
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:GGCX-KCMF1 (FusionGDB2 ID:33021) |
Fusion Gene Summary for GGCX-KCMF1 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: GGCX-KCMF1 | Fusion gene ID: 33021 | Hgene | Tgene | Gene symbol | GGCX | KCMF1 | Gene ID | 2677 | 56888 |
| Gene name | gamma-glutamyl carboxylase | potassium channel modulatory factor 1 | |
| Synonyms | VKCFD1 | DEBT91|FIGC|PCMF|ZZZ1 | |
| Cytomap | 2p11.2 | 2p11.2 | |
| Type of gene | protein-coding | protein-coding | |
| Description | vitamin K-dependent gamma-carboxylasepeptidyl-glutamate 4-carboxylase | E3 ubiquitin-protein ligase KCMF1FGF-induced in gastric cancerFGF-induced ubiquitin-protein ligase in gastric cancersRING-type E3 ubiquitin transferase KCMF1ZZ-type zinc finger-containing protein 1differentially expressed in branching tubulogenesis 9 | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | P38435 | Q9P0J7 | |
| Ensembl transtripts involved in fusion gene | ENST00000233838, ENST00000430215, ENST00000473665, | ENST00000409785, | |
| Fusion gene scores | * DoF score | 3 X 3 X 2=18 | 5 X 6 X 3=90 |
| # samples | 3 | 8 | |
| ** MAII score | log2(3/18*10)=0.736965594166206 effective Gene in Pan-Cancer Fusion Genes (eGinPCFGs). DoF>8 and MAII>0 | log2(8/90*10)=-0.169925001442312 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: GGCX [Title/Abstract] AND KCMF1 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | KCMF1(85198590)-GGCX(85783383), # samples:2 GGCX(85787938)-KCMF1(85255012), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | KCMF1-GGCX seems lost the major protein functional domain in Hgene partner, which is a essential gene due to the frame-shifted ORF. KCMF1-GGCX seems lost the major protein functional domain in Hgene partner, which is a transcription factor due to the frame-shifted ORF. KCMF1-GGCX seems lost the major protein functional domain in Tgene partner, which is a cell metabolism gene due to the frame-shifted ORF. KCMF1-GGCX seems lost the major protein functional domain in Tgene partner, which is a essential gene due to the frame-shifted ORF. KCMF1-GGCX seems lost the major protein functional domain in Tgene partner, which is a IUPHAR drug target due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
 Fusion gene breakpoints across GGCX (5'-gene) Fusion gene breakpoints across GGCX (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
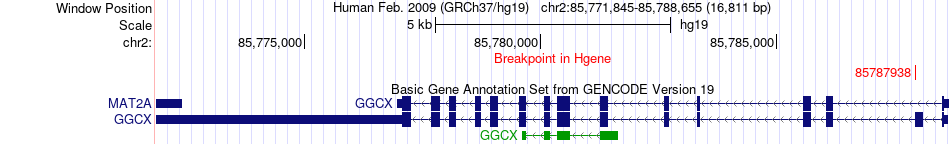 |
 Fusion gene breakpoints across KCMF1 (3'-gene) Fusion gene breakpoints across KCMF1 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
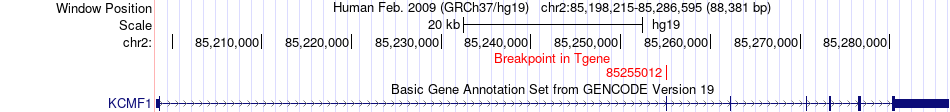 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | PRAD | TCGA-HC-A76X-01A | GGCX | chr2 | 85787938 | - | KCMF1 | chr2 | 85255012 | + |
Top |
Fusion Gene ORF analysis for GGCX-KCMF1 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| In-frame | ENST00000233838 | ENST00000409785 | GGCX | chr2 | 85787938 | - | KCMF1 | chr2 | 85255012 | + |
| intron-3CDS | ENST00000430215 | ENST00000409785 | GGCX | chr2 | 85787938 | - | KCMF1 | chr2 | 85255012 | + |
| intron-3CDS | ENST00000473665 | ENST00000409785 | GGCX | chr2 | 85787938 | - | KCMF1 | chr2 | 85255012 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000233838 | GGCX | chr2 | 85787938 | - | ENST00000409785 | KCMF1 | chr2 | 85255012 | + | 7488 | 295 | 81 | 1424 | 447 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000233838 | ENST00000409785 | GGCX | chr2 | 85787938 | - | KCMF1 | chr2 | 85255012 | + | 0.000522479 | 0.99947757 |
Top |
Fusion Genomic Features for GGCX-KCMF1 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| GGCX | chr2 | 85787937 | - | KCMF1 | chr2 | 85255011 | + | 1.76E-09 | 1 |
| GGCX | chr2 | 85787937 | - | KCMF1 | chr2 | 85255011 | + | 1.76E-09 | 1 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
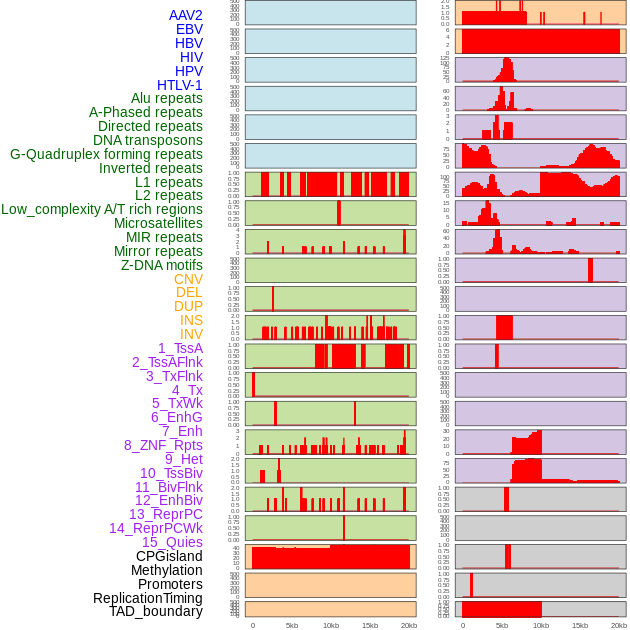 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
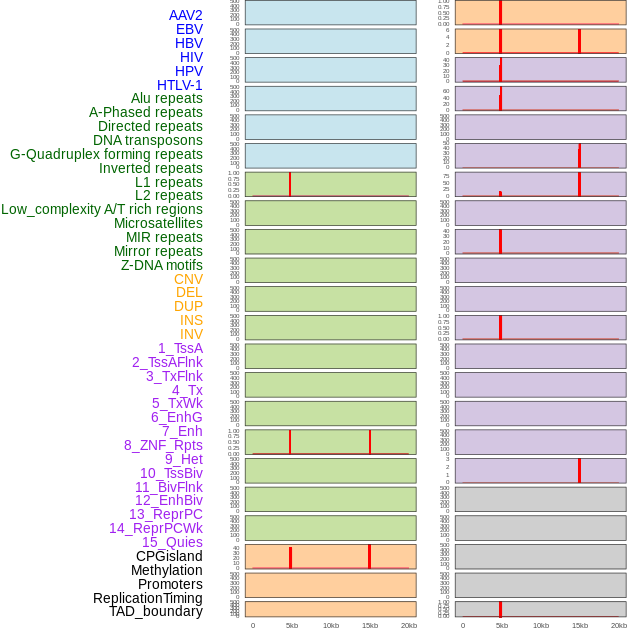 |
Top |
Fusion Protein Features for GGCX-KCMF1 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr2:85198590/chr2:85783383) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| GGCX | KCMF1 |
| FUNCTION: Mediates the vitamin K-dependent carboxylation of glutamate residues to calcium-binding gamma-carboxyglutamate (Gla) residues with the concomitant conversion of the reduced hydroquinone form of vitamin K to vitamin K epoxide. {ECO:0000269|PubMed:17073445}. | FUNCTION: Has intrinsic E3 ubiquitin ligase activity and promotes ubiquitination. {ECO:0000269|PubMed:15581609}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | GGCX | chr2:85787938 | chr2:85255012 | ENST00000233838 | - | 2 | 15 | 2_60 | 71 | 759.0 | Topological domain | Cytoplasmic |
| Tgene | KCMF1 | chr2:85787938 | chr2:85255012 | ENST00000409785 | 0 | 7 | 225_257 | 5 | 382.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | KCMF1 | chr2:85787938 | chr2:85255012 | ENST00000409785 | 0 | 7 | 333_336 | 5 | 382.0 | Compositional bias | Note=Poly-Ser | |
| Tgene | KCMF1 | chr2:85787938 | chr2:85255012 | ENST00000409785 | 0 | 7 | 376_380 | 5 | 382.0 | Compositional bias | Note=Poly-Pro | |
| Tgene | KCMF1 | chr2:85787938 | chr2:85255012 | ENST00000409785 | 0 | 7 | 3_50 | 5 | 382.0 | Zinc finger | ZZ-type | |
| Tgene | KCMF1 | chr2:85787938 | chr2:85255012 | ENST00000409785 | 0 | 7 | 78_101 | 5 | 382.0 | Zinc finger | C2H2-type |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | GGCX | chr2:85787938 | chr2:85255012 | ENST00000233838 | - | 2 | 15 | 135_136 | 71 | 759.0 | Topological domain | Cytoplasmic |
| Hgene | GGCX | chr2:85787938 | chr2:85255012 | ENST00000233838 | - | 2 | 15 | 158_292 | 71 | 759.0 | Topological domain | Lumenal |
| Hgene | GGCX | chr2:85787938 | chr2:85255012 | ENST00000233838 | - | 2 | 15 | 314_361 | 71 | 759.0 | Topological domain | Cytoplasmic |
| Hgene | GGCX | chr2:85787938 | chr2:85255012 | ENST00000233838 | - | 2 | 15 | 383_758 | 71 | 759.0 | Topological domain | Lumenal |
| Hgene | GGCX | chr2:85787938 | chr2:85255012 | ENST00000233838 | - | 2 | 15 | 82_113 | 71 | 759.0 | Topological domain | Lumenal |
| Hgene | GGCX | chr2:85787938 | chr2:85255012 | ENST00000430215 | - | 1 | 14 | 135_136 | 0 | 702.0 | Topological domain | Cytoplasmic |
| Hgene | GGCX | chr2:85787938 | chr2:85255012 | ENST00000430215 | - | 1 | 14 | 158_292 | 0 | 702.0 | Topological domain | Lumenal |
| Hgene | GGCX | chr2:85787938 | chr2:85255012 | ENST00000430215 | - | 1 | 14 | 2_60 | 0 | 702.0 | Topological domain | Cytoplasmic |
| Hgene | GGCX | chr2:85787938 | chr2:85255012 | ENST00000430215 | - | 1 | 14 | 314_361 | 0 | 702.0 | Topological domain | Cytoplasmic |
| Hgene | GGCX | chr2:85787938 | chr2:85255012 | ENST00000430215 | - | 1 | 14 | 383_758 | 0 | 702.0 | Topological domain | Lumenal |
| Hgene | GGCX | chr2:85787938 | chr2:85255012 | ENST00000430215 | - | 1 | 14 | 82_113 | 0 | 702.0 | Topological domain | Lumenal |
| Hgene | GGCX | chr2:85787938 | chr2:85255012 | ENST00000233838 | - | 2 | 15 | 114_134 | 71 | 759.0 | Transmembrane | Helical |
| Hgene | GGCX | chr2:85787938 | chr2:85255012 | ENST00000233838 | - | 2 | 15 | 137_157 | 71 | 759.0 | Transmembrane | Helical |
| Hgene | GGCX | chr2:85787938 | chr2:85255012 | ENST00000233838 | - | 2 | 15 | 293_313 | 71 | 759.0 | Transmembrane | Helical |
| Hgene | GGCX | chr2:85787938 | chr2:85255012 | ENST00000233838 | - | 2 | 15 | 362_382 | 71 | 759.0 | Transmembrane | Helical |
| Hgene | GGCX | chr2:85787938 | chr2:85255012 | ENST00000233838 | - | 2 | 15 | 61_81 | 71 | 759.0 | Transmembrane | Helical |
| Hgene | GGCX | chr2:85787938 | chr2:85255012 | ENST00000430215 | - | 1 | 14 | 114_134 | 0 | 702.0 | Transmembrane | Helical |
| Hgene | GGCX | chr2:85787938 | chr2:85255012 | ENST00000430215 | - | 1 | 14 | 137_157 | 0 | 702.0 | Transmembrane | Helical |
| Hgene | GGCX | chr2:85787938 | chr2:85255012 | ENST00000430215 | - | 1 | 14 | 293_313 | 0 | 702.0 | Transmembrane | Helical |
| Hgene | GGCX | chr2:85787938 | chr2:85255012 | ENST00000430215 | - | 1 | 14 | 362_382 | 0 | 702.0 | Transmembrane | Helical |
| Hgene | GGCX | chr2:85787938 | chr2:85255012 | ENST00000430215 | - | 1 | 14 | 61_81 | 0 | 702.0 | Transmembrane | Helical |
Top |
Fusion Gene Sequence for GGCX-KCMF1 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >33021_33021_1_GGCX-KCMF1_GGCX_chr2_85787938_ENST00000233838_KCMF1_chr2_85255012_ENST00000409785_length(transcript)=7488nt_BP=295nt CGGAGCCTAGGGAAGCAAATTCTCCTGGCGGCCTCCGTTCAGACGCGGCAGCTGTGACCCACCTGCCTCCTCCGCAGAGCAATGGCGGTG TCTGCCGGGTCCGCGCGGACCTCGCCCAGCTCAGATAAAGTACAGAAAGACAAGGCTGAACTGATCTCAGGGCCCAGGCAGGACAGCCGA ATAGGGAAACTCTTGGGTTTTGAGTGGACAGATTTGTCCAGTTGGCGGAGGCTGGTGACCCTGCTGAATCGACCAACGGACCCTGCAAGC TTAGCTGTCTTTCGTTTTCTTTTTGGTGTCAGCTGTGATGCATGTTTAAAAGGAAATTTTCGAGGTCGCAGATATAAGTGTTTAATTTGC TACGATTACGATCTTTGTGCATCTTGTTATGAAAGTGGTGCAACAACAACAAGGCATACAACTGACCACCCAATGCAGTGCATATTAACA AGGGTAGATTTTGATTTATACTATGGTGGGGAAGCTTTCTCTGTAGAGCAGCCACAGTCTTTTACTTGTCCCTATTGTGGAAAAATGGGC TATACGGAGACATCTCTTCAAGAACATGTTACTTCTGAACATGCAGAAACATCAACAGAAGTGATTTGTCCAATATGTGCAGCGTTACCT GGAGGCGATCCTAATCATGTCACGGATGACTTTGCAGCTCATCTTACACTTGAACACAGAGCCCCTAGAGATTTAGATGAATCGAGTGGT GTTCGACATGTACGTAGAATGTTTCACCCTGGCCGGGGATTAGGAGGTCCTCGTGCTCGTAGATCAAACATGCACTTTACTAGCAGTTCT ACTGGTGGACTTTCTTCTTCTCAGAGTTCATATTCTCCAAGCAATAGGGAAGCCATGGATCCTATAGCTGAGCTTTTATCTCAGTTATCA GGAGTGAGACGTTCTGCAGGAGGACAGCTTAATTCCTCTGGCCCTTCCGCTTCTCAGTTACAACAACTGCAGATGCAGCTGCAGCTAGAA CGGCAGCATGCCCAGGCAGCACGGCAACAACTGGAGACCGCACGCAACGCAACCCGGCGTACTAACACAAGCAGTGTCACCACTACAATC ACACAATCCACAGCAACAACCAACATAGCTAATACAGAAAGCAGTCAGCAGACTCTACAGAATTCCCAGTTTCTTTTAACAAGGTTGAAT GATCCTAAAATGTCTGAAACGGAGCGCCAGTCCATGGAAAGCGAGCGTGCAGACCGCAGCCTGTTTGTCCAAGAGCTCCTTCTGTCCACT TTAGTGCGTGAAGAGAGCTCATCCTCAGATGAGGATGATCGGGGGGAGATGGCAGATTTTGGTGCTATGGGCTGTGTAGATATTATGCCT TTAGATGTTGCTTTAGAAAACCTAAATTTAAAAGAGAGTAATAAAGGAAATGAGCCTCCACCACCTCCTCTTTGATGACATCCCAATTCG CAGACAATGTCCTCTGTGCTGTATTTGCCAATGAAAGTGGACAACAACTATCTTGGGTTTGTTTGGTGATTGTAATTTCAGGTCTGTCAC TCTTGTTACATTGTGTACATTCAAAAGGAAGAGAGAAAATATATATGATAATCATTTCCACTTAACTAATTTTTACTTCTAGCAGGTAAA TGTAGGTAGCAGTGCAGGGGTGATCTCTGCTTCCTGTACCTTGACATGCAAAAGGCTCTCCTAATACTCCACATTCAAACTGAAGAGGAA AATTGAAATCTCTAATGAAGCTGCTGTGTGTATTTATGAATATTAATGAATAAAAACTGCTTGGATGGTTTACCTTAACTACTGCATGAG GTTTTTTGCAGCGTGCATGAGTTTTAGTGACCTTGTTATTTAAGAAGTTAAATACAAGGAGTAAAACTTAAAAAAAAAATACAAAGCCCA AAGCTTTCCCAAACATTATTCAATGGTTACACGACGAAGTAGCTTTTGAATAATGTCTGCCTGAATCACCTTTCTTTGTGTGCCTCCTAC GCACAAAGCCAGCTCTGCAGTGGAATCTGGGGATTATAGCCGGGTGTGGCACTCCGCCCTGTGTGACTGTCCTGTCGCCCTGTTAGTCAT CTTGCCTGTGTGGAGCTCAGCCTGTCTCTTTAACTCATCTGTAGAAGACACACCAGTAAAGCTACTGTTGGAATCTGCTGCAGGGGCCTT TGTGTGCCCTAAAAACAAATCCTGTTCATGTTTGTTTAAAGTTTTTACTTTTTGTGGTTGTTTAAAATTTTTTCAATTGTTAAATATGTT TTATTCAGGTGTAGATGAATTTCATTTATTGACTGTTCAACAGAGTTAACCTGAATTATGTTGTCTTTGTTTTTAAAAATCTCACATTCT CAATCATATTTTGCATTATTTATGTATTTGCTTTGTAGTTTGCTGAGACAGATCAGTATCAGGGGAGCTTTGAGGATTTGCCTTCCCAGA TTTGTCAGTATATTACAACCAAATTCTTAATGCTAATTTTAGCACCTTTTATTTATTGGGTTTTTTCTGGCATAAAAAGTAAAGCCTTTT AATTGAATCATGCCACCTATATGCCTATATTATTAATCCTATGTGTAAAAAAAATGTACAGCTTTTTTTGGGTTTGTTTTGGGGTTTGGA AGGGCCGGGTTATTTTTTATTTCCTGTTTCAGTTTTTGTGCATAGACTTTCACAATAGCTCCAAGGCAGGGACAGCGGGTTTGGGGGTTG GGAGGGCAGTTTTTGGAATGTAAATTTAGGACTTTTAAAAAGGTGCGCACAGCTTCTGATAAATTTATAACTAGACTTAACCTAATCATG TCTCGTTCCAGTTCTCTTTTCTCTGAGCCCTTTTCAAAGTCTCCTCTCTTTCTCCTGTCATCCTTTTCCTTTCCTGTCCGTGTATCTCCG TTTCTTCAACATGACAAGCATACAGACTTGAACACCCCTCCGGTGTTCTTCCGAGAACTGTGAAGTCCATGTTCATCCAAATGTAACCAA AAAAGAAGTCACCCTACATGTCTGAAAAACTGTTGCTTCTCCTCTGAAACTTCAAACTCCAACGATTTCCAAATACAATAGCTTTGTTTT CTTTAGTTCTGTAATGGATAATGTTTAAAGGAAAACTTTACACCAGGCTTCTGGTTACACTAGAAGTCAAGCCCATTAGGGATTTTCATT TTTTTTCATTTGGTTGTTGAGAAGTTTCAAAAATCAGTTTTCAAGCTGTGGTCTTTCAAACACATCTGCACATAAGTCACACATTTCAAT AAAGCATTTTCAAGACTGTTGACCACTTGAATTTCTTTGGTGTTAGTGGATTAACCTAACCATTACTCTGAGATTTTATATCTCTAATCC AGTATTTTTTTTCTTTTTATGCTAAACATGGGAGATCCTGGTTTTGTGTGAATGCATTATTTTGGATGTGAGAAATAAATGCCAATTATA TGTACTTTCCCTTTTCTGCACAAATTCTGGGAAAATGTAAAAGCCTTCATTTTTTAACATGGTTTTAGGGAACAACTGTAAGTCCATTCA GAGCAGTTTAATTTAGATGATCACTTTTAACACTGCAGAAGACCTAAGGAGTCATAAGATTCCATCTCATGTAGAATACTGTATATTAAT TATTTTTTACCAGATATTTTACAAATACCTCACCTCATCCTGTATGTAATCAATAATGTATTATTTTAGGGTAGAACGACACAGAATATC AATATAAAAATATATTCAAGCCCTTGAAATTCTTGTGTCCTTTTTCTTTAGTTTCTGAATGAAAATCTTATTACTGGATGTACTATTGAA TAAAAATTAATTGCAAAGACTTTGATGGAGACAAAATCAAAATATACCACTGTTTATTTTGGCAGCACCTTCAAATTTCTAAGGACCAGA TCCTGAATTTTGAGAATTTTTTGTTTCGTTGAGTTTTGAGTGTCTAGGATATAAGTTTATAAATGTTCTACAGTGCTTGTCTTTACTTCC TGTTTAGAAATCATTTAAATCTTTCCCCTCGAGGTGCTCTGGAAGAAGTTTAATTTCAGCTCTGAACAAGTAGAAATAACTGTGCTTCCT TGTATGGCTGCAATCATAAGACAGAATTTCTGTGGTTGGCTACCGTGAAATCTTTGTATTTATATTGCATTGTTTTGTTAAAAAAAAAAA AATCCACAGAAGTAGAACCTAGGTTTGCTACTAACAAAGTAGTGGATTCTTATAATTAGGCTCAGGTAAAGTAAGAGAAAACTTCTCTCC CTGTGGCCCCCAGAGAAATGAAGTAGGATAGCAGATTACACTATTAGAAATAATAGACGTAACAAAAAAAATCCAGCCCATTATCCCCAC CCCCCATTTTCCCTCTCAATATTAGAAATTTCAATAATATGCTGACAGATTTTCAAAGGTCACCAAGGCTTTTGTGTTTTTTTCCGGTCT GTGGGCCACAGTTAAAAGAACGGGTAACAGGTTCAGAGTTCTCTTGATACGTCCAAGCCTTAGTTTCCAGGAAAAAAGCTAGGCTCCTCA CTGCAAATATTAACGCTTTATATATTTACTCCATGGATGTTGGGGTAAGCACATATATCAATAGTTCATAGCTCTAGTTAAGGACAAAGT GGGTTGGGGGGTAGAAGGAGAATACTAGAGAGGTTACGGTAGCAGGTGGCTGCAGAAAAGAATCTTTTGAATGGGATTCCTGTAACAGTG TTCAGTACCAATATTAAGTCTACTGTAAGTGGGGTCTGATATGGCTTACTGATACAGGCATGTGTAAAAAACTAAGTGTGATGGAGGAAT GGGTGCTAAAAGCTCTTAACAGTTTTGTAGAGAAGCCCTGCAGCCGTGTTTACTATATATGACATGCCTGTTTCTTAGTTTGCATGCACA GTTTAAGGGGCACATGCAATATTGGTTTCATGTGCAGTAAAAATTAAAACACGGCTGAGCGTGGTGGCTCACGCCTGTAATCCCAGCACT TTGCGAGGCCGAGGCGGGTGGATCACGAGGTCGAGTTTGAGACCATTTTGGCCAGCATAGTGAAACCCCATCTCTACTAAAAAATACAAA AAGTTAGCTGGGCATGGTGGCAGGCACCTGTAATCCCAGCTACTCGGGAGGCTGAGGCAGGAGAATCGCTTGAACCTGGGAGGCAGAGGT TGCAGTGAGCTGAGATCACGCCGCCATTGCACTCCAGCCCGGGCGACGGTGCAAGACTCTGTCTCAAAAAAAAAAAAAATTAAAACACTA GGTACGAAATTATCCCAGAATTATACGCGAAAGTTGGTGTTCATGCTGGAAAACAGTTACGGTTATCTGTGCTCTTTCTAAAGCTCAACT AGACACTAAAACAGTTCTCTCCTTGAGACCGGAAGTGGGATGGGGGTGGGCAGGTGGAAGGGAAGAGGCTGGGGAGGAGCCTCTCATCTC ATCTCATTTCAGCCTCAACAGTAATTGAAACCCTATGCCTGGCCTGGAGTGGGTAGAGGTTAGGGTTAGAGTGCAGGATGGAGCAGCAGT CTGCAGACTTTCAGGTTGTGACGATGTCATGAAATTCTGTGCTATTAAACATGGCTATGTCTGAGTCAGTCATTACGGTATTCCAAAGCC AAATTCAAGTGACTGTTTTTCTGAATAGTATAAAAATCTAATTCCCTGTGCTTATCGGCTTCAACTGTGCTTACACAACGTTTGTTTCTA ATTCGTACTCTTGCCTCTTGGTTTCTGTAGTTTTTCAACCCATTTCTGTCAGTGTTCTATAAATGCATACATTCGTTATTTAAGGAACCC TGCAGCAGACCTCGAATAAAATGTGACTTGATGTTTGTTGCCACTTCCTCAACTTGGTTAGCAAATTCAAGTTTTGAAAGACGATTCGTC TCAGAGAAGTTGATCCTGAACCTCCACGTCACATTTTAGTCGGTTGCTATTAGGTGGGTAATGCCTAATAACATGGGTTCCTCAAATCAC AAAGGAGAATGCTCTCAAGTCCTCATGTTAACTTTAGTAAGTCAGTTAACTTTACAGCAACTAAAGAGATCAATTAAAAAGAACCCTTAA GTCTGACTGTCCCAGGTAACTTTACTTGCTGTACAGATAGTGCACAGATATGGATGAGGGCTGCACATGGCTGATTTGAAAACCCATGTG GGGCCGGGTGTGGTGGCTCACGCCTGTAATCCCAGCACTTTGGGAGGCCAAGGCGGGTGGATCACGAGGTTAGTTCAGGACCAGCCTGGC CAAGATGGTGAAACCCCATCTCTACTAAAAATACGAAAATTAGCTGGGCTTGGTGGCAGATGCCTGTAATTCCTGCTACTCGGGAGGCTG AGGCAGAGAATTGCTTGAACCCGGGAGGCGGAGGTTGCAGTGAGCCGAGATTGCGCCACTGCACTCCAGCCTGGGCGACACAGTGAGACT CTCCTCAAAACAAAAAAAAAAGAAAGAAAACCCATGTGGGCCGCATGTGCACAGCCTGGGGCTGCTTTCCACAGGCCTGGATAATGATGT TTCTCTTCAAGGAGGCCTTTACTAACCACAGGGAACCTAGCATCTGACTGTGTCCTTGGCAGACAGTGCATGTAGCCTGTAGAATGTTGG CTCCTCTGTCTATAGAAACCATCAGTTCGCTGCTCTCTCATTCATACATACTTTTTGAGTTCCCAGTAGGTACGAAGCTTTGTCTAAACA TTATTTACCAACTGCGATTTTTACAGACTTCCCCTGTAAGAATTTTCTAAAAGAGCCTTTCTCCCTTACACAATTGAAACGTTTTCAAAG GTCATCAAGCCGGTTGCGTTTGGTCAGTCACGTGACCAATCAGAGCTGGTGAGTGAGCTGCTCCCTTAAATCTACCAAATTAGACTGACA GCATTTGGAATCTGCCTCACACATAAGAGATCCATATTCATTTAATAGTGAAGCCAGCCCAGAAGCATCCTCTTCTGTAAGCTGGATGGA GTCCCAGCTCTCTCTCCTTGTATATAAGGAGACCACACCCTGTATATAAGGTTTTCTTGAGGTAGATGCAGCAATATTCCTGGGGTAAGA TTCTATAATACTCCTGTATTGGCAAGTGGACTGGACCAACAGACACAGGCCTCAGCCACTGTCGCTAGGGACTGAAGGAGAGTTCATGGA TTCTTACCCCCTAGTCATGAAAGCTATTCTGGCCAGCCTTGAATCCATTTAATTTTGCCCACGAGAACCTTTCATGACAATTAACAAGAC ACTAGTTTCCCCATAAAAGTCCATTTTTAAATATATAGTATGCTAATTAGCAGTGTCTGTTGAATTGCCCTATCGGATAGCAGCCACCAT GTGTGCTGACCACTCACGATTCAGACTTACCTTGGAAAATATCACACTGACGATACAGTACTTTAGAATAAATTGCATATTGTATCAGGC TCCGAGACTAATTTGTACTGAACCCTTGATATAGATTACCTTTTATAACATCACCACCTGTCATGAAGCTTATCTGTTTTCTGATTTGAA >33021_33021_1_GGCX-KCMF1_GGCX_chr2_85787938_ENST00000233838_KCMF1_chr2_85255012_ENST00000409785_length(amino acids)=447AA_BP=66 MAVSAGSARTSPSSDKVQKDKAELISGPRQDSRIGKLLGFEWTDLSSWRRLVTLLNRPTDPASLAVFRFLFGVSCDACLKGNFRGRRYKC LICYDYDLCASCYESGATTTRHTTDHPMQCILTRVDFDLYYGGEAFSVEQPQSFTCPYCGKMGYTETSLQEHVTSEHAETSTEVICPICA ALPGGDPNHVTDDFAAHLTLEHRAPRDLDESSGVRHVRRMFHPGRGLGGPRARRSNMHFTSSSTGGLSSSQSSYSPSNREAMDPIAELLS QLSGVRRSAGGQLNSSGPSASQLQQLQMQLQLERQHAQAARQQLETARNATRRTNTSSVTTTITQSTATTNIANTESSQQTLQNSQFLLT -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for GGCX-KCMF1 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for GGCX-KCMF1 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for GGCX-KCMF1 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
