
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:GGNBP2-NEBL (FusionGDB2 ID:33031) |
Fusion Gene Summary for GGNBP2-NEBL |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: GGNBP2-NEBL | Fusion gene ID: 33031 | Hgene | Tgene | Gene symbol | GGNBP2 | NEBL | Gene ID | 79893 | 10529 |
| Gene name | gametogenetin binding protein 2 | nebulette | |
| Synonyms | DIF-3|DIF3|LCRG1|LZK1|ZFP403|ZNF403 | C10orf113|LASP2|LNEBL|bA165O3.1 | |
| Cytomap | 17q12 | 10p12.31 | |
| Type of gene | protein-coding | protein-coding | |
| Description | gametogenetin-binding protein 2C3HC4-type zinc finger proteinlaryngeal carcinoma related gene 1laryngeal carcinoma-related protein 1zinc finger protein 403 | nebuletteLIM and SH3 protein 2LIM-nebuletteactin-binding Z-disk protein | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | Q9H3C7 | O76041 | |
| Ensembl transtripts involved in fusion gene | ENST00000304718, ENST00000485685, | ENST00000377119, ENST00000464278, ENST00000377122, ENST00000377159, ENST00000417816, | |
| Fusion gene scores | * DoF score | 6 X 7 X 6=252 | 12 X 11 X 6=792 |
| # samples | 6 | 13 | |
| ** MAII score | log2(6/252*10)=-2.0703893278914 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(13/792*10)=-2.60698880705116 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: GGNBP2 [Title/Abstract] AND NEBL [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | GGNBP2(34901666)-NEBL(21101869), # samples:3 | ||
| Anticipated loss of major functional domain due to fusion event. | GGNBP2-NEBL seems lost the major protein functional domain in Hgene partner, which is a essential gene due to the frame-shifted ORF. GGNBP2-NEBL seems lost the major protein functional domain in Hgene partner, which is a tumor suppressor due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
 Fusion gene breakpoints across GGNBP2 (5'-gene) Fusion gene breakpoints across GGNBP2 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
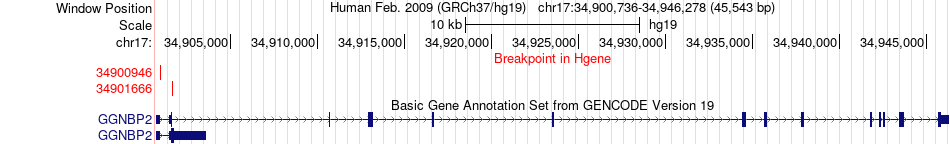 |
 Fusion gene breakpoints across NEBL (3'-gene) Fusion gene breakpoints across NEBL (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
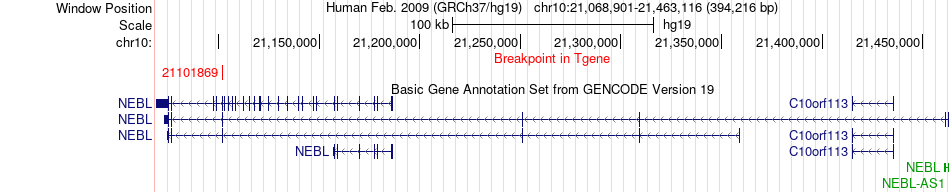 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | CESC | TCGA-C5-A3HD-01B | GGNBP2 | chr17 | 34900946 | + | NEBL | chr10 | 21101869 | - |
| ChimerDB4 | CESC | TCGA-C5-A3HD-01B | GGNBP2 | chr17 | 34901666 | - | NEBL | chr10 | 21101869 | - |
| ChimerDB4 | CESC | TCGA-C5-A3HD-01B | GGNBP2 | chr17 | 34901666 | + | NEBL | chr10 | 21101869 | - |
Top |
Fusion Gene ORF analysis for GGNBP2-NEBL |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-intron | ENST00000304718 | ENST00000377119 | GGNBP2 | chr17 | 34901666 | + | NEBL | chr10 | 21101869 | - |
| 5CDS-intron | ENST00000304718 | ENST00000464278 | GGNBP2 | chr17 | 34901666 | + | NEBL | chr10 | 21101869 | - |
| 5CDS-intron | ENST00000485685 | ENST00000377119 | GGNBP2 | chr17 | 34901666 | + | NEBL | chr10 | 21101869 | - |
| 5CDS-intron | ENST00000485685 | ENST00000464278 | GGNBP2 | chr17 | 34901666 | + | NEBL | chr10 | 21101869 | - |
| 5UTR-3CDS | ENST00000304718 | ENST00000377122 | GGNBP2 | chr17 | 34900946 | + | NEBL | chr10 | 21101869 | - |
| 5UTR-3CDS | ENST00000304718 | ENST00000377159 | GGNBP2 | chr17 | 34900946 | + | NEBL | chr10 | 21101869 | - |
| 5UTR-3CDS | ENST00000304718 | ENST00000417816 | GGNBP2 | chr17 | 34900946 | + | NEBL | chr10 | 21101869 | - |
| 5UTR-3CDS | ENST00000485685 | ENST00000377122 | GGNBP2 | chr17 | 34900946 | + | NEBL | chr10 | 21101869 | - |
| 5UTR-3CDS | ENST00000485685 | ENST00000377159 | GGNBP2 | chr17 | 34900946 | + | NEBL | chr10 | 21101869 | - |
| 5UTR-3CDS | ENST00000485685 | ENST00000417816 | GGNBP2 | chr17 | 34900946 | + | NEBL | chr10 | 21101869 | - |
| 5UTR-intron | ENST00000304718 | ENST00000377119 | GGNBP2 | chr17 | 34900946 | + | NEBL | chr10 | 21101869 | - |
| 5UTR-intron | ENST00000304718 | ENST00000464278 | GGNBP2 | chr17 | 34900946 | + | NEBL | chr10 | 21101869 | - |
| 5UTR-intron | ENST00000485685 | ENST00000377119 | GGNBP2 | chr17 | 34900946 | + | NEBL | chr10 | 21101869 | - |
| 5UTR-intron | ENST00000485685 | ENST00000464278 | GGNBP2 | chr17 | 34900946 | + | NEBL | chr10 | 21101869 | - |
| Frame-shift | ENST00000485685 | ENST00000377122 | GGNBP2 | chr17 | 34901666 | + | NEBL | chr10 | 21101869 | - |
| Frame-shift | ENST00000485685 | ENST00000377159 | GGNBP2 | chr17 | 34901666 | + | NEBL | chr10 | 21101869 | - |
| Frame-shift | ENST00000485685 | ENST00000417816 | GGNBP2 | chr17 | 34901666 | + | NEBL | chr10 | 21101869 | - |
| In-frame | ENST00000304718 | ENST00000377122 | GGNBP2 | chr17 | 34901666 | + | NEBL | chr10 | 21101869 | - |
| In-frame | ENST00000304718 | ENST00000377159 | GGNBP2 | chr17 | 34901666 | + | NEBL | chr10 | 21101869 | - |
| In-frame | ENST00000304718 | ENST00000417816 | GGNBP2 | chr17 | 34901666 | + | NEBL | chr10 | 21101869 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000304718 | GGNBP2 | chr17 | 34901666 | + | ENST00000377122 | NEBL | chr10 | 21101869 | - | 6882 | 409 | 217 | 1107 | 296 |
| ENST00000304718 | GGNBP2 | chr17 | 34901666 | + | ENST00000417816 | NEBL | chr10 | 21101869 | - | 2706 | 409 | 217 | 864 | 215 |
| ENST00000304718 | GGNBP2 | chr17 | 34901666 | + | ENST00000377159 | NEBL | chr10 | 21101869 | - | 1141 | 409 | 217 | 864 | 215 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000304718 | ENST00000377122 | GGNBP2 | chr17 | 34901666 | + | NEBL | chr10 | 21101869 | - | 0.000326463 | 0.9996736 |
| ENST00000304718 | ENST00000417816 | GGNBP2 | chr17 | 34901666 | + | NEBL | chr10 | 21101869 | - | 0.000854117 | 0.99914587 |
| ENST00000304718 | ENST00000377159 | GGNBP2 | chr17 | 34901666 | + | NEBL | chr10 | 21101869 | - | 0.003626789 | 0.99637324 |
Top |
Fusion Genomic Features for GGNBP2-NEBL |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
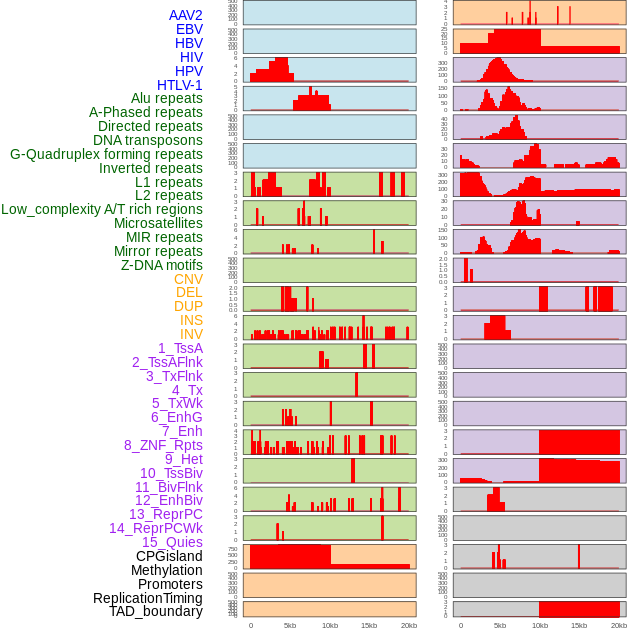 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for GGNBP2-NEBL |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr17:34901666/chr10:21101869) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| GGNBP2 | NEBL |
| FUNCTION: May be involved in spermatogenesis. | FUNCTION: Binds to actin and plays an important role in the assembly of the Z-disk. May functionally link sarcomeric actin to the desmin intermediate filaments in the heart muscle sarcomeres (PubMed:27733623). Isoform 2 might play a role in the assembly of focal adhesion (PubMed:15004028). {ECO:0000269|PubMed:15004028, ECO:0000269|PubMed:27733623}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | NEBL | chr17:34901666 | chr10:21101869 | ENST00000377122 | 22 | 28 | 954_1014 | 782 | 1015.0 | Domain | SH3 | |
| Tgene | NEBL | chr17:34901666 | chr10:21101869 | ENST00000417816 | 3 | 7 | 954_1014 | 119 | 271.0 | Domain | SH3 | |
| Tgene | NEBL | chr17:34901666 | chr10:21101869 | ENST00000377122 | 22 | 28 | 836_953 | 782 | 1015.0 | Region | Note=Linker | |
| Tgene | NEBL | chr17:34901666 | chr10:21101869 | ENST00000417816 | 3 | 7 | 836_953 | 119 | 271.0 | Region | Note=Linker | |
| Tgene | NEBL | chr17:34901666 | chr10:21101869 | ENST00000377122 | 22 | 28 | 795_830 | 782 | 1015.0 | Repeat | Note=Nebulin 23 | |
| Tgene | NEBL | chr17:34901666 | chr10:21101869 | ENST00000417816 | 3 | 7 | 137_172 | 119 | 271.0 | Repeat | Note=Nebulin 4 | |
| Tgene | NEBL | chr17:34901666 | chr10:21101869 | ENST00000417816 | 3 | 7 | 173_205 | 119 | 271.0 | Repeat | Note=Nebulin 5 | |
| Tgene | NEBL | chr17:34901666 | chr10:21101869 | ENST00000417816 | 3 | 7 | 206_241 | 119 | 271.0 | Repeat | Note=Nebulin 6 | |
| Tgene | NEBL | chr17:34901666 | chr10:21101869 | ENST00000417816 | 3 | 7 | 242_278 | 119 | 271.0 | Repeat | Note=Nebulin 7 | |
| Tgene | NEBL | chr17:34901666 | chr10:21101869 | ENST00000417816 | 3 | 7 | 279_313 | 119 | 271.0 | Repeat | Note=Nebulin 8 | |
| Tgene | NEBL | chr17:34901666 | chr10:21101869 | ENST00000417816 | 3 | 7 | 314_348 | 119 | 271.0 | Repeat | Note=Nebulin 9 | |
| Tgene | NEBL | chr17:34901666 | chr10:21101869 | ENST00000417816 | 3 | 7 | 349_385 | 119 | 271.0 | Repeat | Note=Nebulin 10 | |
| Tgene | NEBL | chr17:34901666 | chr10:21101869 | ENST00000417816 | 3 | 7 | 386_422 | 119 | 271.0 | Repeat | Note=Nebulin 11 | |
| Tgene | NEBL | chr17:34901666 | chr10:21101869 | ENST00000417816 | 3 | 7 | 423_459 | 119 | 271.0 | Repeat | Note=Nebulin 12 | |
| Tgene | NEBL | chr17:34901666 | chr10:21101869 | ENST00000417816 | 3 | 7 | 460_496 | 119 | 271.0 | Repeat | Note=Nebulin 13 | |
| Tgene | NEBL | chr17:34901666 | chr10:21101869 | ENST00000417816 | 3 | 7 | 497_533 | 119 | 271.0 | Repeat | Note=Nebulin 14 | |
| Tgene | NEBL | chr17:34901666 | chr10:21101869 | ENST00000417816 | 3 | 7 | 534_569 | 119 | 271.0 | Repeat | Note=Nebulin 15 | |
| Tgene | NEBL | chr17:34901666 | chr10:21101869 | ENST00000417816 | 3 | 7 | 570_599 | 119 | 271.0 | Repeat | Note=Nebulin 16 | |
| Tgene | NEBL | chr17:34901666 | chr10:21101869 | ENST00000417816 | 3 | 7 | 600_635 | 119 | 271.0 | Repeat | Note=Nebulin 17 | |
| Tgene | NEBL | chr17:34901666 | chr10:21101869 | ENST00000417816 | 3 | 7 | 636_666 | 119 | 271.0 | Repeat | Note=Nebulin 18 | |
| Tgene | NEBL | chr17:34901666 | chr10:21101869 | ENST00000417816 | 3 | 7 | 667_693 | 119 | 271.0 | Repeat | Note=Nebulin 19 | |
| Tgene | NEBL | chr17:34901666 | chr10:21101869 | ENST00000417816 | 3 | 7 | 694_728 | 119 | 271.0 | Repeat | Note=Nebulin 20 | |
| Tgene | NEBL | chr17:34901666 | chr10:21101869 | ENST00000417816 | 3 | 7 | 729_759 | 119 | 271.0 | Repeat | Note=Nebulin 21 | |
| Tgene | NEBL | chr17:34901666 | chr10:21101869 | ENST00000417816 | 3 | 7 | 760_794 | 119 | 271.0 | Repeat | Note=Nebulin 22 | |
| Tgene | NEBL | chr17:34901666 | chr10:21101869 | ENST00000417816 | 3 | 7 | 795_830 | 119 | 271.0 | Repeat | Note=Nebulin 23 |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | NEBL | chr17:34901666 | chr10:21101869 | ENST00000377122 | 22 | 28 | 100_136 | 782 | 1015.0 | Repeat | Note=Nebulin 3 | |
| Tgene | NEBL | chr17:34901666 | chr10:21101869 | ENST00000377122 | 22 | 28 | 137_172 | 782 | 1015.0 | Repeat | Note=Nebulin 4 | |
| Tgene | NEBL | chr17:34901666 | chr10:21101869 | ENST00000377122 | 22 | 28 | 173_205 | 782 | 1015.0 | Repeat | Note=Nebulin 5 | |
| Tgene | NEBL | chr17:34901666 | chr10:21101869 | ENST00000377122 | 22 | 28 | 206_241 | 782 | 1015.0 | Repeat | Note=Nebulin 6 | |
| Tgene | NEBL | chr17:34901666 | chr10:21101869 | ENST00000377122 | 22 | 28 | 242_278 | 782 | 1015.0 | Repeat | Note=Nebulin 7 | |
| Tgene | NEBL | chr17:34901666 | chr10:21101869 | ENST00000377122 | 22 | 28 | 279_313 | 782 | 1015.0 | Repeat | Note=Nebulin 8 | |
| Tgene | NEBL | chr17:34901666 | chr10:21101869 | ENST00000377122 | 22 | 28 | 29_63 | 782 | 1015.0 | Repeat | Note=Nebulin 1 | |
| Tgene | NEBL | chr17:34901666 | chr10:21101869 | ENST00000377122 | 22 | 28 | 314_348 | 782 | 1015.0 | Repeat | Note=Nebulin 9 | |
| Tgene | NEBL | chr17:34901666 | chr10:21101869 | ENST00000377122 | 22 | 28 | 349_385 | 782 | 1015.0 | Repeat | Note=Nebulin 10 | |
| Tgene | NEBL | chr17:34901666 | chr10:21101869 | ENST00000377122 | 22 | 28 | 386_422 | 782 | 1015.0 | Repeat | Note=Nebulin 11 | |
| Tgene | NEBL | chr17:34901666 | chr10:21101869 | ENST00000377122 | 22 | 28 | 423_459 | 782 | 1015.0 | Repeat | Note=Nebulin 12 | |
| Tgene | NEBL | chr17:34901666 | chr10:21101869 | ENST00000377122 | 22 | 28 | 460_496 | 782 | 1015.0 | Repeat | Note=Nebulin 13 | |
| Tgene | NEBL | chr17:34901666 | chr10:21101869 | ENST00000377122 | 22 | 28 | 497_533 | 782 | 1015.0 | Repeat | Note=Nebulin 14 | |
| Tgene | NEBL | chr17:34901666 | chr10:21101869 | ENST00000377122 | 22 | 28 | 534_569 | 782 | 1015.0 | Repeat | Note=Nebulin 15 | |
| Tgene | NEBL | chr17:34901666 | chr10:21101869 | ENST00000377122 | 22 | 28 | 570_599 | 782 | 1015.0 | Repeat | Note=Nebulin 16 | |
| Tgene | NEBL | chr17:34901666 | chr10:21101869 | ENST00000377122 | 22 | 28 | 600_635 | 782 | 1015.0 | Repeat | Note=Nebulin 17 | |
| Tgene | NEBL | chr17:34901666 | chr10:21101869 | ENST00000377122 | 22 | 28 | 636_666 | 782 | 1015.0 | Repeat | Note=Nebulin 18 | |
| Tgene | NEBL | chr17:34901666 | chr10:21101869 | ENST00000377122 | 22 | 28 | 64_99 | 782 | 1015.0 | Repeat | Note=Nebulin 2 | |
| Tgene | NEBL | chr17:34901666 | chr10:21101869 | ENST00000377122 | 22 | 28 | 667_693 | 782 | 1015.0 | Repeat | Note=Nebulin 19 | |
| Tgene | NEBL | chr17:34901666 | chr10:21101869 | ENST00000377122 | 22 | 28 | 694_728 | 782 | 1015.0 | Repeat | Note=Nebulin 20 | |
| Tgene | NEBL | chr17:34901666 | chr10:21101869 | ENST00000377122 | 22 | 28 | 729_759 | 782 | 1015.0 | Repeat | Note=Nebulin 21 | |
| Tgene | NEBL | chr17:34901666 | chr10:21101869 | ENST00000377122 | 22 | 28 | 760_794 | 782 | 1015.0 | Repeat | Note=Nebulin 22 | |
| Tgene | NEBL | chr17:34901666 | chr10:21101869 | ENST00000417816 | 3 | 7 | 100_136 | 119 | 271.0 | Repeat | Note=Nebulin 3 | |
| Tgene | NEBL | chr17:34901666 | chr10:21101869 | ENST00000417816 | 3 | 7 | 29_63 | 119 | 271.0 | Repeat | Note=Nebulin 1 | |
| Tgene | NEBL | chr17:34901666 | chr10:21101869 | ENST00000417816 | 3 | 7 | 64_99 | 119 | 271.0 | Repeat | Note=Nebulin 2 |
Top |
Fusion Gene Sequence for GGNBP2-NEBL |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >33031_33031_1_GGNBP2-NEBL_GGNBP2_chr17_34901666_ENST00000304718_NEBL_chr10_21101869_ENST00000377122_length(transcript)=6882nt_BP=409nt ACCTTCCCCTCGTCCCCCGCCCTCCTTCTTCCACTCCCCGCGGCGCGAGCGGCTGACTGCCCGTAGAGGAAACGACATTCGGAGCTGCGC TCCCGCCCAGGCCGGCCCTGACGCGGGCCTCGTCAGCCAGTAACAGGGAGCAGAGGTGGGAGTTAGCGAGGCGACCACGAAAACGGTGAA GGTCGGAACCGACAGCCTCCTCCGAGAAGGGCAGGAGCTGGGAGGAGGCGGCAGCGGCGGCGGCAGAAACAGCAGCGGCGGCGGCGGCGG CAGCTGGGAGGAGGTGGTGACGGTGGCAACGGCAGCGTCGGGGACGATGGCGCGACTCGTGGCAGTGTGCAGGGACGGGGAGGAGGAGTT CCCCTTCGAGAGGAGGCAGATTCCCCTCTACATAGACGACACCCTGACGGTAAAATACCATGAAGATTTTGAAAAAACAAAGGGGAGAGG CTTTACTCCCGTCGTGGACGATCCTGTGACAGAGAGAGTGAGGAAGAACACCCAGGTGGTCAGCGATGCTGCCTATAAAGGGGTCCACCC TCACATCGTGGAGATGGACAGGAGACCTGGAATCATTGTTGACCTCAAAGTTTGGCGCACAGATCCTGGCTCCATCTTCGACCTTGATCC CCTGGAAGACAATATTCAGTCTAGAAGTCTCCATATGCTCTCTGAAAAGGCGAGTCACTATAGGCGACACTGGTCTCGATCCCATTCCAG CAGTACTTTCGGTACAGGTCTCGGAGACGACAGGTCAGAAATCTCCGAGATTTACCCTAGCTTTTCATGCTGCAGTGAGGTAACAAGACC GTCTGATGAAGGAGCACCTGTTCTTCCCGGAGCCTATCAGCAAAGCCATTCCCAAGGCTATGGCTACATGCACCAGACCAGTGTGTCATC CATGAGATCAATGCAGCATTCACCAAATCTAAGGACCTACCGAGCCATGTACGATTACAGTGCCCAGGATGAAGACGAGGTCTCCTTTAG AGACGGCGACTACATCGTCAACGTGCAGCCTATTGACGATGGCTGGATGTACGGCACAGTGCAGAGAACAGGGAGAACAGGAATGCTCCC AGCGAATTACATTGAGTTTGTTAATTAATTATTTCTCCCTGCCCTTTGAGCTTTATTCTAATGTATCCCAAACCTAATCTTTTTAAAAGA TAGAAGATACTTTTAAGACAACTTGGCCATTATTTTACAATGATGTATCCTTCCTTTGACAATTAGACACACAGGTACCAGGAAGAAGGA ATGACCTCTGGGCTGAAAACAGCAGCATTTTCAGTAATTCCTACAAACAAAAATCTTTGTGTCTGGACACCTGGTGCTGCTAATTGTGTT CATGGTTTCCTTTGATTGGCTATTGAACCCTTCTGGGAAATGTATTTTTGTAGACTTTAATAGAGAAGTTGATTGTCCCTTAAATGTAGT GTGTGTTTGAAACTTCTTAGCTGTCACTTTGGAATCACCCCAAGCCAATTCTCTTAACTCTGTAATGCAGCCAATAATTTCAAACCCGTT TTGCTTTTGAGTCATGAGGCAATTTCCAATATTAGTGAAAATTGCCCAATATAATAAGTGTAAACAGTGGCAGAAGGACAGTCTGGTTAA AATTATATTGACTGGTGGCCTTAGGGATCTAGAAACTTCTACTAAACAGAGAAATTTCCTTGTTCCCTAGGCTGACTGGTATCTATTTAT TTCTCATTTGTACCAAGGCATCTCCTACTCTCCATTTATATTCTATGGACCCAAGTCTATGCTCAGTTCCACAGAATGTCAGGACCAAAT AACTTCACAGCTACTCTGCAAAGGGCAAATTATAATGTCATTGATATAATTTCCCTAGTAGCATTTACCCTGTTGCATGTCATGTAGATT CAAGCTTCTGTAACATAGGCAGCTGCACTGCGCGTTCCTATTATTGAAGCAAAAAGGGTGACTGATACCTAAAAGCCCTTTCTTCCTCTA GTCGCCAGCTCATCAGAAAAACATACTTTGAAAAGATGCTTGAGATTTTCCTGCTGCATCGCACTCTAGTTTTGAAGGATTTACATCTTA GGAAATAACATGTATACTCTAGTAAATAAGCGATTTAGGTGTTCCATTGAACAGCTTTGATTAACTTAATGCCACCATTGATTTCAAAGT GAAGAAAATGTAACAGAAGCCAGTGAAGCAATGGAAGCTGGAGTGTGACTGGAAAAATACTCAGCAAACAAAGTTACCAATTCCATACAG AGATGATCTGGTATCTTCTTTTGGAAAATGGTATTCAAATTCTGGAATGGAAATCTAGCCACCAAAACGGGTTAATCAAAAGACGTCCTT TTCCATTTTTTTTTGCTTTTATTTTCTAAATCATTTTTAAGGGAATGAAACAGGAATGTCATCAGAGATTTTTTAGTACAGGCCCAAGAG CCTGTTCTCTAAGAAAGAAATTGTTGCCATGTTTTGATTTTCGAATAAGTGACTTTGCAGGCTTTATGCTAGCCCTTGCTGGTGGGTCTT GAAATTTCATCCAGAGTCTGCAGTCCAGGTCACCAAGCCAGCGGCACCCGTCGGCAACCCTGTGTTTTTCTGATTGTGCCGTTTACTGTG ACCTGCAACGGGGTGGCATTCACTTAGGGTCTGACTTCACAGCTATGACAAAACCGAAAAAGCAAAACTGCAAAAAAGTACTAAGATGTA CGGGTCTTGGGGATATCTGCCTTATATGTTATATTCAAGGAAATTAACAAAACATCCTGTAAAACATCGTTTAAGGAAACGTTTACTAGT CCAAAGGCCAAAGCTAATTTATTTCCACTTTAGAAAAGTTAGCACATGCTTTTGAAAATCTGTGATTTCATTTTATTAGGCTAAAAGGGT AAATAGGCTTTATTACACTGAAGCTGCATCTATATGTCACTGACATAAAGTTGAAAAAATAAATGCAGGCAAATAACTAGAGACTTCTTT TAAGGGGGTTTGGCTGGTTTTCTCTCACTGAAATGGCCAGTCGTGATTAAAGTGATAAAACCCCATATCTGTTTTGGTATATTGTACACA AACCTACAAAAATAAACTGAACTTGCAATATTTTTGCAAAAAAATCTGTCGTTAAAACTGAGGATAAAATACCTGCTCAATTTTATTTTA CTAAGTATATATTTACATTTCACCCAGGCAGGCCATTTTCTTTTGTGATTATAAGAAAGAGTAGTTGTTGATTAAATTTTCAGACTAAAT ATAGGACAGGTACAATTTTGGATAAATAGCACATTTATAAGAACCGCAATGAAAACTGACTTGAAATAATGCTTGTAATCAGGAAAGTAA TTTCATCCACCGATTTCAAAACCAGATTCACTGAGCATAAAAGTCAATACATATTTGAGGAATAAGTCTCCTAAAATTTTAAGCTTCACG TAATAATGTTTGCATAGCAAAATATTTCTGCTTCAAGCCTTTAGGAATTAAGATCTGATCAGAATTTAACTAAAGGGTAGTTGTTTTACA ATGAAGACTAAAACTGAACAAGATGTTGCATGCTCTTGAGGCCATAATTTGGTAGTGTTGGCAGTTGTTAATAAAGCTTGTCAGGATGTT AAGCATCTCAGGAGAAATATTGGAAAATTATATGTATAAAACCAAAGTGCTATTTTTAAAAGCATCATTTAAAAAAAAATGACATGCCTG AACAACTTTTCCACTTTCCACGTGCTTCCCTCCCACCTTTGGTTTGGCAACAGGTATCTCGTGCATGAAGCTGACAGCTAAAGAAGATTT TAAAAATTGAGTTAAAGATGACTGTGTAAATGTCCAAGCACAGAGAGCATGCACCTGACTTTCTAAAGTTTGATGTGTTCTCAAGCCTGA CAGAAGCACAAGGAACAGTTTGATACACTTTTAAAAGGTTCTGAAAACAAAGCTGTATAGGGATCCTCTCTCTCTTGAGCAAAGTATAGC AACAGAATATATTGCTTTTGTTGTAAGCTTTTGTAGTACATGTTTTTACTAATAATTCTTGTTCTCTAGAAAGCTTTCTATTTCTAACCT ATGGCAAAATGAATCCTTCATGTCTTCTTGTTATTGTTTACACACTTGCAGTGTAGCCCAGTTTGAAATATTTATTTGGTTATCAACTGC CCATGGAGGAGGCTCTTGATGATCCCAGGTCTCCTCGACCTCCATACACCACACAGGCATTTGTAAGCACAGTTTCCACAAGCACCTTGT AGGAATATGGATAAGATTAGACCAGCCCCTCTCTGTCCACTGGGTTTATTTCTTGAAGAAGATGCAGATCTGGTTTTTCCAATGTGCCAC AGTCTTTCCTTATCCTCTCCATGCTGAGCTTGACAACACTCTGGGAATGAGGAACAAGACTTTTTCTAAAAAGATAGTGGAAGTTCAAGG GATGTACCTCGTTTTCAGGTTCATCCATCTCCAGTGGAATGTTTTCAATAAAAGATGAAGAAAATGTGTGTGATCTTTAATAACACATCC CTATAGAAAGTGGATAAAAGATATACCAAAACTGTAATACAGATATATACAAATATAGGTGCCTTTTTGATTACTCTTGTTTGTCTAGTA TGCTCTTGGAAAGAAAACCAAGCAAGCAAGTTGCTGCCTATTCTATAGTAATATTTTATTACACATGATTGATATTTTTGTGGTAGGGAA GTGGGATGCTCCTCAGATATTAAAGGTGTTAGCTGATTGTATTTTATCTCTAAAGATTTAGAACTTTAGAAAATGCCGACTTCTTCCATC TATTTCTGAAAGGTTCTTTGTGGATTTATATAGAGTTGAGCTATATAAACATTAACTTTAGATTTGGGATTTAAAATGCCTATTGTAAGA TAGAATAATTGTGAGGCTGGATTCACTACACAAGATGAACTTCACTTCATAAATTAATTATACCTTAGCGATTTGCTTCTGATAATCTAA AAGTGGCTAGATTGTGGTTGTTTTGGTTAAGGTGATATGGAGGTGGGAGAGCTTTTAGTTAAGTAAGAAGCTATGTAAACTGACAAGGAT GCTAAAATAAAAGTCTCTGAAGTATTCCATGCCTTTTGGACCCTTTCCTCGCAACTAACTGTCAACTGTTGATCAAAAAAGTCAAGGCAT TGTATGTTGCTTCTGTGGTTATTATTCTGTGATGCTTAGACTACTTGAACCCATAAACTTGGAAGAATCTTTGAGCAAATTTTCTCAGTT GTCTGTATGACTTCAGTATATTCCTGGGAATGCCATAGGATTTTTTGTGCTTGATACATGGTATCCAGTTTGCATAGTATCACTTCTTTG TAATCCAGTTGCTGTTAAGAATGATGTACTTTAAAGGAAAAGAGAAAACTGCATCACAGTCCCATTCTCCAGTGTCCATGCAATGAATTG CTGAGCATTTAGGAAGCAGCACCAAGTCTATTACAGGCATGGTGTGAAACTTGATGTTTGACCTGTGATCAAAATTGAACCATTGTACAG TTTGGCTTCTGTTTGCTTCAAAATATGTAGAATTGTGGTTGATGATTAATTTGCGAGACTAACTTTGAGAGTGTAACAGTTTTGAAGAAA ACATTGAATGTTTTGCAAATGAAGGGGCTTCACGGAATGTTACAATGTTACTAATATAATTTGGCTTTTGTTATGCAAATTGTTAACACC AGCTATTAAAATATATTTTAGTAGAAATGCTTTAATTCATATTTTTTTCCTCTACACTGTGAATCTTTAAGCCTTGGTGGACTAGAGCAA CATCGTGCTGCCCAAAGGACTAACCTATGCAAACTAGTTCACATTTTAGTGGATGTCGCAGTTAATGTGTAATAAGACATTATTTCCCCT GCATAATGTACAACAGCATTGAAATGACACATTAAGCCTAGCATCACATTGTATAGTACAGTCACTCACAAACCCTTCAAGGCTACCCTA ATCATTAACATTAATATTTGTTTAAAAGCAAATCACCGATTTATCTATTGAAACTACTTAAATGACGGCAAACCAGGAATGACAGATGGC TGTGTCAGCAATGGCTTTAATGTGTTCCCTGCAAGTGGTCTCCTATGATAGAACTGCGTTCTCAAATGCACTCTCTTCAGGGTCTTAATA TTCTGTGTTTTCTCTCTGTATTTGTAAAACATTATAACACATTAATTTCCTATCTCTACACATTTGGTTTGCTTAAATAAATGCAGGATA TAAAAAAAATGGTTCACTTCTTGGCTCTCACCGTGGTTTCTTGGAGCATGGGTTGTTAGATGCAAGCAATGCACCCTAATAATACCCCGG GTCTGAGATTTAACATGACAACTCACATCAAATCGCATCAGAGGTGTGTGCTGCCTTCAGTGCATTTACATTGGTGAATCAGTCAAGATA TTTTCCTCCCCCAAATAAACTTAGTTGTAAGTGATAACAATATTATGCTTCTCCAAGCTCAGTATCTTTCTGATTTTATATCAAAGTACC GCAACAATGCATCATTGTAGTTAATTTATTTCAAGAATAAATTCCTCATATGTCCTCAATAGTACAATTCTAATTTTCTTCTATTCATAA GATGAAAGAAATGGTTTGGAGCATAGAATAGAAAGTGCACAAATTGAGTACATAAAATGGGAAGCAACTGATTTCTCAGCTAAGAAAGGC TCATTTATCACAGAACACAATTGCTTTTCTCCCCCCACTACGCTTCCCATAATTGAAAAAGTGAGTCCCTATTTTTCACACTCATATAAA TCTATGCGATTTGGATGCTAGTCTTATTGTATTATTTTGTAAAACTTTCTCTTTGGCTCATAATCCTTCCTAATTGTAAATTGATAAACT >33031_33031_1_GGNBP2-NEBL_GGNBP2_chr17_34901666_ENST00000304718_NEBL_chr10_21101869_ENST00000377122_length(amino acids)=296AA_BP=64 MGGGGSGGGRNSSGGGGGSWEEVVTVATAASGTMARLVAVCRDGEEEFPFERRQIPLYIDDTLTVKYHEDFEKTKGRGFTPVVDDPVTER VRKNTQVVSDAAYKGVHPHIVEMDRRPGIIVDLKVWRTDPGSIFDLDPLEDNIQSRSLHMLSEKASHYRRHWSRSHSSSTFGTGLGDDRS EISEIYPSFSCCSEVTRPSDEGAPVLPGAYQQSHSQGYGYMHQTSVSSMRSMQHSPNLRTYRAMYDYSAQDEDEVSFRDGDYIVNVQPID -------------------------------------------------------------- >33031_33031_2_GGNBP2-NEBL_GGNBP2_chr17_34901666_ENST00000304718_NEBL_chr10_21101869_ENST00000377159_length(transcript)=1141nt_BP=409nt ACCTTCCCCTCGTCCCCCGCCCTCCTTCTTCCACTCCCCGCGGCGCGAGCGGCTGACTGCCCGTAGAGGAAACGACATTCGGAGCTGCGC TCCCGCCCAGGCCGGCCCTGACGCGGGCCTCGTCAGCCAGTAACAGGGAGCAGAGGTGGGAGTTAGCGAGGCGACCACGAAAACGGTGAA GGTCGGAACCGACAGCCTCCTCCGAGAAGGGCAGGAGCTGGGAGGAGGCGGCAGCGGCGGCGGCAGAAACAGCAGCGGCGGCGGCGGCGG CAGCTGGGAGGAGGTGGTGACGGTGGCAACGGCAGCGTCGGGGACGATGGCGCGACTCGTGGCAGTGTGCAGGGACGGGGAGGAGGAGTT CCCCTTCGAGAGGAGGCAGATTCCCCTCTACATAGACGACACCCTGACGGTAAAATACCATGAAGATTTTGAAAAAACAAAGGGGAGAGG CTTTACTCCCGTCGTGGACGATCCTGTGACAGAGAGAGTGAGGAAGAACACCCAGGTGGTCAGCGATGCTGCCTATAAAGGGGTCCACCC TCACATCGTGGAGATGGACAGGAGACCTGGAATCATTGTTGCACCTGTTCTTCCCGGAGCCTATCAGCAAAGCCATTCCCAAGGCTATGG CTACATGCACCAGACCAGTGTGTCATCCATGAGATCAATGCAGCATTCACCAAATCTAAGGACCTACCGAGCCATGTACGATTACAGTGC CCAGGATGAAGACGAGGTCTCCTTTAGAGACGGCGACTACATCGTCAACGTGCAGCCTATTGACGATGGCTGGATGTACGGCACAGTGCA GAGAACAGGGAGAACAGGAATGCTCCCAGCGAATTACATTGAGTTTGTTAATTAATTATTTCTCCCTGCCCTTTGAGCTTTATTCTAATG TATCCCAAACCTAATCTTTTTAAAAGATAGAAGATACTTTTAAGACAACTTGGCCATTATTTTACAATGATGTATCCTTCCTTTGACAAT TAGACACACAGGTACCAGGAAGAAGGAATGACCTCTGGGCTGAAAACAGCAGCATTTTCAGTAATTCCTACAAACAAAAATCTTTGTGTC >33031_33031_2_GGNBP2-NEBL_GGNBP2_chr17_34901666_ENST00000304718_NEBL_chr10_21101869_ENST00000377159_length(amino acids)=215AA_BP=64 MGGGGSGGGRNSSGGGGGSWEEVVTVATAASGTMARLVAVCRDGEEEFPFERRQIPLYIDDTLTVKYHEDFEKTKGRGFTPVVDDPVTER VRKNTQVVSDAAYKGVHPHIVEMDRRPGIIVAPVLPGAYQQSHSQGYGYMHQTSVSSMRSMQHSPNLRTYRAMYDYSAQDEDEVSFRDGD -------------------------------------------------------------- >33031_33031_3_GGNBP2-NEBL_GGNBP2_chr17_34901666_ENST00000304718_NEBL_chr10_21101869_ENST00000417816_length(transcript)=2706nt_BP=409nt ACCTTCCCCTCGTCCCCCGCCCTCCTTCTTCCACTCCCCGCGGCGCGAGCGGCTGACTGCCCGTAGAGGAAACGACATTCGGAGCTGCGC TCCCGCCCAGGCCGGCCCTGACGCGGGCCTCGTCAGCCAGTAACAGGGAGCAGAGGTGGGAGTTAGCGAGGCGACCACGAAAACGGTGAA GGTCGGAACCGACAGCCTCCTCCGAGAAGGGCAGGAGCTGGGAGGAGGCGGCAGCGGCGGCGGCAGAAACAGCAGCGGCGGCGGCGGCGG CAGCTGGGAGGAGGTGGTGACGGTGGCAACGGCAGCGTCGGGGACGATGGCGCGACTCGTGGCAGTGTGCAGGGACGGGGAGGAGGAGTT CCCCTTCGAGAGGAGGCAGATTCCCCTCTACATAGACGACACCCTGACGGTAAAATACCATGAAGATTTTGAAAAAACAAAGGGGAGAGG CTTTACTCCCGTCGTGGACGATCCTGTGACAGAGAGAGTGAGGAAGAACACCCAGGTGGTCAGCGATGCTGCCTATAAAGGGGTCCACCC TCACATCGTGGAGATGGACAGGAGACCTGGAATCATTGTTGCACCTGTTCTTCCCGGAGCCTATCAGCAAAGCCATTCCCAAGGCTATGG CTACATGCACCAGACCAGTGTGTCATCCATGAGATCAATGCAGCATTCACCAAATCTAAGGACCTACCGAGCCATGTACGATTACAGTGC CCAGGATGAAGACGAGGTCTCCTTTAGAGACGGCGACTACATCGTCAACGTGCAGCCTATTGACGATGGCTGGATGTACGGCACAGTGCA GAGAACAGGGAGAACAGGAATGCTCCCAGCGAATTACATTGAGTTTGTTAATTAATTATTTCTCCCTGCCCTTTGAGCTTTATTCTAATG TATCCCAAACCTAATCTTTTTAAAAGATAGAAGATACTTTTAAGACAACTTGGCCATTATTTTACAATGATGTATCCTTCCTTTGACAAT TAGACACACAGGTACCAGGAAGAAGGAATGACCTCTGGGCTGAAAACAGCAGCATTTTCAGTAATTCCTACAAACAAAAATCTTTGTGTC TGGACACCTGGTGCTGCTAATTGTGTTCATGGTTTCCTTTGATTGGCTATTGAACCCTTCTGGGAAATGTATTTTTGTAGACTTTAATAG AGAAGTTGATTGTCCCTTAAATGTAGTGTGTGTTTGAAACTTCTTAGCTGTCACTTTGGAATCACCCCAAGCCAATTCTCTTAACTCTGT AATGCAGCCAATAATTTCAAACCCGTTTTGCTTTTGAGTCATGAGGCAATTTCCAATATTAGTGAAAATTGCCCAATATAATAAGTGTAA ACAGTGGCAGAAGGACAGTCTGGTTAAAATTATATTGACTGGTGGCCTTAGGGATCTAGAAACTTCTACTAAACAGAGAAATTTCCTTGT TCCCTAGGCTGACTGGTATCTATTTATTTCTCATTTGTACCAAGGCATCTCCTACTCTCCATTTATATTCTATGGACCCAAGTCTATGCT CAGTTCCACAGAATGTCAGGACCAAATAACTTCACAGCTACTCTGCAAAGGGCAAATTATAATGTCATTGATATAATTTCCCTAGTAGCA TTTACCCTGTTGCATGTCATGTAGATTCAAGCTTCTGTAACATAGGCAGCTGCACTGCGCGTTCCTATTATTGAAGCAAAAAGGGTGACT GATACCTAAAAGCCCTTTCTTCCTCTAGTCGCCAGCTCATCAGAAAAACATACTTTGAAAAGATGCTTGAGATTTTCCTGCTGCATCGCA CTCTAGTTTTGAAGGATTTACATCTTAGGAAATAACATGTATACTCTAGTAAATAAGCGATTTAGGTGTTCCATTGAACAGCTTTGATTA ACTTAATGCCACCATTGATTTCAAAGTGAAGAAAATGTAACAGAAGCCAGTGAAGCAATGGAAGCTGGAGTGTGACTGGAAAAATACTCA GCAAACAAAGTTACCAATTCCATACAGAGATGATCTGGTATCTTCTTTTGGAAAATGGTATTCAAATTCTGGAATGGAAATCTAGCCACC AAAACGGGTTAATCAAAAGACGTCCTTTTCCATTTTTTTTTGCTTTTATTTTCTAAATCATTTTTAAGGGAATGAAACAGGAATGTCATC AGAGATTTTTTAGTACAGGCCCAAGAGCCTGTTCTCTAAGAAAGAAATTGTTGCCATGTTTTGATTTTCGAATAAGTGACTTTGCAGGCT TTATGCTAGCCCTTGCTGGTGGGTCTTGAAATTTCATCCAGAGTCTGCAGTCCAGGTCACCAAGCCAGCGGCACCCGTCGGCAACCCTGT GTTTTTCTGATTGTGCCGTTTACTGTGACCTGCAACGGGGTGGCATTCACTTAGGGTCTGACTTCACAGCTATGACAAAACCGAAAAAGC AAAACTGCAAAAAAGTACTAAGATGTACGGGTCTTGGGGATATCTGCCTTATATGTTATATTCAAGGAAATTAACAAAACATCCTGTAAA ACATCGTTTAAGGAAACGTTTACTAGTCCAAAGGCCAAAGCTAATTTATTTCCACTTTAGAAAAGTTAGCACATGCTTTTGAAAATCTGT GATTTCATTTTATTAGGCTAAAAGGGTAAATAGGCTTTATTACACTGAAGCTGCATCTATATGTCACTGACATAAAGTTGAAAAAATAAA >33031_33031_3_GGNBP2-NEBL_GGNBP2_chr17_34901666_ENST00000304718_NEBL_chr10_21101869_ENST00000417816_length(amino acids)=215AA_BP=64 MGGGGSGGGRNSSGGGGGSWEEVVTVATAASGTMARLVAVCRDGEEEFPFERRQIPLYIDDTLTVKYHEDFEKTKGRGFTPVVDDPVTER VRKNTQVVSDAAYKGVHPHIVEMDRRPGIIVAPVLPGAYQQSHSQGYGYMHQTSVSSMRSMQHSPNLRTYRAMYDYSAQDEDEVSFRDGD -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for GGNBP2-NEBL |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for GGNBP2-NEBL |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for GGNBP2-NEBL |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
