
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:GLB1-PSD2 (FusionGDB2 ID:33214) |
Fusion Gene Summary for GLB1-PSD2 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: GLB1-PSD2 | Fusion gene ID: 33214 | Hgene | Tgene | Gene symbol | GLB1 | PSD2 | Gene ID | 2720 | 84249 |
| Gene name | galactosidase beta 1 | pleckstrin and Sec7 domain containing 2 | |
| Synonyms | EBP|ELNR1|MPS4B | EFA6C | |
| Cytomap | 3p22.3 | 5q31.2 | |
| Type of gene | protein-coding | protein-coding | |
| Description | beta-galactosidaseacid beta-galactosidaseelastin binding proteinelastin receptor 1, 67kDalactase | PH and SEC7 domain-containing protein 2exchange factor for ADP-ribosylation factor guanine nucleotide factor 6 Cexchange factor for ARF6 Cpleckstrin homology and SEC7 domain-containing protein 2 | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | P16278 | . | |
| Ensembl transtripts involved in fusion gene | ENST00000399402, ENST00000307363, ENST00000307377, ENST00000445488, ENST00000497796, | ENST00000274710, | |
| Fusion gene scores | * DoF score | 9 X 9 X 7=567 | 6 X 6 X 6=216 |
| # samples | 9 | 8 | |
| ** MAII score | log2(9/567*10)=-2.65535182861255 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(8/216*10)=-1.43295940727611 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: GLB1 [Title/Abstract] AND PSD2 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | GLB1(33094983)-PSD2(139215308), # samples:3 | ||
| Anticipated loss of major functional domain due to fusion event. | GLB1-PSD2 seems lost the major protein functional domain in Hgene partner, which is a cell metabolism gene due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | GLB1 | GO:0044262 | cellular carbohydrate metabolic process | 11927518 |
 Fusion gene breakpoints across GLB1 (5'-gene) Fusion gene breakpoints across GLB1 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
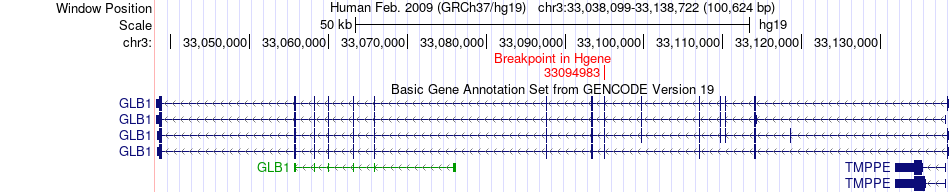 |
 Fusion gene breakpoints across PSD2 (3'-gene) Fusion gene breakpoints across PSD2 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
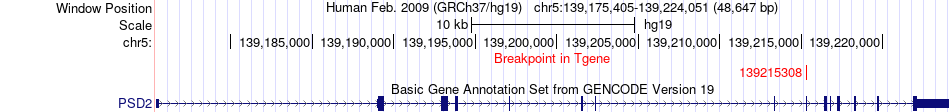 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | PRAD | TCGA-KK-A7B1-01A | GLB1 | chr3 | 33094983 | - | PSD2 | chr5 | 139215308 | + |
| ChimerDB4 | PRAD | TCGA-KK-A7B1 | GLB1 | chr3 | 33094983 | - | PSD2 | chr5 | 139215308 | + |
Top |
Fusion Gene ORF analysis for GLB1-PSD2 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| Frame-shift | ENST00000399402 | ENST00000274710 | GLB1 | chr3 | 33094983 | - | PSD2 | chr5 | 139215308 | + |
| In-frame | ENST00000307363 | ENST00000274710 | GLB1 | chr3 | 33094983 | - | PSD2 | chr5 | 139215308 | + |
| In-frame | ENST00000307377 | ENST00000274710 | GLB1 | chr3 | 33094983 | - | PSD2 | chr5 | 139215308 | + |
| In-frame | ENST00000445488 | ENST00000274710 | GLB1 | chr3 | 33094983 | - | PSD2 | chr5 | 139215308 | + |
| intron-3CDS | ENST00000497796 | ENST00000274710 | GLB1 | chr3 | 33094983 | - | PSD2 | chr5 | 139215308 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000307363 | GLB1 | chr3 | 33094983 | - | ENST00000274710 | PSD2 | chr5 | 139215308 | + | 3886 | 937 | 109 | 1893 | 594 |
| ENST00000445488 | GLB1 | chr3 | 33094983 | - | ENST00000274710 | PSD2 | chr5 | 139215308 | + | 3946 | 997 | 25 | 1953 | 642 |
| ENST00000307377 | GLB1 | chr3 | 33094983 | - | ENST00000274710 | PSD2 | chr5 | 139215308 | + | 3409 | 460 | 25 | 1416 | 463 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000307363 | ENST00000274710 | GLB1 | chr3 | 33094983 | - | PSD2 | chr5 | 139215308 | + | 0.002392453 | 0.9976076 |
| ENST00000445488 | ENST00000274710 | GLB1 | chr3 | 33094983 | - | PSD2 | chr5 | 139215308 | + | 0.001850516 | 0.9981495 |
| ENST00000307377 | ENST00000274710 | GLB1 | chr3 | 33094983 | - | PSD2 | chr5 | 139215308 | + | 0.000859237 | 0.99914074 |
Top |
Fusion Genomic Features for GLB1-PSD2 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| GLB1 | chr3 | 33094982 | - | PSD2 | chr5 | 139215307 | + | 3.58E-09 | 1 |
| GLB1 | chr3 | 33094982 | - | PSD2 | chr5 | 139215307 | + | 3.58E-09 | 1 |
| GLB1 | chr3 | 33094982 | - | PSD2 | chr5 | 139215307 | + | 3.58E-09 | 1 |
| GLB1 | chr3 | 33094982 | - | PSD2 | chr5 | 139215307 | + | 3.58E-09 | 1 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
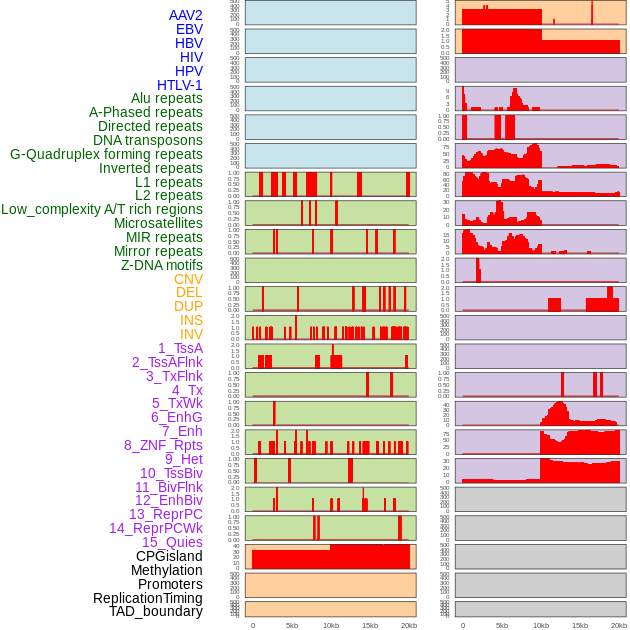 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
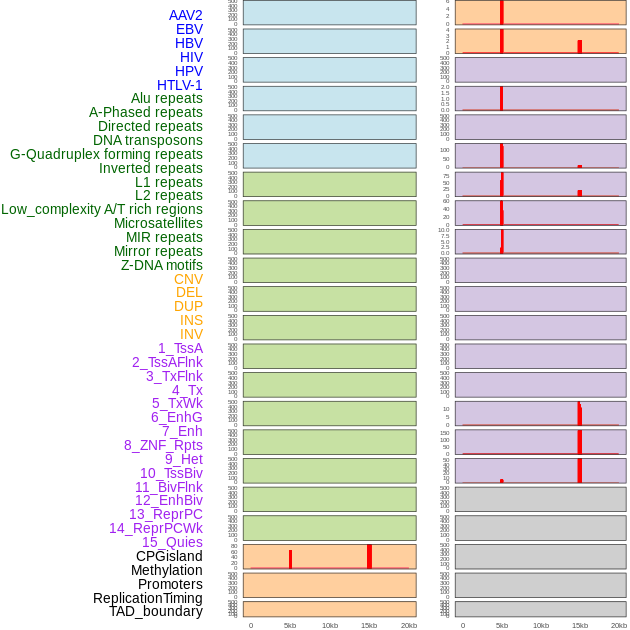 |
Top |
Fusion Protein Features for GLB1-PSD2 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr3:33094983/chr5:139215308) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| GLB1 | . |
| FUNCTION: [Isoform 1]: Cleaves beta-linked terminal galactosyl residues from gangliosides, glycoproteins, and glycosaminoglycans. {ECO:0000269|PubMed:15714521, ECO:0000269|PubMed:19472408, ECO:0000269|PubMed:2511208, ECO:0000269|PubMed:25936995, ECO:0000269|PubMed:8200356}.; FUNCTION: [Isoform 2]: Has no beta-galactosidase catalytic activity, but plays functional roles in the formation of extracellular elastic fibers (elastogenesis) and in the development of connective tissue. Seems to be identical to the elastin-binding protein (EBP), a major component of the non-integrin cell surface receptor expressed on fibroblasts, smooth muscle cells, chondroblasts, leukocytes, and certain cancer cell types. In elastin producing cells, associates with tropoelastin intracellularly and functions as a recycling molecular chaperone which facilitates the secretions of tropoelastin and its assembly into elastic fibers. {ECO:0000269|PubMed:10841810, ECO:0000269|PubMed:8922281}. | FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | PSD2 | chr3:33094983 | chr5:139215308 | ENST00000274710 | 7 | 15 | 651_680 | 453 | 772.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | PSD2 | chr3:33094983 | chr5:139215308 | ENST00000274710 | 7 | 15 | 512_625 | 453 | 772.0 | Domain | PH | |
| Tgene | PSD2 | chr3:33094983 | chr5:139215308 | ENST00000274710 | 7 | 15 | 622_639 | 453 | 772.0 | Transmembrane | Helical |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | PSD2 | chr3:33094983 | chr5:139215308 | ENST00000274710 | 7 | 15 | 260_462 | 453 | 772.0 | Domain | SEC7 |
Top |
Fusion Gene Sequence for GLB1-PSD2 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >33214_33214_1_GLB1-PSD2_GLB1_chr3_33094983_ENST00000307363_PSD2_chr5_139215308_ENST00000274710_length(transcript)=3886nt_BP=937nt CACAGCGGGGCGGGGCGCGGGGCTGAGCAGGCGCGAGGGCTGGCTGCTGGGCCGACGAGGGGCGGTGCCAGGCCGTGGGTCCTTAGTCAA GTGACGCGAAGCGGCCGGCCTGGGCGCCGACTGCAGAGCCGGGAGGCTGGTGGTCATGCCGGGGTTCCTGGTTCGCATCCTCCCTCTGTT GCTGGTTCTGCTGCTTCTGGGCCCTACGCGCGGCTTGCGCAATGCCACCCAGAGGATGTTTGAAATTGACTATAGCCGGGACTCCTTCCT CAAGGATGGCCAGCCATTTCGCTACATCTCAGGAAGCATTCACTACTCCCGTGTGCCCCGCTTCTACTGGAAGGACCGGCTGCTGAAGAT GAAGATGGCTGGGCTGAACGCCATCCAGACGTATGTGCCCTGGAACTTTCATGAGCCCTGGCCAGGACAGTACCAGTTTTCTGAGGACCA TGATGTGGAATATTTTCTTCGGCTGGCTCATGAGCTGGGACTGCTGGTTATCCTGAGGCCCGGGCCCTACATCTGTGCAGAGTGGGAAAT GGGAGGATTACCTGCTTGGCTGCTAGAGAAAGAGTCTATTCTTCTCCGCTCCTCCGACCCAGATTACCTGGCAGCTGTGGACAAGTGGTT GGGAGTCCTTCTGCCCAAGATGAAGCCTCTCCTCTATCAGAATGGAGGGCCAGTTATAACAGTGCAGGTTGAAAATGAATATGGCAGCTA CTTTGCCTGTGATTTTGACTACCTGCGCTTCCTGCAGAAGCGCTTTCGCCACCATCTGGGGGATGATGTGGTTCTGTTTACCACTGATGG AGCACATAAAACATTCCTGAAATGTGGGGCCCTGCAGGGCCTCTACACCACGGTGGACTTTGGAACAGGCAGCAACATCACAGATGCTTT CCTAAGCCAGAGGAAGTGTGAGCCCAAAGGACCCTTGACCCTTTACAACTCCATCAAGAATGAAAAGCTGGAATGGGCCATTGATGAGGA TGAGCTGAGGAAATCCCTGTCTGAGCTGGTGGATGACAAGTTCGGGACAGGCACGAAGAAGGTGACGCGAATCCTGGATGGTGGCAACCC CTTCCTGGATGTCCCACAGGCGCTCAGTGCCACCACCTACAAGCACGGCGTCCTGACCCGGAAGACTCACGCTGACATGGATGGCAAGAG GACGCCCCGTGGGAGGCGTGGCTGGAAGAAATTCTACGCAGTGCTCAAAGGGACCATCCTGTACCTGCAGAAGGATGAGTACAGGCCTGA CAAAGCTCTATCGGAGGGTGACCTGAAGAACGCCATTCGCGTGCATCACGCTCTGGCCACCAGGGCCTCTGACTACAGCAAGAAGTCCAA CGTGCTGAAGCTTAAGACAGCCGACTGGAGGGTATTCCTCTTCCAGGCACCGAGCAAGGAAGAAATGCTGTCCTGGATCCTCAGGATCAA CCTGGTGGCAGCCATCTTCTCTGCCCCGGCCTTCCCAGCCGCTGTCAGCTCCATGAAGAAGTTCTGTCGGCCCCTGCTGCCCTCCTGCAC CACCCGCCTCTGCCAGGAGGAGCAACTGCGGTCTCATGAGAATAAGTTGAGGCAGCTGACTGCGGAGCTGGCCGAACACAGGTGTCACCC AGTCGAGAGGGGCATCAAGTCCAAGGAGGCCGAGGAGTACCGGTTGAAGGAGCACTATCTCACCTTCGAGAAAAGCCGTTATGAGACCTA TATCCACCTCCTGGCTATGAAAATCAAAGTGGGCTCAGATGATCTGGAGCGGATTGAGGCCCGGCTGGCCACTCTGGAAGGGGATGACCC TTCTCTCCGGAAGACACATTCAAGCCCTGCCCTCAGCCAGGGCCATGTGACTGGCAGCAAAACCACAAAGGATGCCACTGGGCCTGATAC TTAGCTGACATGGATTTGCAGACCCCAGGGTGGGCAGATGTCTCCAGTGGGGTCAGTGAGCACAATTCCAGCCAGGGGCCACTTGGACCA AGCTCCAGTCAGTTGATGGGCAGCTAGAGGGGTGCAGAAAGCCTGTGGGCCCAGGAGATGGAGATGCCGTTTGTGGCGTTGATCTCCTTG CGTCCTTGGGCATCTCCGGGCATCAGACCCTCTCCCTGGCCCTTGTTTTCCTCTCCACCATGGAGCCTCATTTTGTAGGCCAGTTGTGTG CATGCTCTAGACACCACCTCGCTGGAGAAGCTGGAAGGGCTGTTGTCTTCCCAGGTCTTTCTCTTCTCATCAAGCTCCTCTCCTCATCTT TTTTGTGTGTGAGGGCAGGTCTTGACTCTAGGTCTCAGCTGGAACCCCACCCTTTCTCCTCCTCCTTCCTCTGAGTTGACCAGCAGCAGG TCTGCCGACCACCAGCACCATCCTCTCCTCCCAGCAGCCTCCAGAACCATGCCCAGGTCTCCTGCCTCACATCACAATAATCTGGGACCC AGGCTTGTGCCCTTTCAGTGTAAAGCTGACTCCATCACATGTGCATCCACTTCTTTTCATCCATTGAGATCACACTGCCTCCTTTTTATA CAGACACAAATATACATCTATAAGAATAATATATACATAAGGAACCCCTGAAAGATGGTTTTGGAACTGGAATCAGTTAGAGGATGAAAT CAGATAAAGGAAAAGCCTATTTTGGAGCTTCCCCTGTTAGGAAGGATGGCTGCACCTGGCCCCCTGGCATTCCTGACGCTCTAGGAGGGA AGGGGGAGGCAGTGCTGGCCTCCCTTGCCCTGTTTTTCCCTCTTCCAGCTGACCTGTGACTTATACTGCTCTTACCGATGATACTTTTGG AAAAAATAGAGCGTGTATGCACCGCCCCGTTTGTCCCATGGATATCCTGGGGTGTGAGTCGGATGGGACCACGGCCCTGTTTATATTTGG GTCTTTATGTTGGTGCTGCCAGGTCTCTGAGCTCCAGAGGTGGCCTCTTGGACAGATCTACTGCTATAGGAATAAAAGACACTCTGTCTC GCAAATGGCTGCTTGTCAACAAGCCCAAAGATGCTTGTCGGAGGACGGTTATGGAAGCCCTTAATTCTTGGTTGTGGGAAAAGGTGGAAT GACAAGTTATTGATTGTTTTTCTGTCGCTATTTCTTTCATTTGTCTAGTGAATCAGAAAGGCTTAGCCAAGGCCACATCTGGGAAGAGTG GAGAAATTTGCCACTTGACGATCACGGATTAGCTAGCACCTTTAAGCCCTGCATTTCTCCAACTGACAAGTGGGTGGGGGTGATGGCACA TTCAGTGTGGCTATGAAGAGCGAATCCTCTCTATTGTTTAAATAGATTACTGTAGTTTGGCCAGGAATTTGGCGTCAGTGGTAACACACT TAGTTAATAAAATAAGCCAGGCTTGCAACTAAGTATCTAACTTTACAGGCCCACTCACATTTGAGGCAAGGGGCTATTGAGTATGTGGAG AGATGTAGTGATTTAAATTCAGATTATTTAAGTTGGATCAGCTGAAGTGTGTTTTAGACCCAAACCATCTGGCCCCTTCGTTTTGCTCAG AGGAAGTAAATGTTCACTTAAATGAAATTGAAAACGCCATGTGGCACCACAAAAGAGCTCTCTGTACTTTCCCCATGCTGCCTCAAAAGT TCTGTGAGTTTCGGGGTCAGTGTCCCACCCTTCACTTCCCGAGGGCGGGTGAGTGGAGAGCAGAGCCAGGAGCTCTGGCAGCTGTGGACA GATGTGCTTCCTGAGCATGGGTTGTGCCTCCCATCAGTAAAAAAATGTTTAGTTCACTTCCTTAATTGTATAATTATTTATTTGTAAATT ATATACATGTACTACTGTACTAAAATATTATGTACATTATAAAACATACACAAAAATAGAAATTTAAAAAAGATGAGATGAAAATAAATC >33214_33214_1_GLB1-PSD2_GLB1_chr3_33094983_ENST00000307363_PSD2_chr5_139215308_ENST00000274710_length(amino acids)=594AA_BP=276 MGADCRAGRLVVMPGFLVRILPLLLVLLLLGPTRGLRNATQRMFEIDYSRDSFLKDGQPFRYISGSIHYSRVPRFYWKDRLLKMKMAGLN AIQTYVPWNFHEPWPGQYQFSEDHDVEYFLRLAHELGLLVILRPGPYICAEWEMGGLPAWLLEKESILLRSSDPDYLAAVDKWLGVLLPK MKPLLYQNGGPVITVQVENEYGSYFACDFDYLRFLQKRFRHHLGDDVVLFTTDGAHKTFLKCGALQGLYTTVDFGTGSNITDAFLSQRKC EPKGPLTLYNSIKNEKLEWAIDEDELRKSLSELVDDKFGTGTKKVTRILDGGNPFLDVPQALSATTYKHGVLTRKTHADMDGKRTPRGRR GWKKFYAVLKGTILYLQKDEYRPDKALSEGDLKNAIRVHHALATRASDYSKKSNVLKLKTADWRVFLFQAPSKEEMLSWILRINLVAAIF SAPAFPAAVSSMKKFCRPLLPSCTTRLCQEEQLRSHENKLRQLTAELAEHRCHPVERGIKSKEAEEYRLKEHYLTFEKSRYETYIHLLAM -------------------------------------------------------------- >33214_33214_2_GLB1-PSD2_GLB1_chr3_33094983_ENST00000307377_PSD2_chr5_139215308_ENST00000274710_length(transcript)=3409nt_BP=460nt AGTCAAGTGACGCGAAGCGGCCGGCCTGGGCGCCGACTGCAGAGCCGGGAGGCTGGTGGTCATGCCGGGGTTCCTGGTTCGCATCCTCCC TCTGTTGCTGGTTCTGCTGCTTCTGGGCCCTACGCGCGGCTTGCGCAATGCCACCCAGAGGATGTTTGAAATTGACTATAGCCGGGACTC CTTCCTCAAGGATGGCCAGCCATTTCGCTACATCTCAGGAAGCATTCACTACTCCCGTGTGCCCCGCTTCTACTGGAAGGACCGGCTGCT GAAGATGAAGATGGCTGGGCTGAACGCCATCCAGACATTACCTGGCAGCTGTGGACAAGTGGTTGGGAGTCCTTCTGCCCAAGATGAAGC CTCTCCTCTATCAGAATGGAGGGCCAGTTATAACAGTGCAGGCAGCAACATCACAGATGCTTTCCTAAGCCAGAGGAAGTGTGAGCCCAA AGGACCCTTGACCCTTTACAACTCCATCAAGAATGAAAAGCTGGAATGGGCCATTGATGAGGATGAGCTGAGGAAATCCCTGTCTGAGCT GGTGGATGACAAGTTCGGGACAGGCACGAAGAAGGTGACGCGAATCCTGGATGGTGGCAACCCCTTCCTGGATGTCCCACAGGCGCTCAG TGCCACCACCTACAAGCACGGCGTCCTGACCCGGAAGACTCACGCTGACATGGATGGCAAGAGGACGCCCCGTGGGAGGCGTGGCTGGAA GAAATTCTACGCAGTGCTCAAAGGGACCATCCTGTACCTGCAGAAGGATGAGTACAGGCCTGACAAAGCTCTATCGGAGGGTGACCTGAA GAACGCCATTCGCGTGCATCACGCTCTGGCCACCAGGGCCTCTGACTACAGCAAGAAGTCCAACGTGCTGAAGCTTAAGACAGCCGACTG GAGGGTATTCCTCTTCCAGGCACCGAGCAAGGAAGAAATGCTGTCCTGGATCCTCAGGATCAACCTGGTGGCAGCCATCTTCTCTGCCCC GGCCTTCCCAGCCGCTGTCAGCTCCATGAAGAAGTTCTGTCGGCCCCTGCTGCCCTCCTGCACCACCCGCCTCTGCCAGGAGGAGCAACT GCGGTCTCATGAGAATAAGTTGAGGCAGCTGACTGCGGAGCTGGCCGAACACAGGTGTCACCCAGTCGAGAGGGGCATCAAGTCCAAGGA GGCCGAGGAGTACCGGTTGAAGGAGCACTATCTCACCTTCGAGAAAAGCCGTTATGAGACCTATATCCACCTCCTGGCTATGAAAATCAA AGTGGGCTCAGATGATCTGGAGCGGATTGAGGCCCGGCTGGCCACTCTGGAAGGGGATGACCCTTCTCTCCGGAAGACACATTCAAGCCC TGCCCTCAGCCAGGGCCATGTGACTGGCAGCAAAACCACAAAGGATGCCACTGGGCCTGATACTTAGCTGACATGGATTTGCAGACCCCA GGGTGGGCAGATGTCTCCAGTGGGGTCAGTGAGCACAATTCCAGCCAGGGGCCACTTGGACCAAGCTCCAGTCAGTTGATGGGCAGCTAG AGGGGTGCAGAAAGCCTGTGGGCCCAGGAGATGGAGATGCCGTTTGTGGCGTTGATCTCCTTGCGTCCTTGGGCATCTCCGGGCATCAGA CCCTCTCCCTGGCCCTTGTTTTCCTCTCCACCATGGAGCCTCATTTTGTAGGCCAGTTGTGTGCATGCTCTAGACACCACCTCGCTGGAG AAGCTGGAAGGGCTGTTGTCTTCCCAGGTCTTTCTCTTCTCATCAAGCTCCTCTCCTCATCTTTTTTGTGTGTGAGGGCAGGTCTTGACT CTAGGTCTCAGCTGGAACCCCACCCTTTCTCCTCCTCCTTCCTCTGAGTTGACCAGCAGCAGGTCTGCCGACCACCAGCACCATCCTCTC CTCCCAGCAGCCTCCAGAACCATGCCCAGGTCTCCTGCCTCACATCACAATAATCTGGGACCCAGGCTTGTGCCCTTTCAGTGTAAAGCT GACTCCATCACATGTGCATCCACTTCTTTTCATCCATTGAGATCACACTGCCTCCTTTTTATACAGACACAAATATACATCTATAAGAAT AATATATACATAAGGAACCCCTGAAAGATGGTTTTGGAACTGGAATCAGTTAGAGGATGAAATCAGATAAAGGAAAAGCCTATTTTGGAG CTTCCCCTGTTAGGAAGGATGGCTGCACCTGGCCCCCTGGCATTCCTGACGCTCTAGGAGGGAAGGGGGAGGCAGTGCTGGCCTCCCTTG CCCTGTTTTTCCCTCTTCCAGCTGACCTGTGACTTATACTGCTCTTACCGATGATACTTTTGGAAAAAATAGAGCGTGTATGCACCGCCC CGTTTGTCCCATGGATATCCTGGGGTGTGAGTCGGATGGGACCACGGCCCTGTTTATATTTGGGTCTTTATGTTGGTGCTGCCAGGTCTC TGAGCTCCAGAGGTGGCCTCTTGGACAGATCTACTGCTATAGGAATAAAAGACACTCTGTCTCGCAAATGGCTGCTTGTCAACAAGCCCA AAGATGCTTGTCGGAGGACGGTTATGGAAGCCCTTAATTCTTGGTTGTGGGAAAAGGTGGAATGACAAGTTATTGATTGTTTTTCTGTCG CTATTTCTTTCATTTGTCTAGTGAATCAGAAAGGCTTAGCCAAGGCCACATCTGGGAAGAGTGGAGAAATTTGCCACTTGACGATCACGG ATTAGCTAGCACCTTTAAGCCCTGCATTTCTCCAACTGACAAGTGGGTGGGGGTGATGGCACATTCAGTGTGGCTATGAAGAGCGAATCC TCTCTATTGTTTAAATAGATTACTGTAGTTTGGCCAGGAATTTGGCGTCAGTGGTAACACACTTAGTTAATAAAATAAGCCAGGCTTGCA ACTAAGTATCTAACTTTACAGGCCCACTCACATTTGAGGCAAGGGGCTATTGAGTATGTGGAGAGATGTAGTGATTTAAATTCAGATTAT TTAAGTTGGATCAGCTGAAGTGTGTTTTAGACCCAAACCATCTGGCCCCTTCGTTTTGCTCAGAGGAAGTAAATGTTCACTTAAATGAAA TTGAAAACGCCATGTGGCACCACAAAAGAGCTCTCTGTACTTTCCCCATGCTGCCTCAAAAGTTCTGTGAGTTTCGGGGTCAGTGTCCCA CCCTTCACTTCCCGAGGGCGGGTGAGTGGAGAGCAGAGCCAGGAGCTCTGGCAGCTGTGGACAGATGTGCTTCCTGAGCATGGGTTGTGC CTCCCATCAGTAAAAAAATGTTTAGTTCACTTCCTTAATTGTATAATTATTTATTTGTAAATTATATACATGTACTACTGTACTAAAATA >33214_33214_2_GLB1-PSD2_GLB1_chr3_33094983_ENST00000307377_PSD2_chr5_139215308_ENST00000274710_length(amino acids)=463AA_BP=145 MGADCRAGRLVVMPGFLVRILPLLLVLLLLGPTRGLRNATQRMFEIDYSRDSFLKDGQPFRYISGSIHYSRVPRFYWKDRLLKMKMAGLN AIQTLPGSCGQVVGSPSAQDEASPLSEWRASYNSAGSNITDAFLSQRKCEPKGPLTLYNSIKNEKLEWAIDEDELRKSLSELVDDKFGTG TKKVTRILDGGNPFLDVPQALSATTYKHGVLTRKTHADMDGKRTPRGRRGWKKFYAVLKGTILYLQKDEYRPDKALSEGDLKNAIRVHHA LATRASDYSKKSNVLKLKTADWRVFLFQAPSKEEMLSWILRINLVAAIFSAPAFPAAVSSMKKFCRPLLPSCTTRLCQEEQLRSHENKLR QLTAELAEHRCHPVERGIKSKEAEEYRLKEHYLTFEKSRYETYIHLLAMKIKVGSDDLERIEARLATLEGDDPSLRKTHSSPALSQGHVT -------------------------------------------------------------- >33214_33214_3_GLB1-PSD2_GLB1_chr3_33094983_ENST00000445488_PSD2_chr5_139215308_ENST00000274710_length(transcript)=3946nt_BP=997nt AGTCAAGTGACGCGAAGCGGCCGGCCTGGGCGCCGACTGCAGAGCCGGGAGGCTGGTGGTCATGCCGGGGTTCCTGGTTCGCATCCTCCC TCTGTTGCTGGTTCTGCTGCTTCTGGGCCCTACGCGCGGCTTGCGCTCTCGATTCCTTCCTTGGGCTTTTCATCTGCCACGGCAAGCGCC TAAGTCTCAGCTGCCGCTTCACAAAAGAGGTACCAAGACTGCACCGAATGAGCACGCAAGCAGCAACAGGAGTGGGAGAAGAAGGAGGCG GCAGCAGTGGAATGCCACCCAGAGGATGTTTGAAATTGACTATAGCCGGGACTCCTTCCTCAAGGATGGCCAGCCATTTCGCTACATCTC AGGAAGCATTCACTACTCCCGTGTGCCCCGCTTCTACTGGAAGGACCGGCTGCTGAAGATGAAGATGGCTGGGCTGAACGCCATCCAGAC GTATGTGCCCTGGAACTTTCATGAGCCCTGGCCAGGACAGTACCAGTTTTCTGAGGACCATGATGTGGAATATTTTCTTCGGCTGGCTCA TGAGCTGGGACTGCTGGTTATCCTGAGGCCCGGGCCCTACATCTGTGCAGAGTGGGAAATGGGAGGATTACCTGCTTGGCTGCTAGAGAA AGAGTCTATTCTTCTCCGCTCCTCCGACCCAGATTACCTGGCAGCTGTGGACAAGTGGTTGGGAGTCCTTCTGCCCAAGATGAAGCCTCT CCTCTATCAGAATGGAGGGCCAGTTATAACAGTGCAGGTTGAAAATGAATATGGCAGCTACTTTGCCTGTGATTTTGACTACCTGCGCTT CCTGCAGAAGCGCTTTCGCCACCATCTGGGGGATGATGTGGTTCTGTTTACCACTGATGGAGCACATAAAACATTCCTGAAATGTGGGGC CCTGCAGGGCCTCTACACCACGGTGGACTTTGGAACAGGCAGCAACATCACAGATGCTTTCCTAAGCCAGAGGAAGTGTGAGCCCAAAGG ACCCTTGACCCTTTACAACTCCATCAAGAATGAAAAGCTGGAATGGGCCATTGATGAGGATGAGCTGAGGAAATCCCTGTCTGAGCTGGT GGATGACAAGTTCGGGACAGGCACGAAGAAGGTGACGCGAATCCTGGATGGTGGCAACCCCTTCCTGGATGTCCCACAGGCGCTCAGTGC CACCACCTACAAGCACGGCGTCCTGACCCGGAAGACTCACGCTGACATGGATGGCAAGAGGACGCCCCGTGGGAGGCGTGGCTGGAAGAA ATTCTACGCAGTGCTCAAAGGGACCATCCTGTACCTGCAGAAGGATGAGTACAGGCCTGACAAAGCTCTATCGGAGGGTGACCTGAAGAA CGCCATTCGCGTGCATCACGCTCTGGCCACCAGGGCCTCTGACTACAGCAAGAAGTCCAACGTGCTGAAGCTTAAGACAGCCGACTGGAG GGTATTCCTCTTCCAGGCACCGAGCAAGGAAGAAATGCTGTCCTGGATCCTCAGGATCAACCTGGTGGCAGCCATCTTCTCTGCCCCGGC CTTCCCAGCCGCTGTCAGCTCCATGAAGAAGTTCTGTCGGCCCCTGCTGCCCTCCTGCACCACCCGCCTCTGCCAGGAGGAGCAACTGCG GTCTCATGAGAATAAGTTGAGGCAGCTGACTGCGGAGCTGGCCGAACACAGGTGTCACCCAGTCGAGAGGGGCATCAAGTCCAAGGAGGC CGAGGAGTACCGGTTGAAGGAGCACTATCTCACCTTCGAGAAAAGCCGTTATGAGACCTATATCCACCTCCTGGCTATGAAAATCAAAGT GGGCTCAGATGATCTGGAGCGGATTGAGGCCCGGCTGGCCACTCTGGAAGGGGATGACCCTTCTCTCCGGAAGACACATTCAAGCCCTGC CCTCAGCCAGGGCCATGTGACTGGCAGCAAAACCACAAAGGATGCCACTGGGCCTGATACTTAGCTGACATGGATTTGCAGACCCCAGGG TGGGCAGATGTCTCCAGTGGGGTCAGTGAGCACAATTCCAGCCAGGGGCCACTTGGACCAAGCTCCAGTCAGTTGATGGGCAGCTAGAGG GGTGCAGAAAGCCTGTGGGCCCAGGAGATGGAGATGCCGTTTGTGGCGTTGATCTCCTTGCGTCCTTGGGCATCTCCGGGCATCAGACCC TCTCCCTGGCCCTTGTTTTCCTCTCCACCATGGAGCCTCATTTTGTAGGCCAGTTGTGTGCATGCTCTAGACACCACCTCGCTGGAGAAG CTGGAAGGGCTGTTGTCTTCCCAGGTCTTTCTCTTCTCATCAAGCTCCTCTCCTCATCTTTTTTGTGTGTGAGGGCAGGTCTTGACTCTA GGTCTCAGCTGGAACCCCACCCTTTCTCCTCCTCCTTCCTCTGAGTTGACCAGCAGCAGGTCTGCCGACCACCAGCACCATCCTCTCCTC CCAGCAGCCTCCAGAACCATGCCCAGGTCTCCTGCCTCACATCACAATAATCTGGGACCCAGGCTTGTGCCCTTTCAGTGTAAAGCTGAC TCCATCACATGTGCATCCACTTCTTTTCATCCATTGAGATCACACTGCCTCCTTTTTATACAGACACAAATATACATCTATAAGAATAAT ATATACATAAGGAACCCCTGAAAGATGGTTTTGGAACTGGAATCAGTTAGAGGATGAAATCAGATAAAGGAAAAGCCTATTTTGGAGCTT CCCCTGTTAGGAAGGATGGCTGCACCTGGCCCCCTGGCATTCCTGACGCTCTAGGAGGGAAGGGGGAGGCAGTGCTGGCCTCCCTTGCCC TGTTTTTCCCTCTTCCAGCTGACCTGTGACTTATACTGCTCTTACCGATGATACTTTTGGAAAAAATAGAGCGTGTATGCACCGCCCCGT TTGTCCCATGGATATCCTGGGGTGTGAGTCGGATGGGACCACGGCCCTGTTTATATTTGGGTCTTTATGTTGGTGCTGCCAGGTCTCTGA GCTCCAGAGGTGGCCTCTTGGACAGATCTACTGCTATAGGAATAAAAGACACTCTGTCTCGCAAATGGCTGCTTGTCAACAAGCCCAAAG ATGCTTGTCGGAGGACGGTTATGGAAGCCCTTAATTCTTGGTTGTGGGAAAAGGTGGAATGACAAGTTATTGATTGTTTTTCTGTCGCTA TTTCTTTCATTTGTCTAGTGAATCAGAAAGGCTTAGCCAAGGCCACATCTGGGAAGAGTGGAGAAATTTGCCACTTGACGATCACGGATT AGCTAGCACCTTTAAGCCCTGCATTTCTCCAACTGACAAGTGGGTGGGGGTGATGGCACATTCAGTGTGGCTATGAAGAGCGAATCCTCT CTATTGTTTAAATAGATTACTGTAGTTTGGCCAGGAATTTGGCGTCAGTGGTAACACACTTAGTTAATAAAATAAGCCAGGCTTGCAACT AAGTATCTAACTTTACAGGCCCACTCACATTTGAGGCAAGGGGCTATTGAGTATGTGGAGAGATGTAGTGATTTAAATTCAGATTATTTA AGTTGGATCAGCTGAAGTGTGTTTTAGACCCAAACCATCTGGCCCCTTCGTTTTGCTCAGAGGAAGTAAATGTTCACTTAAATGAAATTG AAAACGCCATGTGGCACCACAAAAGAGCTCTCTGTACTTTCCCCATGCTGCCTCAAAAGTTCTGTGAGTTTCGGGGTCAGTGTCCCACCC TTCACTTCCCGAGGGCGGGTGAGTGGAGAGCAGAGCCAGGAGCTCTGGCAGCTGTGGACAGATGTGCTTCCTGAGCATGGGTTGTGCCTC CCATCAGTAAAAAAATGTTTAGTTCACTTCCTTAATTGTATAATTATTTATTTGTAAATTATATACATGTACTACTGTACTAAAATATTA >33214_33214_3_GLB1-PSD2_GLB1_chr3_33094983_ENST00000445488_PSD2_chr5_139215308_ENST00000274710_length(amino acids)=642AA_BP=324 MGADCRAGRLVVMPGFLVRILPLLLVLLLLGPTRGLRSRFLPWAFHLPRQAPKSQLPLHKRGTKTAPNEHASSNRSGRRRRRQQWNATQR MFEIDYSRDSFLKDGQPFRYISGSIHYSRVPRFYWKDRLLKMKMAGLNAIQTYVPWNFHEPWPGQYQFSEDHDVEYFLRLAHELGLLVIL RPGPYICAEWEMGGLPAWLLEKESILLRSSDPDYLAAVDKWLGVLLPKMKPLLYQNGGPVITVQVENEYGSYFACDFDYLRFLQKRFRHH LGDDVVLFTTDGAHKTFLKCGALQGLYTTVDFGTGSNITDAFLSQRKCEPKGPLTLYNSIKNEKLEWAIDEDELRKSLSELVDDKFGTGT KKVTRILDGGNPFLDVPQALSATTYKHGVLTRKTHADMDGKRTPRGRRGWKKFYAVLKGTILYLQKDEYRPDKALSEGDLKNAIRVHHAL ATRASDYSKKSNVLKLKTADWRVFLFQAPSKEEMLSWILRINLVAAIFSAPAFPAAVSSMKKFCRPLLPSCTTRLCQEEQLRSHENKLRQ LTAELAEHRCHPVERGIKSKEAEEYRLKEHYLTFEKSRYETYIHLLAMKIKVGSDDLERIEARLATLEGDDPSLRKTHSSPALSQGHVTG -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for GLB1-PSD2 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for GLB1-PSD2 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for GLB1-PSD2 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
