569650 | ENST00000369889 | 1 | 4 | 101_183 | 43 | 343.0 | Compositional bias | Note=Pro-rich | |
| Tgene | WBP1L | chr10:104103939 | chr10:104569650 | ENST00000369889 | 1 | 4 | 335_342 | 43 | 343.0 | Compositional bias | Note=Poly-Ser | |
| Tgene | WBP1L | chr10:104103939 | chr10:104569650 | ENST00000448841 | 1 | 4 | 101_183 | 64 | 364.0 | Compositional bias | Note=Pro-rich | |
| Tgene | WBP1L | chr10:104103939 | chr10:104569650 | ENST00000448841 | 1 | 4 | 335_342 | 64 | 364.0 | Compositional bias | Note=Poly-Ser | |
| Tgene | WBP1L | chr10:104103939 | chr10:104569650 | ENST00000369889 | 1 | 4 | 42_62 | 43 | 343.0 | Transmembrane | Helical |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | GBF1 | chr10:104103939 | chr10:104569650 | ENST00000369983 | + | 4 | 40 | 1751_1854 | 98 | 1860.0 | Compositional bias | Note=Pro-rich |
| Hgene | GBF1 | chr10:104103939 | chr10:104569650 | ENST00000369983 | + | 4 | 40 | 692_882 | 98 | 1860.0 | Domain | SEC7 |
| Hgene | GBF1 | chr10:104103939 | chr10:104569650 | ENST00000369983 | + | 4 | 40 | 886_1370 | 98 | 1860.0 | Region | Phosphatidylinositol-phosphate binding%3B required for translocation to the leading edge and for ARF1 activation upon GPCR signaling |
| Tgene | WBP1L | chr10:104103939 | chr10:104569650 | ENST00000448841 | 1 | 4 | 42_62 | 64 | 364.0 | Transmembrane | Helical |
Top |
Fusion Gene Sequence for GBF1-WBP1L |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >32730_32730_1_GBF1-WBP1L_GBF1_chr10_104103939_ENST00000369983_WBP1L_chr10_104569650_ENST00000369889_length(transcript)=4387nt_BP=555nt AGTTACCACGTCAGTGAGCTGACAGGGAAGGTACCCGGCTCTACTGCCCGTCCCGGACGACTCATCCTGGCGGACTGTGCAGTCCCACCC TGACTTCGGCCTCAATTTCCAGGAAACAGGCTCCTTCTCTTCTCCCATCTGCTACCAGAGCCGGGAGAGCTGCTCGGAGACGCCTCCGGG GTGCGGGCTGGACATGAGCAGCGGCTGCCGGTCCTGGGACTAGGCCCCGCCATTTTGGATCCGCTGACAGGTTTGCCAAGATGGTGGATA AGAATATTTACATCATTCAAGGGGAGATTAACATTGTGGTTGGGGCCATCAAACGAAATGCCCGATGGAGCACCCATACACCACTGGATG AAGAACGGGATCCTCTGCTGCATAGTTTCGGTCATCTAAAGGAGGTTTTAAACAGTATAACAGAACTCTCAGAAATTGAGCCCAATGTAT TCCTTCGACCTTTTCTGGAAGTGATTCGCTCTGAAGATACCACTGGCCCTATCACTGGACTGGCACTCACCTCTGTCAACAAGTTCCTGT CCTATGCACTCATAGGGTTCTGGCTGGTGTGGACCATCATCATCATCCTGAGCTGCTGCTGTGTTTGCCACCACCGCCGAGCCAAGCACC GCCTTCAGGCCCAGCAGCGGCAACATGAAATCAACCTGATCGCTTACCGAGAAGCCCACAATTACTCAGCGCTGCCATTTTATTTCAGGT TTTTGCCAAACTATTTACTACCTCCTTATGAGGAAGTGGTGAACCGACCTCCAACTCCTCCCCCACCATACAGTGCCTTCCAGCTACAGC AGCAGCAGCTGCTGCCTCCACAGTGTGGCCCTGCAGGTGGCAGTCCCCCGGGCATCGATCCCACCAGGGGATCCCAGGGGGCACAGAGCA GCCCCTTGTCTGAGCCCAGCAGAAGCAGCACAAGACCCCCAAGCATCGCTGACCCTGATCCCTCTGACCTACCAGTTGACCGAGCAGCCA CCAAAGCCCCAGGGATGGAGCCCAGTGGCTCTGTGGCTGGCCTGGGGGAGCTGGACCCGGGGGCCTTCCTGGACAAAGATGCAGAATGTA GGGAGGAGCTGCTGAAAGATGACAGCTCTGAACACGGCGCACCCGACAGCAAAGAGAAGACGCCTGGGAGACATCGCCGCTTCACAGGTG ACTCGGGCATTGAAGTGTGTGTGTGCAACCGGGGCCACCATGACGATGACCTCAAAGAGTTCAACACACTCATCGATGATGCTCTGGATG GGCCCCTGGACTTCTGCGACAGCTGCCATGTGCGGCCCCCTGGTGATGAGGAGGAAGGCCTCTGTCAGTCCTCTGAGGAGCAGGCTCGAG AGCCTGGGCACCCGCACCTGCCACGGCCGCCCGCATGCCTGCTGCTGAACACCATCAACGAGCAGGACTCTCCCAACTCCCAGAGCAGCA GCTCCCCCAGCTAGAGCAGGTCCTGCCAGCACCCAGCAACTTGGCAAAGCAACCAGGGTAGGGGAGAACCACGAGAGAAGCATTAAGTGA CTTTCAAAGACTTTCAGAGTACAGCCACTTGGTTCCTTTTTGTTTGTTTTCCTTCTCCTCTCCTGCATTTTCCTCCATCTCCAGGTACAG TTCGGGGTGTGGATGCCTCTTCCTCCACAAGGGCACAGTGTTGTGGAGGGCTAAGTTGGTTCTGTGACTCATTCCTCATACCCTAACTCC ATCTCCTTTCTTTAAAGTCAAATCTCACCTACCTGTTTGGGTCAGAGAGATGTGTTTTAAAAGCCCCCAAGGAAGGAGGCTGGGACTGTG CCCTGACATGATTCTTGGTGATGGAATAGGTTTGTGCTCTGATTCTAGTTTAAGAGAACGTTGCTGTATCTCAGTCCAGGAGAGGCAGCC CATCTTGGCCCTGGATGAAGAAGGAAACCCACAGAGGCCCAGGGCTTGTCATTGGGCTGCCAGTGTCTGCCAAGCCAGCATTGAGCTAAT CCTGTGGGAGGATGAGAGCTACTGGGCCGTTGTATGATAGGTTGGTAGGGGCTTGTTGATCTGTCAAATTCCAGGTGACAAGATCTATGC ACCCCATGCGTCCTTGAGGGGCCTCTTCCCCGCAGGCTCTGGCTGGCCGCAGGCTGGTTCTGGTGTGAAAGGTTATACTGCCTTTTCTTT GTTTGTTTGTTTTTTTCTCTAAAAACAAACAGCAAAAGACAGCTGAAAACAAGAACTTCACCGGTGGGCAGGCAAGAATTCTCTTCTGGA AAATGACGTTTGTGGCTCTTTCCCAAGTTGGCCTTCAAAGAGCCTGCCTGCTGTTGAGCCAGAAGATGTCTCGTGTGAAGGCTGGGGTGG CGGCTGTCTTGGAACCTCTGTGAGCAGGAGGCCCTAAGCCGCAGCAGTGGATAGAGGTGCAGCTCTCTGCCTCTCTGCCCTTTGGTCTGT GTTCACAGGTGACCCGTGTCAGCCTGCATCGCAAGCACACACCCTGCGGGCCTTCAAGTCTCACTGTTCCGTATGAGGAAACAGACAGCG GACTGAGGAAGCGATGGCCCCAGAGAAAGGGCCCCTGTAGCCTGGCTCTCACACAGTATTTTATCTTTGATTCTGAATAAATATTTTTTG TGGGGTTTTTTTTTTTTTTTTGGTGGCAGTTGTTTGTTTTAAACTGACCACTTGGAAGAAACACCTTGGTTATCTGTGGTTTTCATGCCT TGTCCCTGCCTCTACCCCCACCCCTTTTGAGTCGGGTGACTCATTTTTCTGTGTAGAGACTCGGTGGCCCAGGCAGGAGGTGAAAGCAGC CATCCGGAAGGCCCTGGGGACCCTTGTGCCTGTTGCTCGCCTTCAGGTCACCAGCTGAGCTGCGATAGGAAAATCTGAATGGAGGCAGCA AACAGCCAAAACAAACATTCCCCACCCGGCCCTGTGCATATGAAGTCTTTCTTCCCCCAACTCTTGAACGATGATGATATTCAGACGAAG CATTGATGTTATGGAAGAAAGAAAGAAACAAACAAAATATATATATATATGTCCAAAAACAGACAAATCCAAGGGTGTGAGGTAAACGAG TGTCTGCATTTAGATTCCACAAAACCAAAATCCATGTTGAACAAAGTTAAGTCCGTACACAGTGACTTTTTGGGTGAGCCGTGTGTGTCT GTCTGTTGTGTGTGTGCCTCAAGCCCTGTTTTCCTGTGAAGATACTTTGAGTGGCAGCCATTCTCTCCACGTGAACCACACGTCTGGAGC ACAGACAGGCCTCTCAAGGTCATTGATCTTACGCATTTACTGTTTACCGAACAAATGTCTGACTGTGTACTCGGGTGTACTCCGCAGCAT TGTCGACTGCAGTCCCCTGTGTTTGCCAGAGATACTGTGCTCGAAGTAGAGGTTTTACTCTACTCATCACTGCGATTTGCACATTGCTCC GTGGACACTCGGAGGCCTGCGTTCTGTTCCCTATAAATGGAAGCGTGCTCTGAGCCTGTCTGCCTCCCTCGGCTGCTGCTGGTCCTCAGT ACCAGCGCCCGGGGGTGTCCACAACCACTTGGGACAGAAGAAGGTGGAATTTCAGACAGAAGCTTGACTGGGTCTTCAATGACAGGCTTG GACTAGCTGTGGCCCAGACATCGGCCCTGCCCAGAATTGCCAGGAGGAGGCTTTGCAGGCTCTAGAGGAGCCGCAGGGCCTGCCTGCCTC TGGTGAGTCCAACAGGCACAAGCAAGCTGGCGTGTGGCCAGAGGTAGCCGGAGTGTGTCACAGCCCCTCAGATGCCTTTCCTTCCACCTT TTTTTATTTTTTAAGAATCCCAAATAACTCACTGAAGTGTCTCAAAGGCGAACAAGTTTTACCAAAATGAATCCTTTTTCAGTTAACAGA TCAAATGGATGAGTTCTGAGCCTCTCAAGTTCCTTTCCCCAGTTAGAGTGGGGAACTGGGCAAGTGTTAACTGTGGGACTCACTGCAGCG TCCTATCCTAAAGGCACGAGAAGACGGAAATGCAACCTGCGGAGCTGGGCTTGGTTCCCAGGTCACAGTTTGGCCCCCGCTACAGGATGC TGCCCTGCTCAGAGAGAGATTTAATAGGGAGCTGAAGGAATCGTTAGGGGGCCAGGGAGATGTGACTGAGGCTGGCTTTCCACGTGAATG AGACGGGGTCGGTGGAGGGTTTGGTGCTACAGCCAGTCAGAAGATTTGCAAATGCGAACACATTCCTGTGTGAGGCACGTTACCCTTTGT CAGTTATTGTGAATATGTGTATTTTAAGCAATAAGATTCAGCTGGTCAGACTTTTCTGGGCAGTCTCAGTGACGCATTTCCTGTGCTGTG >32730_32730_1_GBF1-WBP1L_GBF1_chr10_104103939_ENST00000369983_WBP1L_chr10_104569650_ENST00000369889_length(amino acids)=397AA_BP=0 MVDKNIYIIQGEINIVVGAIKRNARWSTHTPLDEERDPLLHSFGHLKEVLNSITELSEIEPNVFLRPFLEVIRSEDTTGPITGLALTSVN KFLSYALIGFWLVWTIIIILSCCCVCHHRRAKHRLQAQQRQHEINLIAYREAHNYSALPFYFRFLPNYLLPPYEEVVNRPPTPPPPYSAF QLQQQQLLPPQCGPAGGSPPGIDPTRGSQGAQSSPLSEPSRSSTRPPSIADPDPSDLPVDRAATKAPGMEPSGSVAGLGELDPGAFLDKD AECREELLKDDSSEHGAPDSKEKTPGRHRRFTGDSGIEVCVCNRGHHDDDLKEFNTLIDDALDGPLDFCDSCHVRPPGDEEEGLCQSSEE -------------------------------------------------------------- >32730_32730_2_GBF1-WBP1L_GBF1_chr10_104103939_ENST00000369983_WBP1L_chr10_104569650_ENST00000448841_length(transcript)=4363nt_BP=555nt AGTTACCACGTCAGTGAGCTGACAGGGAAGGTACCCGGCTCTACTGCCCGTCCCGGACGACTCATCCTGGCGGACTGTGCAGTCCCACCC TGACTTCGGCCTCAATTTCCAGGAAACAGGCTCCTTCTCTTCTCCCATCTGCTACCAGAGCCGGGAGAGCTGCTCGGAGACGCCTCCGGG GTGCGGGCTGGACATGAGCAGCGGCTGCCGGTCCTGGGACTAGGCCCCGCCATTTTGGATCCGCTGACAGGTTTGCCAAGATGGTGGATA AGAATATTTACATCATTCAAGGGGAGATTAACATTGTGGTTGGGGCCATCAAACGAAATGCCCGATGGAGCACCCATACACCACTGGATG AAGAACGGGATCCTCTGCTGCATAGTTTCGGTCATCTAAAGGAGGTTTTAAACAGTATAACAGAACTCTCAGAAATTGAGCCCAATGTAT TCCTTCGACCTTTTCTGGAAGTGATTCGCTCTGAAGATACCACTGGCCCTATCACTGGACTGGCACTCACCTCTGTCAACAAGTTCCTGT CCTATGCACTCATAGGGTTCTGGCTGGTGTGGACCATCATCATCATCCTGAGCTGCTGCTGTGTTTGCCACCACCGCCGAGCCAAGCACC GCCTTCAGGCCCAGCAGCGGCAACATGAAATCAACCTGATCGCTTACCGAGAAGCCCACAATTACTCAGCGCTGCCATTTTATTTCAGGT TTTTGCCAAACTATTTACTACCTCCTTATGAGGAAGTGGTGAACCGACCTCCAACTCCTCCCCCACCATACAGTGCCTTCCAGCTACAGC AGCAGCAGCTGCTGCCTCCACAGTGTGGCCCTGCAGGTGGCAGTCCCCCGGGCATCGATCCCACCAGGGGATCCCAGGGGGCACAGAGCA GCCCCTTGTCTGAGCCCAGCAGAAGCAGCACAAGACCCCCAAGCATCGCTGACCCTGATCCCTCTGACCTACCAGTTGACCGAGCAGCCA CCAAAGCCCCAGGGATGGAGCCCAGTGGCTCTGTGGCTGGCCTGGGGGAGCTGGACCCGGGGGCCTTCCTGGACAAAGATGCAGAATGTA GGGAGGAGCTGCTGAAAGATGACAGCTCTGAACACGGCGCACCCGACAGCAAAGAGAAGACGCCTGGGAGACATCGCCGCTTCACAGGTG ACTCGGGCATTGAAGTGTGTGTGTGCAACCGGGGCCACCATGACGATGACCTCAAAGAGTTCAACACACTCATCGATGATGCTCTGGATG GGCCCCTGGACTTCTGCGACAGCTGCCATGTGCGGCCCCCTGGTGATGAGGAGGAAGGCCTCTGTCAGTCCTCTGAGGAGCAGGCTCGAG AGCCTGGGCACCCGCACCTGCCACGGCCGCCCGCATGCCTGCTGCTGAACACCATCAACGAGCAGGACTCTCCCAACTCCCAGAGCAGCA GCTCCCCCAGCTAGAGCAGGTCCTGCCAGCACCCAGCAACTTGGCAAAGCAACCAGGGTAGGGGAGAACCACGAGAGAAGCATTAAGTGA CTTTCAAAGACTTTCAGAGTACAGCCACTTGGTTCCTTTTTGTTTGTTTTCCTTCTCCTCTCCTGCATTTTCCTCCATCTCCAGGTACAG TTCGGGGTGTGGATGCCTCTTCCTCCACAAGGGCACAGTGTTGTGGAGGGCTAAGTTGGTTCTGTGACTCATTCCTCATACCCTAACTCC ATCTCCTTTCTTTAAAGTCAAATCTCACCTACCTGTTTGGGTCAGAGAGATGTGTTTTAAAAGCCCCCAAGGAAGGAGGCTGGGACTGTG CCCTGACATGATTCTTGGTGATGGAATAGGTTTGTGCTCTGATTCTAGTTTAAGAGAACGTTGCTGTATCTCAGTCCAGGAGAGGCAGCC CATCTTGGCCCTGGATGAAGAAGGAAACCCACAGAGGCCCAGGGCTTGTCATTGGGCTGCCAGTGTCTGCCAAGCCAGCATTGAGCTAAT CCTGTGGGAGGATGAGAGCTACTGGGCCGTTGTATGATAGGTTGGTAGGGGCTTGTTGATCTGTCAAATTCCAGGTGACAAGATCTATGC ACCCCATGCGTCCTTGAGGGGCCTCTTCCCCGCAGGCTCTGGCTGGCCGCAGGCTGGTTCTGGTGTGAAAGGTTATACTGCCTTTTCTTT GTTTGTTTGTTTTTTTCTCTAAAAACAAACAGCAAAAGACAGCTGAAAACAAGAACTTCACCGGTGGGCAGGCAAGAATTCTCTTCTGGA AAATGACGTTTGTGGCTCTTTCCCAAGTTGGCCTTCAAAGAGCCTGCCTGCTGTTGAGCCAGAAGATGTCTCGTGTGAAGGCTGGGGTGG CGGCTGTCTTGGAACCTCTGTGAGCAGGAGGCCCTAAGCCGCAGCAGTGGATAGAGGTGCAGCTCTCTGCCTCTCTGCCCTTTGGTCTGT GTTCACAGGTGACCCGTGTCAGCCTGCATCGCAAGCACACACCCTGCGGGCCTTCAAGTCTCACTGTTCCGTATGAGGAAACAGACAGCG GACTGAGGAAGCGATGGCCCCAGAGAAAGGGCCCCTGTAGCCTGGCTCTCACACAGTATTTTATCTTTGATTCTGAATAAATATTTTTTG TGGGGTTTTTTTTTTTTTTTTGGTGGCAGTTGTTTGTTTTAAACTGACCACTTGGAAGAAACACCTTGGTTATCTGTGGTTTTCATGCCT TGTCCCTGCCTCTACCCCCACCCCTTTTGAGTCGGGTGACTCATTTTTCTGTGTAGAGACTCGGTGGCCCAGGCAGGAGGTGAAAGCAGC CATCCGGAAGGCCCTGGGGACCCTTGTGCCTGTTGCTCGCCTTCAGGTCACCAGCTGAGCTGCGATAGGAAAATCTGAATGGAGGCAGCA AACAGCCAAAACAAACATTCCCCACCCGGCCCTGTGCATATGAAGTCTTTCTTCCCCCAACTCTTGAACGATGATGATATTCAGACGAAG CATTGATGTTATGGAAGAAAGAAAGAAACAAACAAAATATATATATATATGTCCAAAAACAGACAAATCCAAGGGTGTGAGGTAAACGAG TGTCTGCATTTAGATTCCACAAAACCAAAATCCATGTTGAACAAAGTTAAGTCCGTACACAGTGACTTTTTGGGTGAGCCGTGTGTGTCT GTCTGTTGTGTGTGTGCCTCAAGCCCTGTTTTCCTGTGAAGATACTTTGAGTGGCAGCCATTCTCTCCACGTGAACCACACGTCTGGAGC ACAGACAGGCCTCTCAAGGTCATTGATCTTACGCATTTACTGTTTACCGAACAAATGTCTGACTGTGTACTCGGGTGTACTCCGCAGCAT TGTCGACTGCAGTCCCCTGTGTTTGCCAGAGATACTGTGCTCGAAGTAGAGGTTTTACTCTACTCATCACTGCGATTTGCACATTGCTCC GTGGACACTCGGAGGCCTGCGTTCTGTTCCCTATAAATGGAAGCGTGCTCTGAGCCTGTCTGCCTCCCTCGGCTGCTGCTGGTCCTCAGT ACCAGCGCCCGGGGGTGTCCACAACCACTTGGGACAGAAGAAGGTGGAATTTCAGACAGAAGCTTGACTGGGTCTTCAATGACAGGCTTG GACTAGCTGTGGCCCAGACATCGGCCCTGCCCAGAATTGCCAGGAGGAGGCTTTGCAGGCTCTAGAGGAGCCGCAGGGCCTGCCTGCCTC TGGTGAGTCCAACAGGCACAAGCAAGCTGGCGTGTGGCCAGAGGTAGCCGGAGTGTGTCACAGCCCCTCAGATGCCTTTCCTTCCACCTT TTTTTATTTTTTAAGAATCCCAAATAACTCACTGAAGTGTCTCAAAGGCGAACAAGTTTTACCAAAATGAATCCTTTTTCAGTTAACAGA TCAAATGGATGAGTTCTGAGCCTCTCAAGTTCCTTTCCCCAGTTAGAGTGGGGAACTGGGCAAGTGTTAACTGTGGGACTCACTGCAGCG TCCTATCCTAAAGGCACGAGAAGACGGAAATGCAACCTGCGGAGCTGGGCTTGGTTCCCAGGTCACAGTTTGGCCCCCGCTACAGGATGC TGCCCTGCTCAGAGAGAGATTTAATAGGGAGCTGAAGGAATCGTTAGGGGGCCAGGGAGATGTGACTGAGGCTGGCTTTCCACGTGAATG AGACGGGGTCGGTGGAGGGTTTGGTGCTACAGCCAGTCAGAAGATTTGCAAATGCGAACACATTCCTGTGTGAGGCACGTTACCCTTTGT CAGTTATTGTGAATATGTGTATTTTAAGCAATAAGATTCAGCTGGTCAGACTTTTCTGGGCAGTCTCAGTGACGCATTTCCTGTGCTGTG >32730_32730_2_GBF1-WBP1L_GBF1_chr10_104103939_ENST00000369983_WBP1L_chr10_104569650_ENST00000448841_length(amino acids)=397AA_BP=0 MVDKNIYIIQGEINIVVGAIKRNARWSTHTPLDEERDPLLHSFGHLKEVLNSITELSEIEPNVFLRPFLEVIRSEDTTGPITGLALTSVN KFLSYALIGFWLVWTIIIILSCCCVCHHRRAKHRLQAQQRQHEINLIAYREAHNYSALPFYFRFLPNYLLPPYEEVVNRPPTPPPPYSAF QLQQQQLLPPQCGPAGGSPPGIDPTRGSQGAQSSPLSEPSRSSTRPPSIADPDPSDLPVDRAATKAPGMEPSGSVAGLGELDPGAFLDKD AECREELLKDDSSEHGAPDSKEKTPGRHRRFTGDSGIEVCVCNRGHHDDDLKEFNTLIDDALDGPLDFCDSCHVRPPGDEEEGLCQSSEE -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for GBF1-WBP1L |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
| Hgene | GBF1 | chr10:104103939 | chr10:104569650 | ENST00000369983 | + | 4 | 40 | 1_380 | 98.33333333333333 | 1860.0 | RAB1B |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for GBF1-WBP1L |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for GBF1-WBP1L |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
