
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:GLT8D2-YY1 (FusionGDB2 ID:33369) |
Fusion Gene Summary for GLT8D2-YY1 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: GLT8D2-YY1 | Fusion gene ID: 33369 | Hgene | Tgene | Gene symbol | GLT8D2 | YY1 | Gene ID | 83468 | 7528 |
| Gene name | glycosyltransferase 8 domain containing 2 | YY1 transcription factor | |
| Synonyms | - | DELTA|GADEVS|INO80S|NF-E1|UCRBP|YIN-YANG-1 | |
| Cytomap | 12q23.3 | 14q32.2 | |
| Type of gene | protein-coding | protein-coding | |
| Description | glycosyltransferase 8 domain-containing protein 2 | transcriptional repressor protein YY1INO80 complex subunit SYY-1Yin and Yang 1 proteindelta transcription factor | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | Q9H1C3 | YY1AP1 | |
| Ensembl transtripts involved in fusion gene | ENST00000360814, ENST00000546436, ENST00000548660, ENST00000547583, | ENST00000262238, | |
| Fusion gene scores | * DoF score | 3 X 3 X 2=18 | 12 X 7 X 7=588 |
| # samples | 3 | 13 | |
| ** MAII score | log2(3/18*10)=0.736965594166206 effective Gene in Pan-Cancer Fusion Genes (eGinPCFGs). DoF>8 and MAII>0 | log2(13/588*10)=-2.17730453180791 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: GLT8D2 [Title/Abstract] AND YY1 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | GLT8D2(104396913)-YY1(100741035), # samples:3 | ||
| Anticipated loss of major functional domain due to fusion event. | GLT8D2-YY1 seems lost the major protein functional domain in Tgene partner, which is a epigenetic factor due to the frame-shifted ORF. GLT8D2-YY1 seems lost the major protein functional domain in Tgene partner, which is a transcription factor due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Tgene | YY1 | GO:0000122 | negative regulation of transcription by RNA polymerase II | 9857059|16260628 |
| Tgene | YY1 | GO:0032688 | negative regulation of interferon-beta production | 16260628 |
 Fusion gene breakpoints across GLT8D2 (5'-gene) Fusion gene breakpoints across GLT8D2 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
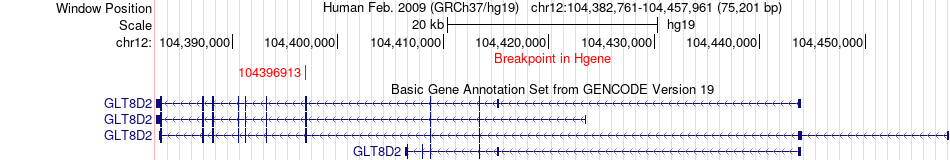 |
 Fusion gene breakpoints across YY1 (3'-gene) Fusion gene breakpoints across YY1 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
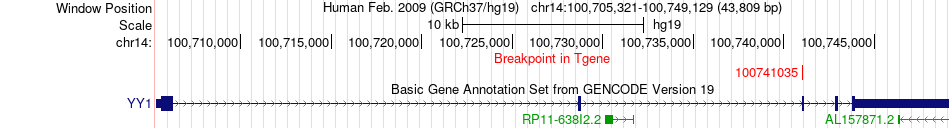 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | SARC | TCGA-DX-A2IZ-01A | GLT8D2 | chr12 | 104396913 | - | YY1 | chr14 | 100741035 | + |
Top |
Fusion Gene ORF analysis for GLT8D2-YY1 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| Frame-shift | ENST00000360814 | ENST00000262238 | GLT8D2 | chr12 | 104396913 | - | YY1 | chr14 | 100741035 | + |
| Frame-shift | ENST00000546436 | ENST00000262238 | GLT8D2 | chr12 | 104396913 | - | YY1 | chr14 | 100741035 | + |
| In-frame | ENST00000548660 | ENST00000262238 | GLT8D2 | chr12 | 104396913 | - | YY1 | chr14 | 100741035 | + |
| intron-3CDS | ENST00000547583 | ENST00000262238 | GLT8D2 | chr12 | 104396913 | - | YY1 | chr14 | 100741035 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000548660 | GLT8D2 | chr12 | 104396913 | - | ENST00000262238 | YY1 | chr14 | 100741035 | + | 6536 | 941 | 657 | 1343 | 228 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000548660 | ENST00000262238 | GLT8D2 | chr12 | 104396913 | - | YY1 | chr14 | 100741035 | + | 0.000720368 | 0.9992797 |
Top |
Fusion Genomic Features for GLT8D2-YY1 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| GLT8D2 | chr12 | 104396912 | - | YY1 | chr14 | 100741034 | + | 3.01E-07 | 0.99999964 |
| GLT8D2 | chr12 | 104396912 | - | YY1 | chr14 | 100741034 | + | 3.01E-07 | 0.99999964 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
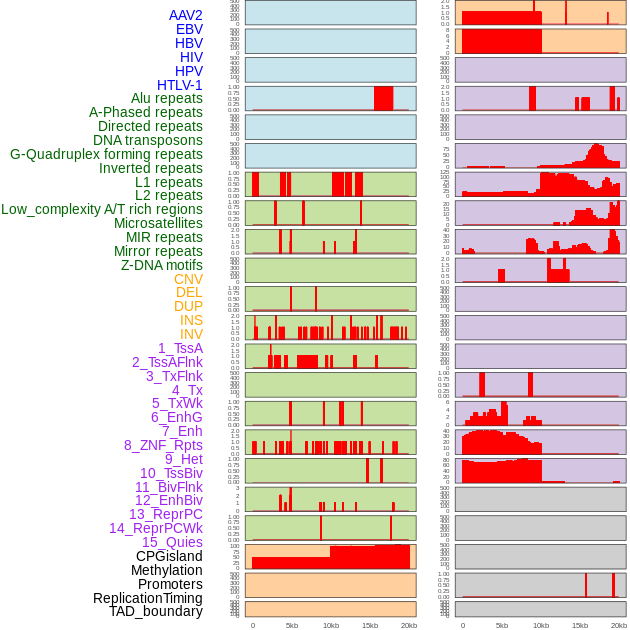 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
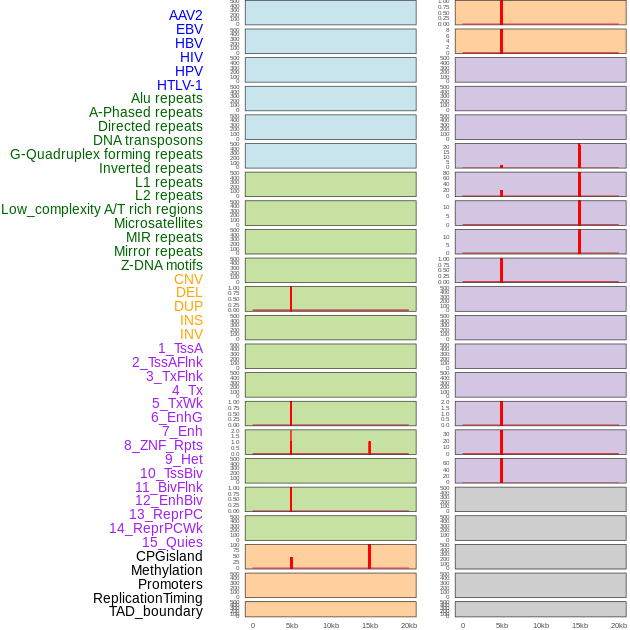 |
Top |
Fusion Protein Features for GLT8D2-YY1 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr12:104396913/chr14:100741035) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| GLT8D2 | YY1 |
| 796 |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | GLT8D2 | chr12:104396913 | chr14:100741035 | ENST00000360814 | - | 5 | 11 | 1_6 | 94 | 350.0 | Topological domain | Cytoplasmic |
| Hgene | GLT8D2 | chr12:104396913 | chr14:100741035 | ENST00000546436 | - | 4 | 10 | 1_6 | 94 | 350.0 | Topological domain | Cytoplasmic |
| Hgene | GLT8D2 | chr12:104396913 | chr14:100741035 | ENST00000548660 | - | 5 | 11 | 1_6 | 94 | 350.0 | Topological domain | Cytoplasmic |
| Hgene | GLT8D2 | chr12:104396913 | chr14:100741035 | ENST00000360814 | - | 5 | 11 | 7_24 | 94 | 350.0 | Transmembrane | Helical%3B Signal-anchor for type II membrane protein |
| Hgene | GLT8D2 | chr12:104396913 | chr14:100741035 | ENST00000546436 | - | 4 | 10 | 7_24 | 94 | 350.0 | Transmembrane | Helical%3B Signal-anchor for type II membrane protein |
| Hgene | GLT8D2 | chr12:104396913 | chr14:100741035 | ENST00000548660 | - | 5 | 11 | 7_24 | 94 | 350.0 | Transmembrane | Helical%3B Signal-anchor for type II membrane protein |
| Tgene | YY1 | chr12:104396913 | chr14:100741035 | ENST00000262238 | 1 | 5 | 295_414 | 280 | 415.0 | Region | Note=Binding to DNA | |
| Tgene | YY1 | chr12:104396913 | chr14:100741035 | ENST00000262238 | 1 | 5 | 333_371 | 280 | 415.0 | Region | Note=Involved in repression of activated transcription | |
| Tgene | YY1 | chr12:104396913 | chr14:100741035 | ENST00000262238 | 1 | 5 | 371_397 | 280 | 415.0 | Region | Note=Involved in masking transactivation domain | |
| Tgene | YY1 | chr12:104396913 | chr14:100741035 | ENST00000262238 | 1 | 5 | 296_320 | 280 | 415.0 | Zinc finger | C2H2-type 1 | |
| Tgene | YY1 | chr12:104396913 | chr14:100741035 | ENST00000262238 | 1 | 5 | 325_347 | 280 | 415.0 | Zinc finger | C2H2-type 2 | |
| Tgene | YY1 | chr12:104396913 | chr14:100741035 | ENST00000262238 | 1 | 5 | 353_377 | 280 | 415.0 | Zinc finger | C2H2-type 3 | |
| Tgene | YY1 | chr12:104396913 | chr14:100741035 | ENST00000262238 | 1 | 5 | 383_407 | 280 | 415.0 | Zinc finger | C2H2-type 4 |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | GLT8D2 | chr12:104396913 | chr14:100741035 | ENST00000360814 | - | 5 | 11 | 25_349 | 94 | 350.0 | Topological domain | Lumenal |
| Hgene | GLT8D2 | chr12:104396913 | chr14:100741035 | ENST00000546436 | - | 4 | 10 | 25_349 | 94 | 350.0 | Topological domain | Lumenal |
| Hgene | GLT8D2 | chr12:104396913 | chr14:100741035 | ENST00000548660 | - | 5 | 11 | 25_349 | 94 | 350.0 | Topological domain | Lumenal |
| Tgene | YY1 | chr12:104396913 | chr14:100741035 | ENST00000262238 | 1 | 5 | 159_170 | 280 | 415.0 | Compositional bias | Note=Gly/Ser-rich | |
| Tgene | YY1 | chr12:104396913 | chr14:100741035 | ENST00000262238 | 1 | 5 | 43_53 | 280 | 415.0 | Compositional bias | Note=Asp/Glu-rich (acidic) | |
| Tgene | YY1 | chr12:104396913 | chr14:100741035 | ENST00000262238 | 1 | 5 | 54_69 | 280 | 415.0 | Compositional bias | Note=Gly-rich | |
| Tgene | YY1 | chr12:104396913 | chr14:100741035 | ENST00000262238 | 1 | 5 | 70_80 | 280 | 415.0 | Compositional bias | Note=Poly-His | |
| Tgene | YY1 | chr12:104396913 | chr14:100741035 | ENST00000262238 | 1 | 5 | 257_341 | 280 | 415.0 | Region | Note=Involved in nuclear matrix association |
Top |
Fusion Gene Sequence for GLT8D2-YY1 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >33369_33369_1_GLT8D2-YY1_GLT8D2_chr12_104396913_ENST00000548660_YY1_chr14_100741035_ENST00000262238_length(transcript)=6536nt_BP=941nt ACAAGCAACGTGAGAGAAGCTCGGGTAGAGGTTCGAAGAGCTGGGAGCGCAGGGCGGGGACCTCGCACGGGAGGGTGCGGGCGGGAGCCT GCCTCGCGAGCCCGGGCCACGGCGGCTGCTCCCGACCGTGACTGGGACCCGGGCAGGGAAGGTCGCTACAAAGCCTGCCGGGCGAACCGA AGGCAGCTTTGTGCGGAGCCATGCCAGGAAGCTTGGGAACCGTGAGCCACAAGAGAACAGCGTTGTTTTTTGCATGTTTTGCACTTAGCC AGAGGAAGTCTGGCGCATCCGAGCCGGCCAGCCGAGCACATCTGGAAGTGGTTTCTGGGGCCGCCCCTCTCTGCCAGCGCAACTCCTGGG TTCCCAGCGGCTTCGCGCAGAGGTGGAAGAAACCCGAGACGTTCCGAAGTCAACGCAAGCAAAGGGGAGTGCGGGTCGGGGAGGAATATT CTTTTGGAAACGTAATATTGGCCTTGGGGCTCTCCAGCCCTTTGGGACTTCCAATGGGATCTTAGAAGCAGCCGAAGCAGCGTGAGGGCG GCAGCCCAGGGCCAGCCACGATTTGAACGCTCTGCCTTGCAGCTCTTCTGGACCGAGGAGCCCAAAGCCCTACCCTCACCATTCACCAGG TTACAGTTCTTATCCGCGTGAATACACATGGCTCTGTTACGAAAAATTAATCAGGTGCTGCTGTTCCTTCTGATCGTGACCCTCTGTGTG ATTCTGTATAAGAAAGTTCATAAGGGGACTGTGCCCAAGAATGACGCAGATGATGAATCCGAGACTCCTGAAGAACTGGAAGAAGAGATT CCTGTGGTGATTTGTGCTGCAGCAGGGAGGATGGGTGCCACTATGGCTGCCATCAATAGCATCTACAGCAACACTGACGCCAACATCTTG TTCTATGTAGTGGGACTCCGGAATACTCTGACTCGAATACGAATGAAGCCAAGAAAAATTAAAGAAGATGATGCTCCAAGAACAATAGCT TGCCCTCATAAAGGCTGCACAAAGATGTTCAGGGATAACTCGGCCATGAGAAAACATCTGCACACCCACGGTCCCAGAGTCCACGTCTGT GCAGAATGTGGCAAAGCTTTTGTTGAGAGTTCAAAACTAAAACGACACCAACTGGTTCATACTGGAGAGAAGCCCTTTCAGTGCACGTTC GAAGGCTGTGGGAAACGCTTTTCACTGGACTTCAATTTGCGCACACATGTGCGAATCCATACCGGAGACAGGCCCTATGTGTGCCCCTTC GATGGTTGTAATAAGAAGTTTGCTCAGTCAACTAACCTGAAATCTCACATCTTAACACATGCTAAGGCCAAAAACAACCAGTGAAAAGAA GAGAGAAGACCCTTCTCGACCACGGGAAGCATCTTCCAGAAGTGTGATTGGGAATAAATATGCCTCTCCTTTGTATATTATTTCTAGGAA GAATTTTAAAAATGAATCCTACACACCTAAGGGACATGTTTTGATAAAGTAGTAAAAATTAAAAAAAAAAAACTTTACTAAGATGACATT GCTAAGATGCTCTATCTTGCTCTGTAATCTCGTTTCAAAAACACAGTGTTTTTGTAAAGTGTGGTCCCAACAGGAGGACAATTCATGAAC TTCGCATCAAAAGACAATTCTTTATACAACAGTGCTAAAAATGGGACTTCTTTTCACATTCTTATAAATATGAAGCTCACCTGTTGCTTA CAATTTTTTTAATTTTGTATTTTCCAAGTGTGCATATTGTACACTTTTTTGGGGATATGCTTAGTAATGCTACGTGTGATTTTTCTGGAG GTTGATAACTTTGCTTGCAGTAGATTTTCTTTAAAAGAATGGGCAGTTACATGCATACTTCAAAAGTATTTTCCTGTAAAAAAAAAAAAG TTATATAGGTTTTGTTTGCTATCTTAATTTTGGTTGTATTCTTTGATGTTAACACATTTTGTATAATTGTATCGTATAGCTGTATTGAAT CATGTAGTATCAAATATTAGATGTGATTTAATAGTGTTAATCAATTTAAACCCATTTTAGTCACTTTTTTTTTCCAAAAAAATACTGCCA GATGCTGATGTTCAGTGTAATTTCTTTGCCTGTTCAGTTACAGAAAGTGGTGCTCAGTTGTAGAATGTATTGTACCTTTTAACACCTGAT GTGTACATCCCATGTAACAGAAAGGGCAACAATAAAATAGCAATCCTAAAGCAAGAATATGGCAGAACAAGATCTGTAAGCACAGTCTTA TTTTCTTTTGTTGTCCAGAATACTTATAATTCTTGAGCCTCCCAGAAATTGGAAGCTAAATAAAGCAACTCAAGTTTCCTTTATTTTGCA CTCAATTACAGTGATTATTGATGAAAGCGATGCATGGATATTTTAATACTTCCTACATGTCCTGACTTCTGAAAGAGAGTAGGTAACAGG CATCCCGAGTTCAGGAACTACCTCAGAACACCCCAGGCCAGGTTGGTCATAGGCTGTGATTTTAGCCCCCGGCAAGTGTGAGTGAAGCAT CTGTACCACCGCGCAGGCTGAGCGCCTGCGCAGGGTAAGGTGCCACCTGGCAGTGGGGCACACAGAGGGAAGACCAGGCCTGTCCATCAG CCGGCTGCCTTCAGAGGCAGCTCCAGCAGGACCTTGGCTTGTCTGACAGGAAATGCTTGTGGTCGTTGGTTATTTGGTTTGAGAGCCCTT GTTCCTCCATCTAGTGGAGTCCTTATTAAATGCTAGCAATGTGGCAATTGAGTGCCAGTAGCTTAATTTCATGTTTCTAAATGGACACTG GGTGCAAATCGGCAAGGTGATTAGATTATTAGAAAATTCTGCTCAATGAGTACCTCTCACATTGTAACCTCTTAAATGTACGATTATAAT GTTGGATCTTAGGTCAAATAACAGACTTGAGAATACTTTATAAACTGCCATGATACAGCAAAATCTCATTTATTCAAAGTGCAAACATAC TGTGTGTCTTTCTGTTTTGTGTATGCTTGACTGCAAGATAAATGACAAACTAGCTAATATCCGAGATTTATTTACAAATGGCCAGGGACC CCATCAGGTGAAACACATAGCCCATTGTCTTAGAGCTTCTCCATCGGTCCTGTACAGGAGGATTGGTGACCTACTAACAGTGACCCTGTG GCTCTGTGGAATTTTGAAGTGCCTTTTGTGAATCATGAATGAAACATTTAACTTGAGACCCTTTAAAAAATTCATCCAGAATCTGTTACC CCTTGGGTAGTTTGATGGAACTGAGTTAAGCAGAAGACCCCAGCTGTTTTCTGGGTAAGCCAGGCTTTGCTGTATCTGGCAGTCAGGAGA GAGGTCAGCCCATGGTGGAGGTCTTCAGTGCCGAGGGCTCGGATGAGAACGTGTAGTTCTTTGACTTGGACCTGGGCAGCGTCTTCTATG GATGCCTGCCTTGTGAGCACCCACACCTTCCCTCCCCACTGTCGGGTTGAGTAGCCCCAGCGTGGCTGTGTTAGGAGTCCTTTGCCCAAC ATTGTGCTGCACTGGAGCCTGGCCCTGGCACTGTCCTCAGGACCCTGCCCAAGGGCCCACCGCAGAGCACACCTATGCTATGGGGAGCCC TGCTGGCAGCCCCGAGAGCCATGCCATGGCCTGCAGGAGCCAGGCTCCTGTGTGGATGAAGTCCCTCTTCCTCTGTGCCTTGATCCCTTG GGGGTGCCTTTGGTCATCTCTTCTGTCCTTTCCTGTCTCTGAAATAGTCATCACTCCCCTTGACTCTCTCTGTTCACGTCTTCTCAGTCT GCAGAGTTAACTTCTGTAAGGAGTTTAATCTGGGGTTCCAAGAAAACAAGTTCCTTGTTAACATAGCACTGACTTTGCAACAATAGAAAA CTAACAAATGAGCAACAATATAAAGAGTAGAGGTAGTTCTCATTGGGTGTAACTTCAACCCATTCTGCTTGTGGTTAGAATTTATAATTT TGCTAACGATGTAATTTACTAAGGTTTGAAATGGTATTCTAACAGTGAGTCCATTGTCTTGAGGATTAATCTGATTTATAAGTAATACTG ATAGACATATTTTCGTACATCTGAGCAGAAATAAATGCATGTTTCTAGCATATGTAATATAAAAACTCCAGAGGATAAGTTTACACTTAA CAAGATGTTGTTTTCTATTTTTGAATTTCTTATTATTTCCCCCTTTTTGGTAGTGGAGTTGAGAGGGGGAGCATACTATTTGTATAAAGA ACTGTCACCCCAGAGTGACATTTATTTTCCAAGATGACCCTGAACTCATGCTGTTGGTTTCATATGGTGTTTGTGTGCTTTGCCTAGATT CAAGGCCTGAGGATTTGAGGAAAATCGGGATTTTTTTTTTCCTATTTGGTTTTCATGTAAAAGTTCTAAATGTCACTTATTTTGCTAAAT AAGACCCTTTCACAGGATAGTGTGCGCAATACCTGTCCGTGTTTCCAAGCAGAGTTCAAGTTCATGTCTTCACAAGTAGATGAGAGTCAC TTCCAGTCGGGGGAAGCCCGCCTGAGCAGTGTTTTGAGAGTGCGTACATTGGAGGACTGCCCCACGTCCGTTTGCACTCTGCCTTGCAAA ACTTACTGTGGAGACGTGTCCCAGACTGGTCTCAGACCAGAGTTTGTGGCACAGGAAAACCTGGCCCCCTAAAAAGAAGTTTTGGAATTG GAAGCCTGATAATTTCGGGAGGCTGAGGCAGGAGAACGGCGTGAACCCGGGAGGCGGAGCTTGCAGTGAGCCGAGATCGCACCACTGCAC TCCAGCCTGGGTGACAGAGCAAGACTCCGTCTCCCAAAAAAAAAAAAAAAAAAAAAAAAAAGGAAACCTGACAATTCTACCCACCTCCCC CCACCCCCGACTATAGGCAAAGAGAGTTGATCACTTCCAGGGTCTCTGACTTCTGTGACCTGGAAATCAGCCTGGCTTGGGAAATTCATG TTGTTGATCTTTAATTATTTAATTATTCTTTAATAATCCTTCTATTTTCTTGACATCTGACCCCCAGCAAGAGTAAGGATCCCAGAGGTC AAAACAGTGGTGAGAGCCAAGCTGCTCACAGACACCCACTTGATGCCACAGCCATGCCATGAGGCTTCCTCCCAGCTGCTCTGCTGGCTT GCTGAAGCCAGCTCTGAGTCACAGGGTTGCCTTGGACCCTCGGCTGTAGGGGGCACCTGTCTGGCTCCGGGCCCTTTGTCCTGGATACAC TGACAGTAATGTGAGCGCAGCACTTCAGAGTTCAGGCCCAGCAGGTCCTGGCAAGTGCATTCCACCCGAACTTTTAACCCAAGCGGTGGG GAAGGAAGCCAAAACTCCAAGCTGCACTTTCTTGGGGTTCTGGCCATGCACTTCTTAGCCTTCTCTCTGACTTTACAGGACAAAAGGTAC CCCACGCCTTCAGAATCATGCTTACCTGCACGGCAGCATCTGACTGGATGGCTGGCTAGGTCTCATCCACCCCAAATTACTGACCCTTGA TTTATGTTGTTACTGCTCAGGTTATGCTCAGATGCCCTGGTTGCCTGAAGATGGTTTAAGTGCAGGCCCTTCAGACAAGTCACGTCAGGC AAGAGAGGGCTGGCCTCTGACCCACAAGAGGGCAGATTTGTGGCCAAGAGGCTTCCTGGCATCCACCCCCAGGCCCAAGGGATCAGTGTG GTGCCAGAGAGGATCTTTAAGCTGGGACTGTACTGAGGCTTGATGGGAAGTATGGGCAGCATGTACTTGACACGTTATCACCCAGTCCCC TTGTAAAATCTGGAATGGAGCGTTTAAGCTGAGCGTTAGCATCAGGTCAGCAGGCAAAGCAAACCTCCCACAAACTGGGTGCCGTAAGGT GGGAGGTGACACATGGAGAGCTTCGTCCTGGCCGAGCAGGCACCCAGCCCCATGTCCCCCGCCAGCATTCTGGATGTCAGAGGAGGGTGC GTGTGATGGCGTCACATTCATCTGAAGCACACTGATTGATTTTCCACAAGGTGGCAAGTTTGATCAAATTGACCGCTTTTCATGAGACAC CCAGATGCTGTAAAAAAAAAAAACAGCACTGGGATGTGTTGAGGAGGGGGCAGTTTGCTGTGCTCTGTTCTGTCTGCTGCTCTCTCTGGG GGCCGCACAATGTCCGCACACATCACGGAGGGGAGAAAGGCATCAGTACCAAACGGAACAACCTCTTTTTTCTACATTGTCCATACCCGG ACATGTGAGGCTCTATTATCAACAGGTGGTGAGAAAAATTATGTTTTTATTCGCTTTCTGGTAACTTCTGTAGGCCCTGGCTCAAGGACT TAGCATTTCGTCTCATGTACATCTTTTTCTTAAGTGTTCTTTGCCATTTCTGGAATTGTCCTTGGTTTTTCCTTAGCTCATAGGTCATAG ATGCAGAAATATAGTATTTAAGGCATCCGCATCCAGCATCAGATGGCTTTGCATCCAGAAAAACATTGATAACTCAGTTTGAAGCACCAA >33369_33369_1_GLT8D2-YY1_GLT8D2_chr12_104396913_ENST00000548660_YY1_chr14_100741035_ENST00000262238_length(amino acids)=228AA_BP=93 MALLRKINQVLLFLLIVTLCVILYKKVHKGTVPKNDADDESETPEELEEEIPVVICAAAGRMGATMAAINSIYSNTDANILFYVVGLRNT LTRIRMKPRKIKEDDAPRTIACPHKGCTKMFRDNSAMRKHLHTHGPRVHVCAECGKAFVESSKLKRHQLVHTGEKPFQCTFEGCGKRFSL -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for GLT8D2-YY1 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
| Tgene | YY1 | chr12:104396913 | chr14:100741035 | ENST00000262238 | 1 | 5 | 1_170 | 280.6666666666667 | 415.0 | the SMAD1/SMAD4 complex |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for GLT8D2-YY1 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for GLT8D2-YY1 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
