
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:GLG1-BCAR1 (FusionGDB2 ID:33240) |
Fusion Gene Summary for GLG1-BCAR1 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: GLG1-BCAR1 | Fusion gene ID: 33240 | Hgene | Tgene | Gene symbol | GLG1 | BCAR1 | Gene ID | 2734 | 9564 |
| Gene name | golgi glycoprotein 1 | BCAR1 scaffold protein, Cas family member | |
| Synonyms | CFR-1|ESL-1|MG-160|MG160 | CAS|CAS1|CASS1|CRKAS|P130Cas | |
| Cytomap | 16q23.1 | 16q23.1 | |
| Type of gene | protein-coding | protein-coding | |
| Description | Golgi apparatus protein 1E-selectin ligand 1cysteine-rich fibroblast growth factor receptorgolgi sialoglycoprotein MG-160 | breast cancer anti-estrogen resistance protein 1BCAR1, Cas family scaffold proteinBCAR1, Cas family scaffolding proteinCas scaffolding protein family member 1Crk-associated substrate p130Cas | |
| Modification date | 20200313 | 20200327 | |
| UniProtAcc | Q92896 | P56945 | |
| Ensembl transtripts involved in fusion gene | ENST00000205061, ENST00000422840, ENST00000447066, | ENST00000546196, ENST00000393420, ENST00000393422, ENST00000418647, ENST00000420641, ENST00000535626, ENST00000538440, ENST00000542031, ENST00000566982, ENST00000162330, | |
| Fusion gene scores | * DoF score | 23 X 21 X 12=5796 | 6 X 4 X 4=96 |
| # samples | 29 | 6 | |
| ** MAII score | log2(29/5796*10)=-4.320932789542 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(6/96*10)=-0.678071905112638 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: GLG1 [Title/Abstract] AND BCAR1 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | GLG1(74640555)-BCAR1(75276988), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Tgene | BCAR1 | GO:0007015 | actin filament organization | 16105984 |
| Tgene | BCAR1 | GO:0007229 | integrin-mediated signaling pathway | 8649368 |
| Tgene | BCAR1 | GO:0010595 | positive regulation of endothelial cell migration | 21245381 |
| Tgene | BCAR1 | GO:0016477 | cell migration | 9425168 |
| Tgene | BCAR1 | GO:0030335 | positive regulation of cell migration | 9472046 |
| Tgene | BCAR1 | GO:0050851 | antigen receptor-mediated signaling pathway | 10587647 |
| Tgene | BCAR1 | GO:0050853 | B cell receptor signaling pathway | 9020138 |
| Tgene | BCAR1 | GO:0086100 | endothelin receptor signaling pathway | 19086031 |
 Fusion gene breakpoints across GLG1 (5'-gene) Fusion gene breakpoints across GLG1 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
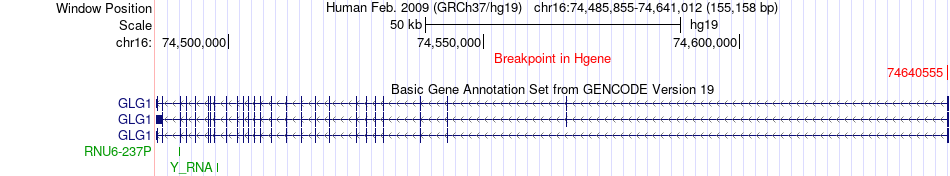 |
 Fusion gene breakpoints across BCAR1 (3'-gene) Fusion gene breakpoints across BCAR1 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
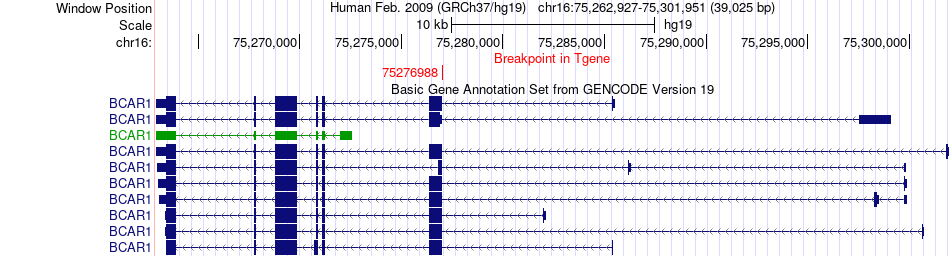 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | PRAD | TCGA-YJ-A8SW-01A | GLG1 | chr16 | 74640555 | - | BCAR1 | chr16 | 75276988 | - |
Top |
Fusion Gene ORF analysis for GLG1-BCAR1 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-5UTR | ENST00000205061 | ENST00000546196 | GLG1 | chr16 | 74640555 | - | BCAR1 | chr16 | 75276988 | - |
| 5CDS-5UTR | ENST00000422840 | ENST00000546196 | GLG1 | chr16 | 74640555 | - | BCAR1 | chr16 | 75276988 | - |
| 5CDS-5UTR | ENST00000447066 | ENST00000546196 | GLG1 | chr16 | 74640555 | - | BCAR1 | chr16 | 75276988 | - |
| 5CDS-intron | ENST00000205061 | ENST00000393420 | GLG1 | chr16 | 74640555 | - | BCAR1 | chr16 | 75276988 | - |
| 5CDS-intron | ENST00000205061 | ENST00000393422 | GLG1 | chr16 | 74640555 | - | BCAR1 | chr16 | 75276988 | - |
| 5CDS-intron | ENST00000205061 | ENST00000418647 | GLG1 | chr16 | 74640555 | - | BCAR1 | chr16 | 75276988 | - |
| 5CDS-intron | ENST00000205061 | ENST00000420641 | GLG1 | chr16 | 74640555 | - | BCAR1 | chr16 | 75276988 | - |
| 5CDS-intron | ENST00000205061 | ENST00000535626 | GLG1 | chr16 | 74640555 | - | BCAR1 | chr16 | 75276988 | - |
| 5CDS-intron | ENST00000205061 | ENST00000538440 | GLG1 | chr16 | 74640555 | - | BCAR1 | chr16 | 75276988 | - |
| 5CDS-intron | ENST00000205061 | ENST00000542031 | GLG1 | chr16 | 74640555 | - | BCAR1 | chr16 | 75276988 | - |
| 5CDS-intron | ENST00000205061 | ENST00000566982 | GLG1 | chr16 | 74640555 | - | BCAR1 | chr16 | 75276988 | - |
| 5CDS-intron | ENST00000422840 | ENST00000393420 | GLG1 | chr16 | 74640555 | - | BCAR1 | chr16 | 75276988 | - |
| 5CDS-intron | ENST00000422840 | ENST00000393422 | GLG1 | chr16 | 74640555 | - | BCAR1 | chr16 | 75276988 | - |
| 5CDS-intron | ENST00000422840 | ENST00000418647 | GLG1 | chr16 | 74640555 | - | BCAR1 | chr16 | 75276988 | - |
| 5CDS-intron | ENST00000422840 | ENST00000420641 | GLG1 | chr16 | 74640555 | - | BCAR1 | chr16 | 75276988 | - |
| 5CDS-intron | ENST00000422840 | ENST00000535626 | GLG1 | chr16 | 74640555 | - | BCAR1 | chr16 | 75276988 | - |
| 5CDS-intron | ENST00000422840 | ENST00000538440 | GLG1 | chr16 | 74640555 | - | BCAR1 | chr16 | 75276988 | - |
| 5CDS-intron | ENST00000422840 | ENST00000542031 | GLG1 | chr16 | 74640555 | - | BCAR1 | chr16 | 75276988 | - |
| 5CDS-intron | ENST00000422840 | ENST00000566982 | GLG1 | chr16 | 74640555 | - | BCAR1 | chr16 | 75276988 | - |
| 5CDS-intron | ENST00000447066 | ENST00000393420 | GLG1 | chr16 | 74640555 | - | BCAR1 | chr16 | 75276988 | - |
| 5CDS-intron | ENST00000447066 | ENST00000393422 | GLG1 | chr16 | 74640555 | - | BCAR1 | chr16 | 75276988 | - |
| 5CDS-intron | ENST00000447066 | ENST00000418647 | GLG1 | chr16 | 74640555 | - | BCAR1 | chr16 | 75276988 | - |
| 5CDS-intron | ENST00000447066 | ENST00000420641 | GLG1 | chr16 | 74640555 | - | BCAR1 | chr16 | 75276988 | - |
| 5CDS-intron | ENST00000447066 | ENST00000535626 | GLG1 | chr16 | 74640555 | - | BCAR1 | chr16 | 75276988 | - |
| 5CDS-intron | ENST00000447066 | ENST00000538440 | GLG1 | chr16 | 74640555 | - | BCAR1 | chr16 | 75276988 | - |
| 5CDS-intron | ENST00000447066 | ENST00000542031 | GLG1 | chr16 | 74640555 | - | BCAR1 | chr16 | 75276988 | - |
| 5CDS-intron | ENST00000447066 | ENST00000566982 | GLG1 | chr16 | 74640555 | - | BCAR1 | chr16 | 75276988 | - |
| In-frame | ENST00000205061 | ENST00000162330 | GLG1 | chr16 | 74640555 | - | BCAR1 | chr16 | 75276988 | - |
| In-frame | ENST00000422840 | ENST00000162330 | GLG1 | chr16 | 74640555 | - | BCAR1 | chr16 | 75276988 | - |
| In-frame | ENST00000447066 | ENST00000162330 | GLG1 | chr16 | 74640555 | - | BCAR1 | chr16 | 75276988 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000205061 | GLG1 | chr16 | 74640555 | - | ENST00000162330 | BCAR1 | chr16 | 75276988 | - | 3540 | 458 | 20 | 3058 | 1012 |
| ENST00000422840 | GLG1 | chr16 | 74640555 | - | ENST00000162330 | BCAR1 | chr16 | 75276988 | - | 3520 | 438 | 0 | 3038 | 1012 |
| ENST00000447066 | GLG1 | chr16 | 74640555 | - | ENST00000162330 | BCAR1 | chr16 | 75276988 | - | 3538 | 456 | 18 | 3056 | 1012 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000205061 | ENST00000162330 | GLG1 | chr16 | 74640555 | - | BCAR1 | chr16 | 75276988 | - | 0.003076244 | 0.99692374 |
| ENST00000422840 | ENST00000162330 | GLG1 | chr16 | 74640555 | - | BCAR1 | chr16 | 75276988 | - | 0.002862095 | 0.9971379 |
| ENST00000447066 | ENST00000162330 | GLG1 | chr16 | 74640555 | - | BCAR1 | chr16 | 75276988 | - | 0.003049239 | 0.99695075 |
Top |
Fusion Genomic Features for GLG1-BCAR1 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
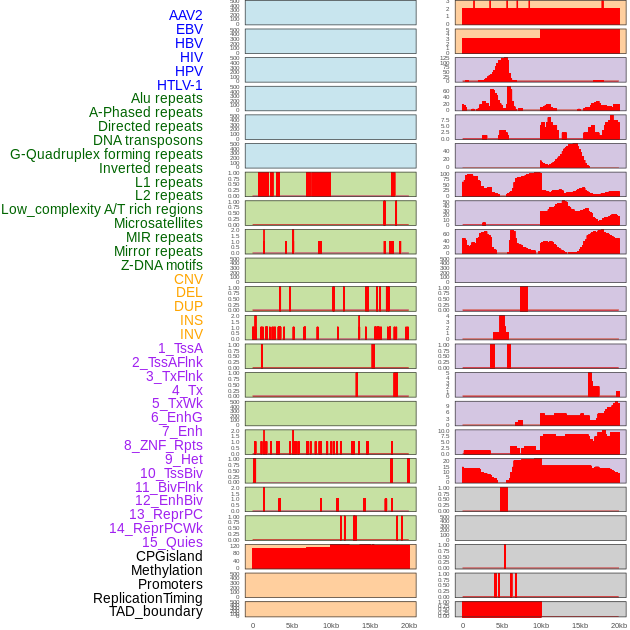 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for GLG1-BCAR1 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr16:74640555/chr16:75276988) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| GLG1 | BCAR1 |
| FUNCTION: Binds fibroblast growth factor and E-selectin (cell-adhesion lectin on endothelial cells mediating the binding of neutrophils). {ECO:0000269|PubMed:8985126}. | FUNCTION: Docking protein which plays a central coordinating role for tyrosine kinase-based signaling related to cell adhesion (PubMed:12832404, PubMed:12432078). Implicated in induction of cell migration and cell branching (PubMed:12432078, PubMed:12832404, PubMed:17038317). Involved in the BCAR3-mediated inhibition of TGFB signaling (By similarity). {ECO:0000250|UniProtKB:Q61140, ECO:0000269|PubMed:12432078, ECO:0000269|PubMed:12832404, ECO:0000269|PubMed:17038317}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | GLG1 | chr16:74640555 | chr16:75276988 | ENST00000205061 | - | 1 | 27 | 54_57 | 146 | 1204.0 | Compositional bias | Note=Poly-Gly |
| Hgene | GLG1 | chr16:74640555 | chr16:75276988 | ENST00000205061 | - | 1 | 27 | 71_86 | 146 | 1204.0 | Compositional bias | Note=Poly-Gln |
| Hgene | GLG1 | chr16:74640555 | chr16:75276988 | ENST00000422840 | - | 1 | 26 | 54_57 | 146 | 1180.0 | Compositional bias | Note=Poly-Gly |
| Hgene | GLG1 | chr16:74640555 | chr16:75276988 | ENST00000422840 | - | 1 | 26 | 71_86 | 146 | 1180.0 | Compositional bias | Note=Poly-Gln |
| Hgene | GLG1 | chr16:74640555 | chr16:75276988 | ENST00000447066 | - | 1 | 26 | 54_57 | 146 | 1193.0 | Compositional bias | Note=Poly-Gly |
| Hgene | GLG1 | chr16:74640555 | chr16:75276988 | ENST00000447066 | - | 1 | 26 | 71_86 | 146 | 1193.0 | Compositional bias | Note=Poly-Gln |
| Hgene | GLG1 | chr16:74640555 | chr16:75276988 | ENST00000205061 | - | 1 | 27 | 116_149 | 146 | 1204.0 | Repeat | Note=Cys-rich GLG1 1 |
| Hgene | GLG1 | chr16:74640555 | chr16:75276988 | ENST00000422840 | - | 1 | 26 | 116_149 | 146 | 1180.0 | Repeat | Note=Cys-rich GLG1 1 |
| Hgene | GLG1 | chr16:74640555 | chr16:75276988 | ENST00000447066 | - | 1 | 26 | 116_149 | 146 | 1193.0 | Repeat | Note=Cys-rich GLG1 1 |
| Tgene | BCAR1 | chr16:74640555 | chr16:75276988 | ENST00000162330 | 0 | 7 | 422_614 | 4 | 871.0 | Compositional bias | Note=Ser-rich | |
| Tgene | BCAR1 | chr16:74640555 | chr16:75276988 | ENST00000162330 | 0 | 7 | 74_87 | 4 | 871.0 | Compositional bias | Note=Pro-rich | |
| Tgene | BCAR1 | chr16:74640555 | chr16:75276988 | ENST00000393420 | 0 | 7 | 422_614 | 4 | 889.0 | Compositional bias | Note=Ser-rich | |
| Tgene | BCAR1 | chr16:74640555 | chr16:75276988 | ENST00000393420 | 0 | 7 | 74_87 | 4 | 889.0 | Compositional bias | Note=Pro-rich | |
| Tgene | BCAR1 | chr16:74640555 | chr16:75276988 | ENST00000393422 | 0 | 7 | 422_614 | 22 | 889.0 | Compositional bias | Note=Ser-rich | |
| Tgene | BCAR1 | chr16:74640555 | chr16:75276988 | ENST00000393422 | 0 | 7 | 74_87 | 22 | 889.0 | Compositional bias | Note=Pro-rich | |
| Tgene | BCAR1 | chr16:74640555 | chr16:75276988 | ENST00000418647 | 1 | 8 | 422_614 | 50 | 917.0 | Compositional bias | Note=Ser-rich | |
| Tgene | BCAR1 | chr16:74640555 | chr16:75276988 | ENST00000418647 | 1 | 8 | 74_87 | 50 | 917.0 | Compositional bias | Note=Pro-rich | |
| Tgene | BCAR1 | chr16:74640555 | chr16:75276988 | ENST00000420641 | 0 | 7 | 422_614 | 22 | 889.0 | Compositional bias | Note=Ser-rich | |
| Tgene | BCAR1 | chr16:74640555 | chr16:75276988 | ENST00000420641 | 0 | 7 | 74_87 | 22 | 889.0 | Compositional bias | Note=Pro-rich | |
| Tgene | BCAR1 | chr16:74640555 | chr16:75276988 | ENST00000535626 | 1 | 8 | 422_614 | 4 | 723.0 | Compositional bias | Note=Ser-rich | |
| Tgene | BCAR1 | chr16:74640555 | chr16:75276988 | ENST00000535626 | 1 | 8 | 74_87 | 4 | 723.0 | Compositional bias | Note=Pro-rich | |
| Tgene | BCAR1 | chr16:74640555 | chr16:75276988 | ENST00000538440 | 0 | 7 | 422_614 | 4 | 871.0 | Compositional bias | Note=Ser-rich | |
| Tgene | BCAR1 | chr16:74640555 | chr16:75276988 | ENST00000538440 | 0 | 7 | 74_87 | 4 | 871.0 | Compositional bias | Note=Pro-rich | |
| Tgene | BCAR1 | chr16:74640555 | chr16:75276988 | ENST00000542031 | 0 | 7 | 422_614 | 2 | 869.0 | Compositional bias | Note=Ser-rich | |
| Tgene | BCAR1 | chr16:74640555 | chr16:75276988 | ENST00000542031 | 0 | 7 | 74_87 | 2 | 869.0 | Compositional bias | Note=Pro-rich | |
| Tgene | BCAR1 | chr16:74640555 | chr16:75276988 | ENST00000162330 | 0 | 7 | 3_65 | 4 | 871.0 | Domain | SH3 | |
| Tgene | BCAR1 | chr16:74640555 | chr16:75276988 | ENST00000393420 | 0 | 7 | 3_65 | 4 | 889.0 | Domain | SH3 | |
| Tgene | BCAR1 | chr16:74640555 | chr16:75276988 | ENST00000535626 | 1 | 8 | 3_65 | 4 | 723.0 | Domain | SH3 | |
| Tgene | BCAR1 | chr16:74640555 | chr16:75276988 | ENST00000538440 | 0 | 7 | 3_65 | 4 | 871.0 | Domain | SH3 | |
| Tgene | BCAR1 | chr16:74640555 | chr16:75276988 | ENST00000542031 | 0 | 7 | 3_65 | 2 | 869.0 | Domain | SH3 | |
| Tgene | BCAR1 | chr16:74640555 | chr16:75276988 | ENST00000162330 | 0 | 7 | 635_643 | 4 | 871.0 | Motif | SH3-binding | |
| Tgene | BCAR1 | chr16:74640555 | chr16:75276988 | ENST00000393420 | 0 | 7 | 635_643 | 4 | 889.0 | Motif | SH3-binding | |
| Tgene | BCAR1 | chr16:74640555 | chr16:75276988 | ENST00000393422 | 0 | 7 | 635_643 | 22 | 889.0 | Motif | SH3-binding | |
| Tgene | BCAR1 | chr16:74640555 | chr16:75276988 | ENST00000418647 | 1 | 8 | 635_643 | 50 | 917.0 | Motif | SH3-binding | |
| Tgene | BCAR1 | chr16:74640555 | chr16:75276988 | ENST00000420641 | 0 | 7 | 635_643 | 22 | 889.0 | Motif | SH3-binding | |
| Tgene | BCAR1 | chr16:74640555 | chr16:75276988 | ENST00000535626 | 1 | 8 | 635_643 | 4 | 723.0 | Motif | SH3-binding | |
| Tgene | BCAR1 | chr16:74640555 | chr16:75276988 | ENST00000538440 | 0 | 7 | 635_643 | 4 | 871.0 | Motif | SH3-binding | |
| Tgene | BCAR1 | chr16:74640555 | chr16:75276988 | ENST00000542031 | 0 | 7 | 635_643 | 2 | 869.0 | Motif | SH3-binding | |
| Tgene | BCAR1 | chr16:74640555 | chr16:75276988 | ENST00000162330 | 0 | 7 | 115_416 | 4 | 871.0 | Region | Substrate for kinases | |
| Tgene | BCAR1 | chr16:74640555 | chr16:75276988 | ENST00000162330 | 0 | 7 | 746_796 | 4 | 871.0 | Region | Note=Divergent helix-loop-helix motif | |
| Tgene | BCAR1 | chr16:74640555 | chr16:75276988 | ENST00000393420 | 0 | 7 | 115_416 | 4 | 889.0 | Region | Substrate for kinases | |
| Tgene | BCAR1 | chr16:74640555 | chr16:75276988 | ENST00000393420 | 0 | 7 | 746_796 | 4 | 889.0 | Region | Note=Divergent helix-loop-helix motif | |
| Tgene | BCAR1 | chr16:74640555 | chr16:75276988 | ENST00000393422 | 0 | 7 | 115_416 | 22 | 889.0 | Region | Substrate for kinases | |
| Tgene | BCAR1 | chr16:74640555 | chr16:75276988 | ENST00000393422 | 0 | 7 | 746_796 | 22 | 889.0 | Region | Note=Divergent helix-loop-helix motif | |
| Tgene | BCAR1 | chr16:74640555 | chr16:75276988 | ENST00000418647 | 1 | 8 | 115_416 | 50 | 917.0 | Region | Substrate for kinases | |
| Tgene | BCAR1 | chr16:74640555 | chr16:75276988 | ENST00000418647 | 1 | 8 | 746_796 | 50 | 917.0 | Region | Note=Divergent helix-loop-helix motif | |
| Tgene | BCAR1 | chr16:74640555 | chr16:75276988 | ENST00000420641 | 0 | 7 | 115_416 | 22 | 889.0 | Region | Substrate for kinases | |
| Tgene | BCAR1 | chr16:74640555 | chr16:75276988 | ENST00000420641 | 0 | 7 | 746_796 | 22 | 889.0 | Region | Note=Divergent helix-loop-helix motif | |
| Tgene | BCAR1 | chr16:74640555 | chr16:75276988 | ENST00000535626 | 1 | 8 | 115_416 | 4 | 723.0 | Region | Substrate for kinases | |
| Tgene | BCAR1 | chr16:74640555 | chr16:75276988 | ENST00000535626 | 1 | 8 | 746_796 | 4 | 723.0 | Region | Note=Divergent helix-loop-helix motif | |
| Tgene | BCAR1 | chr16:74640555 | chr16:75276988 | ENST00000538440 | 0 | 7 | 115_416 | 4 | 871.0 | Region | Substrate for kinases | |
| Tgene | BCAR1 | chr16:74640555 | chr16:75276988 | ENST00000538440 | 0 | 7 | 746_796 | 4 | 871.0 | Region | Note=Divergent helix-loop-helix motif | |
| Tgene | BCAR1 | chr16:74640555 | chr16:75276988 | ENST00000542031 | 0 | 7 | 115_416 | 2 | 869.0 | Region | Substrate for kinases | |
| Tgene | BCAR1 | chr16:74640555 | chr16:75276988 | ENST00000542031 | 0 | 7 | 746_796 | 2 | 869.0 | Region | Note=Divergent helix-loop-helix motif |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | GLG1 | chr16:74640555 | chr16:75276988 | ENST00000205061 | - | 1 | 27 | 1041_1101 | 146 | 1204.0 | Repeat | Note=Cys-rich GLG1 16 |
| Hgene | GLG1 | chr16:74640555 | chr16:75276988 | ENST00000205061 | - | 1 | 27 | 150_212 | 146 | 1204.0 | Repeat | Note=Cys-rich GLG1 2 |
| Hgene | GLG1 | chr16:74640555 | chr16:75276988 | ENST00000205061 | - | 1 | 27 | 215_278 | 146 | 1204.0 | Repeat | Note=Cys-rich GLG1 3 |
| Hgene | GLG1 | chr16:74640555 | chr16:75276988 | ENST00000205061 | - | 1 | 27 | 286_346 | 146 | 1204.0 | Repeat | Note=Cys-rich GLG1 4 |
| Hgene | GLG1 | chr16:74640555 | chr16:75276988 | ENST00000205061 | - | 1 | 27 | 347_413 | 146 | 1204.0 | Repeat | Note=Cys-rich GLG1 5 |
| Hgene | GLG1 | chr16:74640555 | chr16:75276988 | ENST00000205061 | - | 1 | 27 | 414_473 | 146 | 1204.0 | Repeat | Note=Cys-rich GLG1 6 |
| Hgene | GLG1 | chr16:74640555 | chr16:75276988 | ENST00000205061 | - | 1 | 27 | 475_537 | 146 | 1204.0 | Repeat | Note=Cys-rich GLG1 7 |
| Hgene | GLG1 | chr16:74640555 | chr16:75276988 | ENST00000205061 | - | 1 | 27 | 538_604 | 146 | 1204.0 | Repeat | Note=Cys-rich GLG1 8 |
| Hgene | GLG1 | chr16:74640555 | chr16:75276988 | ENST00000205061 | - | 1 | 27 | 609_668 | 146 | 1204.0 | Repeat | Note=Cys-rich GLG1 9 |
| Hgene | GLG1 | chr16:74640555 | chr16:75276988 | ENST00000205061 | - | 1 | 27 | 670_728 | 146 | 1204.0 | Repeat | Note=Cys-rich GLG1 10 |
| Hgene | GLG1 | chr16:74640555 | chr16:75276988 | ENST00000205061 | - | 1 | 27 | 729_788 | 146 | 1204.0 | Repeat | Note=Cys-rich GLG1 11 |
| Hgene | GLG1 | chr16:74640555 | chr16:75276988 | ENST00000205061 | - | 1 | 27 | 796_856 | 146 | 1204.0 | Repeat | Note=Cys-rich GLG1 12 |
| Hgene | GLG1 | chr16:74640555 | chr16:75276988 | ENST00000205061 | - | 1 | 27 | 858_911 | 146 | 1204.0 | Repeat | Note=Cys-rich GLG1 13 |
| Hgene | GLG1 | chr16:74640555 | chr16:75276988 | ENST00000205061 | - | 1 | 27 | 912_979 | 146 | 1204.0 | Repeat | Note=Cys-rich GLG1 14 |
| Hgene | GLG1 | chr16:74640555 | chr16:75276988 | ENST00000205061 | - | 1 | 27 | 980_1035 | 146 | 1204.0 | Repeat | Note=Cys-rich GLG1 15 |
| Hgene | GLG1 | chr16:74640555 | chr16:75276988 | ENST00000422840 | - | 1 | 26 | 1041_1101 | 146 | 1180.0 | Repeat | Note=Cys-rich GLG1 16 |
| Hgene | GLG1 | chr16:74640555 | chr16:75276988 | ENST00000422840 | - | 1 | 26 | 150_212 | 146 | 1180.0 | Repeat | Note=Cys-rich GLG1 2 |
| Hgene | GLG1 | chr16:74640555 | chr16:75276988 | ENST00000422840 | - | 1 | 26 | 215_278 | 146 | 1180.0 | Repeat | Note=Cys-rich GLG1 3 |
| Hgene | GLG1 | chr16:74640555 | chr16:75276988 | ENST00000422840 | - | 1 | 26 | 286_346 | 146 | 1180.0 | Repeat | Note=Cys-rich GLG1 4 |
| Hgene | GLG1 | chr16:74640555 | chr16:75276988 | ENST00000422840 | - | 1 | 26 | 347_413 | 146 | 1180.0 | Repeat | Note=Cys-rich GLG1 5 |
| Hgene | GLG1 | chr16:74640555 | chr16:75276988 | ENST00000422840 | - | 1 | 26 | 414_473 | 146 | 1180.0 | Repeat | Note=Cys-rich GLG1 6 |
| Hgene | GLG1 | chr16:74640555 | chr16:75276988 | ENST00000422840 | - | 1 | 26 | 475_537 | 146 | 1180.0 | Repeat | Note=Cys-rich GLG1 7 |
| Hgene | GLG1 | chr16:74640555 | chr16:75276988 | ENST00000422840 | - | 1 | 26 | 538_604 | 146 | 1180.0 | Repeat | Note=Cys-rich GLG1 8 |
| Hgene | GLG1 | chr16:74640555 | chr16:75276988 | ENST00000422840 | - | 1 | 26 | 609_668 | 146 | 1180.0 | Repeat | Note=Cys-rich GLG1 9 |
| Hgene | GLG1 | chr16:74640555 | chr16:75276988 | ENST00000422840 | - | 1 | 26 | 670_728 | 146 | 1180.0 | Repeat | Note=Cys-rich GLG1 10 |
| Hgene | GLG1 | chr16:74640555 | chr16:75276988 | ENST00000422840 | - | 1 | 26 | 729_788 | 146 | 1180.0 | Repeat | Note=Cys-rich GLG1 11 |
| Hgene | GLG1 | chr16:74640555 | chr16:75276988 | ENST00000422840 | - | 1 | 26 | 796_856 | 146 | 1180.0 | Repeat | Note=Cys-rich GLG1 12 |
| Hgene | GLG1 | chr16:74640555 | chr16:75276988 | ENST00000422840 | - | 1 | 26 | 858_911 | 146 | 1180.0 | Repeat | Note=Cys-rich GLG1 13 |
| Hgene | GLG1 | chr16:74640555 | chr16:75276988 | ENST00000422840 | - | 1 | 26 | 912_979 | 146 | 1180.0 | Repeat | Note=Cys-rich GLG1 14 |
| Hgene | GLG1 | chr16:74640555 | chr16:75276988 | ENST00000422840 | - | 1 | 26 | 980_1035 | 146 | 1180.0 | Repeat | Note=Cys-rich GLG1 15 |
| Hgene | GLG1 | chr16:74640555 | chr16:75276988 | ENST00000447066 | - | 1 | 26 | 1041_1101 | 146 | 1193.0 | Repeat | Note=Cys-rich GLG1 16 |
| Hgene | GLG1 | chr16:74640555 | chr16:75276988 | ENST00000447066 | - | 1 | 26 | 150_212 | 146 | 1193.0 | Repeat | Note=Cys-rich GLG1 2 |
| Hgene | GLG1 | chr16:74640555 | chr16:75276988 | ENST00000447066 | - | 1 | 26 | 215_278 | 146 | 1193.0 | Repeat | Note=Cys-rich GLG1 3 |
| Hgene | GLG1 | chr16:74640555 | chr16:75276988 | ENST00000447066 | - | 1 | 26 | 286_346 | 146 | 1193.0 | Repeat | Note=Cys-rich GLG1 4 |
| Hgene | GLG1 | chr16:74640555 | chr16:75276988 | ENST00000447066 | - | 1 | 26 | 347_413 | 146 | 1193.0 | Repeat | Note=Cys-rich GLG1 5 |
| Hgene | GLG1 | chr16:74640555 | chr16:75276988 | ENST00000447066 | - | 1 | 26 | 414_473 | 146 | 1193.0 | Repeat | Note=Cys-rich GLG1 6 |
| Hgene | GLG1 | chr16:74640555 | chr16:75276988 | ENST00000447066 | - | 1 | 26 | 475_537 | 146 | 1193.0 | Repeat | Note=Cys-rich GLG1 7 |
| Hgene | GLG1 | chr16:74640555 | chr16:75276988 | ENST00000447066 | - | 1 | 26 | 538_604 | 146 | 1193.0 | Repeat | Note=Cys-rich GLG1 8 |
| Hgene | GLG1 | chr16:74640555 | chr16:75276988 | ENST00000447066 | - | 1 | 26 | 609_668 | 146 | 1193.0 | Repeat | Note=Cys-rich GLG1 9 |
| Hgene | GLG1 | chr16:74640555 | chr16:75276988 | ENST00000447066 | - | 1 | 26 | 670_728 | 146 | 1193.0 | Repeat | Note=Cys-rich GLG1 10 |
| Hgene | GLG1 | chr16:74640555 | chr16:75276988 | ENST00000447066 | - | 1 | 26 | 729_788 | 146 | 1193.0 | Repeat | Note=Cys-rich GLG1 11 |
| Hgene | GLG1 | chr16:74640555 | chr16:75276988 | ENST00000447066 | - | 1 | 26 | 796_856 | 146 | 1193.0 | Repeat | Note=Cys-rich GLG1 12 |
| Hgene | GLG1 | chr16:74640555 | chr16:75276988 | ENST00000447066 | - | 1 | 26 | 858_911 | 146 | 1193.0 | Repeat | Note=Cys-rich GLG1 13 |
| Hgene | GLG1 | chr16:74640555 | chr16:75276988 | ENST00000447066 | - | 1 | 26 | 912_979 | 146 | 1193.0 | Repeat | Note=Cys-rich GLG1 14 |
| Hgene | GLG1 | chr16:74640555 | chr16:75276988 | ENST00000447066 | - | 1 | 26 | 980_1035 | 146 | 1193.0 | Repeat | Note=Cys-rich GLG1 15 |
| Hgene | GLG1 | chr16:74640555 | chr16:75276988 | ENST00000205061 | - | 1 | 27 | 1167_1179 | 146 | 1204.0 | Topological domain | Cytoplasmic |
| Hgene | GLG1 | chr16:74640555 | chr16:75276988 | ENST00000205061 | - | 1 | 27 | 30_1145 | 146 | 1204.0 | Topological domain | Extracellular |
| Hgene | GLG1 | chr16:74640555 | chr16:75276988 | ENST00000422840 | - | 1 | 26 | 1167_1179 | 146 | 1180.0 | Topological domain | Cytoplasmic |
| Hgene | GLG1 | chr16:74640555 | chr16:75276988 | ENST00000422840 | - | 1 | 26 | 30_1145 | 146 | 1180.0 | Topological domain | Extracellular |
| Hgene | GLG1 | chr16:74640555 | chr16:75276988 | ENST00000447066 | - | 1 | 26 | 1167_1179 | 146 | 1193.0 | Topological domain | Cytoplasmic |
| Hgene | GLG1 | chr16:74640555 | chr16:75276988 | ENST00000447066 | - | 1 | 26 | 30_1145 | 146 | 1193.0 | Topological domain | Extracellular |
| Hgene | GLG1 | chr16:74640555 | chr16:75276988 | ENST00000205061 | - | 1 | 27 | 1146_1166 | 146 | 1204.0 | Transmembrane | Helical |
| Hgene | GLG1 | chr16:74640555 | chr16:75276988 | ENST00000422840 | - | 1 | 26 | 1146_1166 | 146 | 1180.0 | Transmembrane | Helical |
| Hgene | GLG1 | chr16:74640555 | chr16:75276988 | ENST00000447066 | - | 1 | 26 | 1146_1166 | 146 | 1193.0 | Transmembrane | Helical |
| Tgene | BCAR1 | chr16:74640555 | chr16:75276988 | ENST00000393422 | 0 | 7 | 3_65 | 22 | 889.0 | Domain | SH3 | |
| Tgene | BCAR1 | chr16:74640555 | chr16:75276988 | ENST00000418647 | 1 | 8 | 3_65 | 50 | 917.0 | Domain | SH3 | |
| Tgene | BCAR1 | chr16:74640555 | chr16:75276988 | ENST00000420641 | 0 | 7 | 3_65 | 22 | 889.0 | Domain | SH3 |
Top |
Fusion Gene Sequence for GLG1-BCAR1 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >33240_33240_1_GLG1-BCAR1_GLG1_chr16_74640555_ENST00000205061_BCAR1_chr16_75276988_ENST00000162330_length(transcript)=3540nt_BP=458nt GAGCTCGCCGCGGACTCAAGATGGCGGCGTGTGGACGTGTACGGAGGATGTTCCGCTTGTCGGCGGCGCTGCATCTGCTGCTGCTATTCG CGGCCGGGGCCGAGAAACTCCCCGGCCAGGGCGTCCACAGCCAGGGCCAGGGTCCCGGGGCCAACTTTGTGTCCTTCGTAGGGCAGGCCG GAGGCGGCGGCCCGGCGGGTCAGCAGCTGCCCCAGCTGCCTCAGTCATCGCAGCTTCAGCAGCAACAGCAGCAGCAGCAACAGCAACAGC AGCCTCAGCCGCCGCAGCCGCCTTTCCCGGCGGGTGGGCCTCCGGCCCGGCGGGGAGGAGCGGGGGCTGGTGGGGGCTGGAAGCTGGCGG AGGAAGAGTCCTGCAGGGAGGACGTGACCCGCGTGTGCCCTAAGCACACCTGGAGCAACAACCTGGCGGTGCTCGAGTGCCTGCAGGATG TGAGGGAGAACGTGCTGGCCAAAGCGCTCTATGACAATGTGGCCGAGTCCCCGGATGAGCTCTCCTTCCGCAAGGGTGACATCATGACGG TGCTGGAGCAGGACACGCAGGGCCTGGACGGCTGGTGGCTCTGCTCGCTGCATGGGCGCCAGGGCATCGTGCCTGGGAACCGCCTCAAGA TCTTGGTGGGCATGTATGATAAGAAGCCAGCAGGGCCTGGCCCCGGCCCTCCCGCCACCCCGGCCCAGCCTCAGCCTGGCCTCCATGCCC CAGCGCCTCCGGCCTCCCAGTACACGCCCATGCTCCCCAACACCTACCAGCCCCAGCCAGACAGCGTCTACCTGGTGCCCACTCCCAGCA AGGCTCAGCAAGGCCTCTACCAAGTCCCGGGTCCCAGCCCTCAGTTCCAGTCTCCCCCAGCCAAGCAGACATCCACCTTCTCGAAGCAGA CACCCCATCACCCGTTTCCCAGCCCGGCCACAGACCTGTACCAGGTGCCCCCAGGGCCTGGAGGCCCTGCCCAGGATATTTACCAGGTGC CACCTTCTGCCGGGATGGGGCATGACATCTACCAGGTCCCCCCGTCCATGGACACACGCAGCTGGGAGGGCACGAAGCCCCCGGCAAAGG TGGTGGTGCCCACCCGCGTGGGGCAGGGCTATGTATACGAGGCCGCCCAGCCGGAGCAGGACGAGTACGACATCCCGCGACACCTGCTGG CCCCGGGGCCACAGGACATCTATGATGTGCCCCCGGTTCGGGGGCTGCTTCCCAGCCAGTATGGCCAGGAGGTGTATGACACACCCCCCA TGGCTGTCAAGGGTCCCAATGGCCGAGACCCGTTGCTGGAGGTGTATGACGTGCCCCCCAGTGTGGAGAAGGGCCTGCCACCGTCCAACC ACCACGCAGTCTACGACGTTCCTCCATCGGTGAGCAAGGATGTGCCCGATGGCCCACTGCTGCGTGAGGAGACCTACGATGTGCCCCCCG CCTTCGCCAAGGCCAAGCCCTTTGACCCGGCCCGCACCCCACTGGTACTGGCTGCGCCCCCTCCAGACTCCCCGCCGGCCGAGGACGTGT ATGACGTGCCGCCCCCGGCTCCTGACCTCTACGACGTGCCCCCTGGCTTGCGGCGGCCTGGCCCGGGCACCCTGTACGATGTGCCCCGTG AACGGGTGCTTCCTCCTGAGGTGGCTGATGGTGGCGTGGTCGACAGTGGTGTGTATGCGGTGCCTCCCCCAGCTGAACGTGAAGCCCCGG CAGAGGGCAAGCGCCTGTCGGCCTCCAGCACCGGCAGCACACGCAGCAGCCAGTCTGCGTCCTCCTTGGAGGTGGCAGGGCCGGGCCGGG AACCCCTGGAGCTGGAAGTTGCTGTGGAGGCCCTGGCACGGCTGCAGCAGGGTGTGAGCGCCACCGTTGCCCACCTTCTGGACCTGGCAG GCAGCGCCGGTGCGACTGGGAGCTGGCGTAGCCCCTCTGAGCCACAGGAGCCGCTGGTGCAGGACCTGCAGGCTGCTGTGGCCGCTGTCC AGAGTGCCGTCCACGAGCTGTTGGAGTTTGCCCGCAGCGCGGTGGGCAATGCTGCCCACACATCTGACCGTGCCCTGCATGCCAAGCTTA GCCGGCAGCTGCAGAAGATGGAGGACGTGCACCAGACGCTGGTGGCACATGGTCAGGCCCTCGACGCTGGCCGGGGAGGCTCTGGAGCCA CCCTTGAGGACCTGGACCGGCTGGTGGCCTGCTCGCGGGCTGTGCCCGAGGACGCCAAGCAGCTGGCCTCCTTCCTGCACGGCAATGCCT CACTGCTCTTCAGACGGACCAAGGCCACTGCCCCGGGGCCTGAGGGGGGTGGCACCCTGCACCCCAACCCCACTGACAAGACCAGCAGCA TCCAGTCACGACCCCTGCCCTCACCCCCTAAGTTCACCTCCCAGGACTCGCCAGATGGGCAGTACGAGAACAGCGAGGGGGGCTGGATGG AGGACTATGACTACGTCCACCTACAGGGGAAGGAGGAGTTTGAGAAGACCCAGAAGGAGCTGCTGGAAAAGGGCAGCATCACGCGGCAGG GCAAGAGCCAGCTGGAGTTGCAGCAGCTGAAGCAGTTTGAACGACTGGAACAGGAGGTGTCACGGCCCATAGACCACGACCTGGCCAACT GGACGCCAGCCCAACCCCTGGCCCCGGGGCGAACAGGCGGCCTGGGGCCCTCGGACCGGCAGCTGCTGCTCTTCTACCTGGAGCAGTGTG AGGCCAACCTGACCACACTGACCAACGCCGTGGACGCCTTCTTTACCGCCGTGGCCACCAACCAGCCGCCCAAGATCTTTGTGGCGCACA GCAAGTTCGTCATCCTCAGCGCCCACAAGCTGGTGTTCATCGGGGACACACTGTCACGGCAGGCCAAGGCTGCTGACGTGCGCAGCCAGG TGACCCACTACAGCAACCTGCTGTGCGACCTCCTGCGCGGCATCGTGGCCACCACCAAGGCCGCTGCCTTGCAGTACCCATCGCCTTCCG CGGCCCAGGACATGGTGGAGAGGGTCAAGGAGCTGGGCCACAGCACCCAGCAGTTCCGCCGCGTCCTAGGCCAGCTGGCAGCCGCCTGAG GGTGGTGACCCCAGGAGGGAGGCAGGGGAGGGGTGCGGCGGTCCCAGCTCCCTGGCTCCCATGTCAAGAGTCGCTGTGCCACAGGCTTAG GGACAGGACCCCAGCTCTGCGTCGGTCCTGGTGCCCTGGATGCCCAGGAATCTGTATATATTTATGGCCGGGCAGGGTGTGGGGCCATGC CTCCTCAGGAGCCGAAGCCCAGGGGCCGGCCAGTGGCCTTCCCCAGCATGCACCACGGGCCCGGGTTGGGTCACCAGACGGGGCTGGAGT GTGAGGGTCCTGCAGCCTGCAGGACCTCGTGCCACCCCGAGGGCTGAGCCTGGTCCCACGAGGGTGCCGTGTCCCCTGACAGGGCCAGTG CAGTTTGGTGTGTCCTCCGCCTTACCAGGAGAAGAACCTGAAGAACTATTTTTCGTTATTGGTTTTCCAATCATTTGACTAAGAGTCTCC >33240_33240_1_GLG1-BCAR1_GLG1_chr16_74640555_ENST00000205061_BCAR1_chr16_75276988_ENST00000162330_length(amino acids)=1012AA_BP=146 MAACGRVRRMFRLSAALHLLLLFAAGAEKLPGQGVHSQGQGPGANFVSFVGQAGGGGPAGQQLPQLPQSSQLQQQQQQQQQQQQPQPPQP PFPAGGPPARRGGAGAGGGWKLAEEESCREDVTRVCPKHTWSNNLAVLECLQDVRENVLAKALYDNVAESPDELSFRKGDIMTVLEQDTQ GLDGWWLCSLHGRQGIVPGNRLKILVGMYDKKPAGPGPGPPATPAQPQPGLHAPAPPASQYTPMLPNTYQPQPDSVYLVPTPSKAQQGLY QVPGPSPQFQSPPAKQTSTFSKQTPHHPFPSPATDLYQVPPGPGGPAQDIYQVPPSAGMGHDIYQVPPSMDTRSWEGTKPPAKVVVPTRV GQGYVYEAAQPEQDEYDIPRHLLAPGPQDIYDVPPVRGLLPSQYGQEVYDTPPMAVKGPNGRDPLLEVYDVPPSVEKGLPPSNHHAVYDV PPSVSKDVPDGPLLREETYDVPPAFAKAKPFDPARTPLVLAAPPPDSPPAEDVYDVPPPAPDLYDVPPGLRRPGPGTLYDVPRERVLPPE VADGGVVDSGVYAVPPPAEREAPAEGKRLSASSTGSTRSSQSASSLEVAGPGREPLELEVAVEALARLQQGVSATVAHLLDLAGSAGATG SWRSPSEPQEPLVQDLQAAVAAVQSAVHELLEFARSAVGNAAHTSDRALHAKLSRQLQKMEDVHQTLVAHGQALDAGRGGSGATLEDLDR LVACSRAVPEDAKQLASFLHGNASLLFRRTKATAPGPEGGGTLHPNPTDKTSSIQSRPLPSPPKFTSQDSPDGQYENSEGGWMEDYDYVH LQGKEEFEKTQKELLEKGSITRQGKSQLELQQLKQFERLEQEVSRPIDHDLANWTPAQPLAPGRTGGLGPSDRQLLLFYLEQCEANLTTL TNAVDAFFTAVATNQPPKIFVAHSKFVILSAHKLVFIGDTLSRQAKAADVRSQVTHYSNLLCDLLRGIVATTKAAALQYPSPSAAQDMVE -------------------------------------------------------------- >33240_33240_2_GLG1-BCAR1_GLG1_chr16_74640555_ENST00000422840_BCAR1_chr16_75276988_ENST00000162330_length(transcript)=3520nt_BP=438nt ATGGCGGCGTGTGGACGTGTACGGAGGATGTTCCGCTTGTCGGCGGCGCTGCATCTGCTGCTGCTATTCGCGGCCGGGGCCGAGAAACTC CCCGGCCAGGGCGTCCACAGCCAGGGCCAGGGTCCCGGGGCCAACTTTGTGTCCTTCGTAGGGCAGGCCGGAGGCGGCGGCCCGGCGGGT CAGCAGCTGCCCCAGCTGCCTCAGTCATCGCAGCTTCAGCAGCAACAGCAGCAGCAGCAACAGCAACAGCAGCCTCAGCCGCCGCAGCCG CCTTTCCCGGCGGGTGGGCCTCCGGCCCGGCGGGGAGGAGCGGGGGCTGGTGGGGGCTGGAAGCTGGCGGAGGAAGAGTCCTGCAGGGAG GACGTGACCCGCGTGTGCCCTAAGCACACCTGGAGCAACAACCTGGCGGTGCTCGAGTGCCTGCAGGATGTGAGGGAGAACGTGCTGGCC AAAGCGCTCTATGACAATGTGGCCGAGTCCCCGGATGAGCTCTCCTTCCGCAAGGGTGACATCATGACGGTGCTGGAGCAGGACACGCAG GGCCTGGACGGCTGGTGGCTCTGCTCGCTGCATGGGCGCCAGGGCATCGTGCCTGGGAACCGCCTCAAGATCTTGGTGGGCATGTATGAT AAGAAGCCAGCAGGGCCTGGCCCCGGCCCTCCCGCCACCCCGGCCCAGCCTCAGCCTGGCCTCCATGCCCCAGCGCCTCCGGCCTCCCAG TACACGCCCATGCTCCCCAACACCTACCAGCCCCAGCCAGACAGCGTCTACCTGGTGCCCACTCCCAGCAAGGCTCAGCAAGGCCTCTAC CAAGTCCCGGGTCCCAGCCCTCAGTTCCAGTCTCCCCCAGCCAAGCAGACATCCACCTTCTCGAAGCAGACACCCCATCACCCGTTTCCC AGCCCGGCCACAGACCTGTACCAGGTGCCCCCAGGGCCTGGAGGCCCTGCCCAGGATATTTACCAGGTGCCACCTTCTGCCGGGATGGGG CATGACATCTACCAGGTCCCCCCGTCCATGGACACACGCAGCTGGGAGGGCACGAAGCCCCCGGCAAAGGTGGTGGTGCCCACCCGCGTG GGGCAGGGCTATGTATACGAGGCCGCCCAGCCGGAGCAGGACGAGTACGACATCCCGCGACACCTGCTGGCCCCGGGGCCACAGGACATC TATGATGTGCCCCCGGTTCGGGGGCTGCTTCCCAGCCAGTATGGCCAGGAGGTGTATGACACACCCCCCATGGCTGTCAAGGGTCCCAAT GGCCGAGACCCGTTGCTGGAGGTGTATGACGTGCCCCCCAGTGTGGAGAAGGGCCTGCCACCGTCCAACCACCACGCAGTCTACGACGTT CCTCCATCGGTGAGCAAGGATGTGCCCGATGGCCCACTGCTGCGTGAGGAGACCTACGATGTGCCCCCCGCCTTCGCCAAGGCCAAGCCC TTTGACCCGGCCCGCACCCCACTGGTACTGGCTGCGCCCCCTCCAGACTCCCCGCCGGCCGAGGACGTGTATGACGTGCCGCCCCCGGCT CCTGACCTCTACGACGTGCCCCCTGGCTTGCGGCGGCCTGGCCCGGGCACCCTGTACGATGTGCCCCGTGAACGGGTGCTTCCTCCTGAG GTGGCTGATGGTGGCGTGGTCGACAGTGGTGTGTATGCGGTGCCTCCCCCAGCTGAACGTGAAGCCCCGGCAGAGGGCAAGCGCCTGTCG GCCTCCAGCACCGGCAGCACACGCAGCAGCCAGTCTGCGTCCTCCTTGGAGGTGGCAGGGCCGGGCCGGGAACCCCTGGAGCTGGAAGTT GCTGTGGAGGCCCTGGCACGGCTGCAGCAGGGTGTGAGCGCCACCGTTGCCCACCTTCTGGACCTGGCAGGCAGCGCCGGTGCGACTGGG AGCTGGCGTAGCCCCTCTGAGCCACAGGAGCCGCTGGTGCAGGACCTGCAGGCTGCTGTGGCCGCTGTCCAGAGTGCCGTCCACGAGCTG TTGGAGTTTGCCCGCAGCGCGGTGGGCAATGCTGCCCACACATCTGACCGTGCCCTGCATGCCAAGCTTAGCCGGCAGCTGCAGAAGATG GAGGACGTGCACCAGACGCTGGTGGCACATGGTCAGGCCCTCGACGCTGGCCGGGGAGGCTCTGGAGCCACCCTTGAGGACCTGGACCGG CTGGTGGCCTGCTCGCGGGCTGTGCCCGAGGACGCCAAGCAGCTGGCCTCCTTCCTGCACGGCAATGCCTCACTGCTCTTCAGACGGACC AAGGCCACTGCCCCGGGGCCTGAGGGGGGTGGCACCCTGCACCCCAACCCCACTGACAAGACCAGCAGCATCCAGTCACGACCCCTGCCC TCACCCCCTAAGTTCACCTCCCAGGACTCGCCAGATGGGCAGTACGAGAACAGCGAGGGGGGCTGGATGGAGGACTATGACTACGTCCAC CTACAGGGGAAGGAGGAGTTTGAGAAGACCCAGAAGGAGCTGCTGGAAAAGGGCAGCATCACGCGGCAGGGCAAGAGCCAGCTGGAGTTG CAGCAGCTGAAGCAGTTTGAACGACTGGAACAGGAGGTGTCACGGCCCATAGACCACGACCTGGCCAACTGGACGCCAGCCCAACCCCTG GCCCCGGGGCGAACAGGCGGCCTGGGGCCCTCGGACCGGCAGCTGCTGCTCTTCTACCTGGAGCAGTGTGAGGCCAACCTGACCACACTG ACCAACGCCGTGGACGCCTTCTTTACCGCCGTGGCCACCAACCAGCCGCCCAAGATCTTTGTGGCGCACAGCAAGTTCGTCATCCTCAGC GCCCACAAGCTGGTGTTCATCGGGGACACACTGTCACGGCAGGCCAAGGCTGCTGACGTGCGCAGCCAGGTGACCCACTACAGCAACCTG CTGTGCGACCTCCTGCGCGGCATCGTGGCCACCACCAAGGCCGCTGCCTTGCAGTACCCATCGCCTTCCGCGGCCCAGGACATGGTGGAG AGGGTCAAGGAGCTGGGCCACAGCACCCAGCAGTTCCGCCGCGTCCTAGGCCAGCTGGCAGCCGCCTGAGGGTGGTGACCCCAGGAGGGA GGCAGGGGAGGGGTGCGGCGGTCCCAGCTCCCTGGCTCCCATGTCAAGAGTCGCTGTGCCACAGGCTTAGGGACAGGACCCCAGCTCTGC GTCGGTCCTGGTGCCCTGGATGCCCAGGAATCTGTATATATTTATGGCCGGGCAGGGTGTGGGGCCATGCCTCCTCAGGAGCCGAAGCCC AGGGGCCGGCCAGTGGCCTTCCCCAGCATGCACCACGGGCCCGGGTTGGGTCACCAGACGGGGCTGGAGTGTGAGGGTCCTGCAGCCTGC AGGACCTCGTGCCACCCCGAGGGCTGAGCCTGGTCCCACGAGGGTGCCGTGTCCCCTGACAGGGCCAGTGCAGTTTGGTGTGTCCTCCGC CTTACCAGGAGAAGAACCTGAAGAACTATTTTTCGTTATTGGTTTTCCAATCATTTGACTAAGAGTCTCCATTTAAATAAAGTTTTTAAA >33240_33240_2_GLG1-BCAR1_GLG1_chr16_74640555_ENST00000422840_BCAR1_chr16_75276988_ENST00000162330_length(amino acids)=1012AA_BP=146 MAACGRVRRMFRLSAALHLLLLFAAGAEKLPGQGVHSQGQGPGANFVSFVGQAGGGGPAGQQLPQLPQSSQLQQQQQQQQQQQQPQPPQP PFPAGGPPARRGGAGAGGGWKLAEEESCREDVTRVCPKHTWSNNLAVLECLQDVRENVLAKALYDNVAESPDELSFRKGDIMTVLEQDTQ GLDGWWLCSLHGRQGIVPGNRLKILVGMYDKKPAGPGPGPPATPAQPQPGLHAPAPPASQYTPMLPNTYQPQPDSVYLVPTPSKAQQGLY QVPGPSPQFQSPPAKQTSTFSKQTPHHPFPSPATDLYQVPPGPGGPAQDIYQVPPSAGMGHDIYQVPPSMDTRSWEGTKPPAKVVVPTRV GQGYVYEAAQPEQDEYDIPRHLLAPGPQDIYDVPPVRGLLPSQYGQEVYDTPPMAVKGPNGRDPLLEVYDVPPSVEKGLPPSNHHAVYDV PPSVSKDVPDGPLLREETYDVPPAFAKAKPFDPARTPLVLAAPPPDSPPAEDVYDVPPPAPDLYDVPPGLRRPGPGTLYDVPRERVLPPE VADGGVVDSGVYAVPPPAEREAPAEGKRLSASSTGSTRSSQSASSLEVAGPGREPLELEVAVEALARLQQGVSATVAHLLDLAGSAGATG SWRSPSEPQEPLVQDLQAAVAAVQSAVHELLEFARSAVGNAAHTSDRALHAKLSRQLQKMEDVHQTLVAHGQALDAGRGGSGATLEDLDR LVACSRAVPEDAKQLASFLHGNASLLFRRTKATAPGPEGGGTLHPNPTDKTSSIQSRPLPSPPKFTSQDSPDGQYENSEGGWMEDYDYVH LQGKEEFEKTQKELLEKGSITRQGKSQLELQQLKQFERLEQEVSRPIDHDLANWTPAQPLAPGRTGGLGPSDRQLLLFYLEQCEANLTTL TNAVDAFFTAVATNQPPKIFVAHSKFVILSAHKLVFIGDTLSRQAKAADVRSQVTHYSNLLCDLLRGIVATTKAAALQYPSPSAAQDMVE -------------------------------------------------------------- >33240_33240_3_GLG1-BCAR1_GLG1_chr16_74640555_ENST00000447066_BCAR1_chr16_75276988_ENST00000162330_length(transcript)=3538nt_BP=456nt GCTCGCCGCGGACTCAAGATGGCGGCGTGTGGACGTGTACGGAGGATGTTCCGCTTGTCGGCGGCGCTGCATCTGCTGCTGCTATTCGCG GCCGGGGCCGAGAAACTCCCCGGCCAGGGCGTCCACAGCCAGGGCCAGGGTCCCGGGGCCAACTTTGTGTCCTTCGTAGGGCAGGCCGGA GGCGGCGGCCCGGCGGGTCAGCAGCTGCCCCAGCTGCCTCAGTCATCGCAGCTTCAGCAGCAACAGCAGCAGCAGCAACAGCAACAGCAG CCTCAGCCGCCGCAGCCGCCTTTCCCGGCGGGTGGGCCTCCGGCCCGGCGGGGAGGAGCGGGGGCTGGTGGGGGCTGGAAGCTGGCGGAG GAAGAGTCCTGCAGGGAGGACGTGACCCGCGTGTGCCCTAAGCACACCTGGAGCAACAACCTGGCGGTGCTCGAGTGCCTGCAGGATGTG AGGGAGAACGTGCTGGCCAAAGCGCTCTATGACAATGTGGCCGAGTCCCCGGATGAGCTCTCCTTCCGCAAGGGTGACATCATGACGGTG CTGGAGCAGGACACGCAGGGCCTGGACGGCTGGTGGCTCTGCTCGCTGCATGGGCGCCAGGGCATCGTGCCTGGGAACCGCCTCAAGATC TTGGTGGGCATGTATGATAAGAAGCCAGCAGGGCCTGGCCCCGGCCCTCCCGCCACCCCGGCCCAGCCTCAGCCTGGCCTCCATGCCCCA GCGCCTCCGGCCTCCCAGTACACGCCCATGCTCCCCAACACCTACCAGCCCCAGCCAGACAGCGTCTACCTGGTGCCCACTCCCAGCAAG GCTCAGCAAGGCCTCTACCAAGTCCCGGGTCCCAGCCCTCAGTTCCAGTCTCCCCCAGCCAAGCAGACATCCACCTTCTCGAAGCAGACA CCCCATCACCCGTTTCCCAGCCCGGCCACAGACCTGTACCAGGTGCCCCCAGGGCCTGGAGGCCCTGCCCAGGATATTTACCAGGTGCCA CCTTCTGCCGGGATGGGGCATGACATCTACCAGGTCCCCCCGTCCATGGACACACGCAGCTGGGAGGGCACGAAGCCCCCGGCAAAGGTG GTGGTGCCCACCCGCGTGGGGCAGGGCTATGTATACGAGGCCGCCCAGCCGGAGCAGGACGAGTACGACATCCCGCGACACCTGCTGGCC CCGGGGCCACAGGACATCTATGATGTGCCCCCGGTTCGGGGGCTGCTTCCCAGCCAGTATGGCCAGGAGGTGTATGACACACCCCCCATG GCTGTCAAGGGTCCCAATGGCCGAGACCCGTTGCTGGAGGTGTATGACGTGCCCCCCAGTGTGGAGAAGGGCCTGCCACCGTCCAACCAC CACGCAGTCTACGACGTTCCTCCATCGGTGAGCAAGGATGTGCCCGATGGCCCACTGCTGCGTGAGGAGACCTACGATGTGCCCCCCGCC TTCGCCAAGGCCAAGCCCTTTGACCCGGCCCGCACCCCACTGGTACTGGCTGCGCCCCCTCCAGACTCCCCGCCGGCCGAGGACGTGTAT GACGTGCCGCCCCCGGCTCCTGACCTCTACGACGTGCCCCCTGGCTTGCGGCGGCCTGGCCCGGGCACCCTGTACGATGTGCCCCGTGAA CGGGTGCTTCCTCCTGAGGTGGCTGATGGTGGCGTGGTCGACAGTGGTGTGTATGCGGTGCCTCCCCCAGCTGAACGTGAAGCCCCGGCA GAGGGCAAGCGCCTGTCGGCCTCCAGCACCGGCAGCACACGCAGCAGCCAGTCTGCGTCCTCCTTGGAGGTGGCAGGGCCGGGCCGGGAA CCCCTGGAGCTGGAAGTTGCTGTGGAGGCCCTGGCACGGCTGCAGCAGGGTGTGAGCGCCACCGTTGCCCACCTTCTGGACCTGGCAGGC AGCGCCGGTGCGACTGGGAGCTGGCGTAGCCCCTCTGAGCCACAGGAGCCGCTGGTGCAGGACCTGCAGGCTGCTGTGGCCGCTGTCCAG AGTGCCGTCCACGAGCTGTTGGAGTTTGCCCGCAGCGCGGTGGGCAATGCTGCCCACACATCTGACCGTGCCCTGCATGCCAAGCTTAGC CGGCAGCTGCAGAAGATGGAGGACGTGCACCAGACGCTGGTGGCACATGGTCAGGCCCTCGACGCTGGCCGGGGAGGCTCTGGAGCCACC CTTGAGGACCTGGACCGGCTGGTGGCCTGCTCGCGGGCTGTGCCCGAGGACGCCAAGCAGCTGGCCTCCTTCCTGCACGGCAATGCCTCA CTGCTCTTCAGACGGACCAAGGCCACTGCCCCGGGGCCTGAGGGGGGTGGCACCCTGCACCCCAACCCCACTGACAAGACCAGCAGCATC CAGTCACGACCCCTGCCCTCACCCCCTAAGTTCACCTCCCAGGACTCGCCAGATGGGCAGTACGAGAACAGCGAGGGGGGCTGGATGGAG GACTATGACTACGTCCACCTACAGGGGAAGGAGGAGTTTGAGAAGACCCAGAAGGAGCTGCTGGAAAAGGGCAGCATCACGCGGCAGGGC AAGAGCCAGCTGGAGTTGCAGCAGCTGAAGCAGTTTGAACGACTGGAACAGGAGGTGTCACGGCCCATAGACCACGACCTGGCCAACTGG ACGCCAGCCCAACCCCTGGCCCCGGGGCGAACAGGCGGCCTGGGGCCCTCGGACCGGCAGCTGCTGCTCTTCTACCTGGAGCAGTGTGAG GCCAACCTGACCACACTGACCAACGCCGTGGACGCCTTCTTTACCGCCGTGGCCACCAACCAGCCGCCCAAGATCTTTGTGGCGCACAGC AAGTTCGTCATCCTCAGCGCCCACAAGCTGGTGTTCATCGGGGACACACTGTCACGGCAGGCCAAGGCTGCTGACGTGCGCAGCCAGGTG ACCCACTACAGCAACCTGCTGTGCGACCTCCTGCGCGGCATCGTGGCCACCACCAAGGCCGCTGCCTTGCAGTACCCATCGCCTTCCGCG GCCCAGGACATGGTGGAGAGGGTCAAGGAGCTGGGCCACAGCACCCAGCAGTTCCGCCGCGTCCTAGGCCAGCTGGCAGCCGCCTGAGGG TGGTGACCCCAGGAGGGAGGCAGGGGAGGGGTGCGGCGGTCCCAGCTCCCTGGCTCCCATGTCAAGAGTCGCTGTGCCACAGGCTTAGGG ACAGGACCCCAGCTCTGCGTCGGTCCTGGTGCCCTGGATGCCCAGGAATCTGTATATATTTATGGCCGGGCAGGGTGTGGGGCCATGCCT CCTCAGGAGCCGAAGCCCAGGGGCCGGCCAGTGGCCTTCCCCAGCATGCACCACGGGCCCGGGTTGGGTCACCAGACGGGGCTGGAGTGT GAGGGTCCTGCAGCCTGCAGGACCTCGTGCCACCCCGAGGGCTGAGCCTGGTCCCACGAGGGTGCCGTGTCCCCTGACAGGGCCAGTGCA GTTTGGTGTGTCCTCCGCCTTACCAGGAGAAGAACCTGAAGAACTATTTTTCGTTATTGGTTTTCCAATCATTTGACTAAGAGTCTCCAT >33240_33240_3_GLG1-BCAR1_GLG1_chr16_74640555_ENST00000447066_BCAR1_chr16_75276988_ENST00000162330_length(amino acids)=1012AA_BP=146 MAACGRVRRMFRLSAALHLLLLFAAGAEKLPGQGVHSQGQGPGANFVSFVGQAGGGGPAGQQLPQLPQSSQLQQQQQQQQQQQQPQPPQP PFPAGGPPARRGGAGAGGGWKLAEEESCREDVTRVCPKHTWSNNLAVLECLQDVRENVLAKALYDNVAESPDELSFRKGDIMTVLEQDTQ GLDGWWLCSLHGRQGIVPGNRLKILVGMYDKKPAGPGPGPPATPAQPQPGLHAPAPPASQYTPMLPNTYQPQPDSVYLVPTPSKAQQGLY QVPGPSPQFQSPPAKQTSTFSKQTPHHPFPSPATDLYQVPPGPGGPAQDIYQVPPSAGMGHDIYQVPPSMDTRSWEGTKPPAKVVVPTRV GQGYVYEAAQPEQDEYDIPRHLLAPGPQDIYDVPPVRGLLPSQYGQEVYDTPPMAVKGPNGRDPLLEVYDVPPSVEKGLPPSNHHAVYDV PPSVSKDVPDGPLLREETYDVPPAFAKAKPFDPARTPLVLAAPPPDSPPAEDVYDVPPPAPDLYDVPPGLRRPGPGTLYDVPRERVLPPE VADGGVVDSGVYAVPPPAEREAPAEGKRLSASSTGSTRSSQSASSLEVAGPGREPLELEVAVEALARLQQGVSATVAHLLDLAGSAGATG SWRSPSEPQEPLVQDLQAAVAAVQSAVHELLEFARSAVGNAAHTSDRALHAKLSRQLQKMEDVHQTLVAHGQALDAGRGGSGATLEDLDR LVACSRAVPEDAKQLASFLHGNASLLFRRTKATAPGPEGGGTLHPNPTDKTSSIQSRPLPSPPKFTSQDSPDGQYENSEGGWMEDYDYVH LQGKEEFEKTQKELLEKGSITRQGKSQLELQQLKQFERLEQEVSRPIDHDLANWTPAQPLAPGRTGGLGPSDRQLLLFYLEQCEANLTTL TNAVDAFFTAVATNQPPKIFVAHSKFVILSAHKLVFIGDTLSRQAKAADVRSQVTHYSNLLCDLLRGIVATTKAAALQYPSPSAAQDMVE -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for GLG1-BCAR1 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for GLG1-BCAR1 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for GLG1-BCAR1 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
