
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:GNAS-GAPDH (FusionGDB2 ID:33624) |
Fusion Gene Summary for GNAS-GAPDH |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: GNAS-GAPDH | Fusion gene ID: 33624 | Hgene | Tgene | Gene symbol | GNAS | GAPDH | Gene ID | 2778 | 2597 |
| Gene name | GNAS complex locus | glyceraldehyde-3-phosphate dehydrogenase | |
| Synonyms | AHO|C20orf45|GNAS1|GPSA|GSA|GSP|NESP|PITA3|POH|SCG6|SgVI | G3PD|GAPD|HEL-S-162eP | |
| Cytomap | 20q13.32 | 12p13.31 | |
| Type of gene | protein-coding | protein-coding | |
| Description | protein ALEXprotein GNASprotein SCG6 (secretogranin VI)G protein subunit alpha Sadenylate cyclase-stimulating G alpha proteinalternative gene product encoded by XL-exonextra large alphas proteinguanine nucleotide binding protein (G protein), alpha | glyceraldehyde-3-phosphate dehydrogenaseOCAS, p38 componentOct1 coactivator in S phase, 38 Kd componentaging-associated gene 9 proteinepididymis secretory sperm binding protein Li 162ePpeptidyl-cysteine S-nitrosylase GAPDH | |
| Modification date | 20200329 | 20200327 | |
| UniProtAcc | P63092 | P04406 | |
| Ensembl transtripts involved in fusion gene | ENST00000313949, ENST00000371075, ENST00000464624, ENST00000265620, ENST00000306090, ENST00000354359, ENST00000371085, ENST00000371095, ENST00000371100, ENST00000371102, ENST00000306120, ENST00000371081, ENST00000371098, ENST00000371099, | ENST00000229239, ENST00000396856, ENST00000396858, ENST00000396859, ENST00000396861, | |
| Fusion gene scores | * DoF score | 44 X 25 X 16=17600 | 27 X 29 X 8=6264 |
| # samples | 51 | 31 | |
| ** MAII score | log2(51/17600*10)=-5.10893437155316 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(31/6264*10)=-4.3367440920168 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: GNAS [Title/Abstract] AND GAPDH [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | GNAS(57478846)-GAPDH(6647267), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | GNAS-GAPDH seems lost the major protein functional domain in Hgene partner, which is a cell metabolism gene due to the frame-shifted ORF. GNAS-GAPDH seems lost the major protein functional domain in Hgene partner, which is a CGC due to the frame-shifted ORF. GNAS-GAPDH seems lost the major protein functional domain in Tgene partner, which is a cell metabolism gene due to the frame-shifted ORF. GNAS-GAPDH seems lost the major protein functional domain in Tgene partner, which is a essential gene due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Tgene | GAPDH | GO:0010951 | negative regulation of endopeptidase activity | 22832495 |
| Tgene | GAPDH | GO:0017148 | negative regulation of translation | 23071094 |
| Tgene | GAPDH | GO:0031640 | killing of cells of other organism | 22832495 |
| Tgene | GAPDH | GO:0050715 | positive regulation of cytokine secretion | 22832495 |
| Tgene | GAPDH | GO:0050832 | defense response to fungus | 22832495 |
| Tgene | GAPDH | GO:0051873 | killing by host of symbiont cells | 22832495 |
| Tgene | GAPDH | GO:0052501 | positive regulation by organism of apoptotic process in other organism involved in symbiotic interaction | 22832495 |
| Tgene | GAPDH | GO:0061844 | antimicrobial humoral immune response mediated by antimicrobial peptide | 22832495 |
| Tgene | GAPDH | GO:0071346 | cellular response to interferon-gamma | 15479637 |
 Fusion gene breakpoints across GNAS (5'-gene) Fusion gene breakpoints across GNAS (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
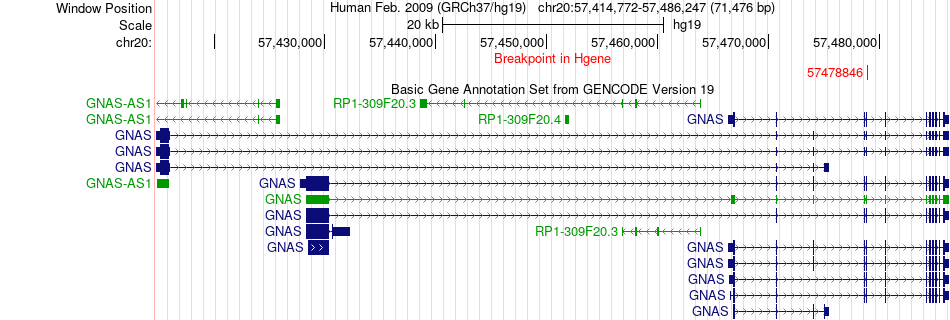 |
 Fusion gene breakpoints across GAPDH (3'-gene) Fusion gene breakpoints across GAPDH (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
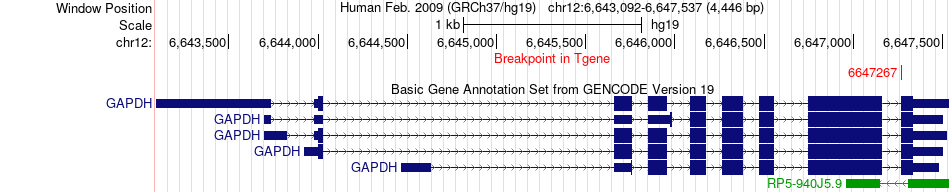 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | CESC | TCGA-IR-A3LI-01A | GNAS | chr20 | 57478846 | + | GAPDH | chr12 | 6647267 | + |
Top |
Fusion Gene ORF analysis for GNAS-GAPDH |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 3UTR-3CDS | ENST00000313949 | ENST00000229239 | GNAS | chr20 | 57478846 | + | GAPDH | chr12 | 6647267 | + |
| 3UTR-3CDS | ENST00000313949 | ENST00000396856 | GNAS | chr20 | 57478846 | + | GAPDH | chr12 | 6647267 | + |
| 3UTR-3CDS | ENST00000313949 | ENST00000396858 | GNAS | chr20 | 57478846 | + | GAPDH | chr12 | 6647267 | + |
| 3UTR-3CDS | ENST00000313949 | ENST00000396859 | GNAS | chr20 | 57478846 | + | GAPDH | chr12 | 6647267 | + |
| 3UTR-3CDS | ENST00000313949 | ENST00000396861 | GNAS | chr20 | 57478846 | + | GAPDH | chr12 | 6647267 | + |
| 3UTR-3CDS | ENST00000371075 | ENST00000229239 | GNAS | chr20 | 57478846 | + | GAPDH | chr12 | 6647267 | + |
| 3UTR-3CDS | ENST00000371075 | ENST00000396856 | GNAS | chr20 | 57478846 | + | GAPDH | chr12 | 6647267 | + |
| 3UTR-3CDS | ENST00000371075 | ENST00000396858 | GNAS | chr20 | 57478846 | + | GAPDH | chr12 | 6647267 | + |
| 3UTR-3CDS | ENST00000371075 | ENST00000396859 | GNAS | chr20 | 57478846 | + | GAPDH | chr12 | 6647267 | + |
| 3UTR-3CDS | ENST00000371075 | ENST00000396861 | GNAS | chr20 | 57478846 | + | GAPDH | chr12 | 6647267 | + |
| 3UTR-3CDS | ENST00000464624 | ENST00000229239 | GNAS | chr20 | 57478846 | + | GAPDH | chr12 | 6647267 | + |
| 3UTR-3CDS | ENST00000464624 | ENST00000396856 | GNAS | chr20 | 57478846 | + | GAPDH | chr12 | 6647267 | + |
| 3UTR-3CDS | ENST00000464624 | ENST00000396858 | GNAS | chr20 | 57478846 | + | GAPDH | chr12 | 6647267 | + |
| 3UTR-3CDS | ENST00000464624 | ENST00000396859 | GNAS | chr20 | 57478846 | + | GAPDH | chr12 | 6647267 | + |
| 3UTR-3CDS | ENST00000464624 | ENST00000396861 | GNAS | chr20 | 57478846 | + | GAPDH | chr12 | 6647267 | + |
| Frame-shift | ENST00000265620 | ENST00000396856 | GNAS | chr20 | 57478846 | + | GAPDH | chr12 | 6647267 | + |
| Frame-shift | ENST00000265620 | ENST00000396858 | GNAS | chr20 | 57478846 | + | GAPDH | chr12 | 6647267 | + |
| Frame-shift | ENST00000265620 | ENST00000396859 | GNAS | chr20 | 57478846 | + | GAPDH | chr12 | 6647267 | + |
| Frame-shift | ENST00000306090 | ENST00000396856 | GNAS | chr20 | 57478846 | + | GAPDH | chr12 | 6647267 | + |
| Frame-shift | ENST00000306090 | ENST00000396858 | GNAS | chr20 | 57478846 | + | GAPDH | chr12 | 6647267 | + |
| Frame-shift | ENST00000306090 | ENST00000396859 | GNAS | chr20 | 57478846 | + | GAPDH | chr12 | 6647267 | + |
| Frame-shift | ENST00000354359 | ENST00000396856 | GNAS | chr20 | 57478846 | + | GAPDH | chr12 | 6647267 | + |
| Frame-shift | ENST00000354359 | ENST00000396858 | GNAS | chr20 | 57478846 | + | GAPDH | chr12 | 6647267 | + |
| Frame-shift | ENST00000354359 | ENST00000396859 | GNAS | chr20 | 57478846 | + | GAPDH | chr12 | 6647267 | + |
| Frame-shift | ENST00000371085 | ENST00000396856 | GNAS | chr20 | 57478846 | + | GAPDH | chr12 | 6647267 | + |
| Frame-shift | ENST00000371085 | ENST00000396858 | GNAS | chr20 | 57478846 | + | GAPDH | chr12 | 6647267 | + |
| Frame-shift | ENST00000371085 | ENST00000396859 | GNAS | chr20 | 57478846 | + | GAPDH | chr12 | 6647267 | + |
| Frame-shift | ENST00000371095 | ENST00000396856 | GNAS | chr20 | 57478846 | + | GAPDH | chr12 | 6647267 | + |
| Frame-shift | ENST00000371095 | ENST00000396858 | GNAS | chr20 | 57478846 | + | GAPDH | chr12 | 6647267 | + |
| Frame-shift | ENST00000371095 | ENST00000396859 | GNAS | chr20 | 57478846 | + | GAPDH | chr12 | 6647267 | + |
| Frame-shift | ENST00000371100 | ENST00000396856 | GNAS | chr20 | 57478846 | + | GAPDH | chr12 | 6647267 | + |
| Frame-shift | ENST00000371100 | ENST00000396858 | GNAS | chr20 | 57478846 | + | GAPDH | chr12 | 6647267 | + |
| Frame-shift | ENST00000371100 | ENST00000396859 | GNAS | chr20 | 57478846 | + | GAPDH | chr12 | 6647267 | + |
| Frame-shift | ENST00000371102 | ENST00000396856 | GNAS | chr20 | 57478846 | + | GAPDH | chr12 | 6647267 | + |
| Frame-shift | ENST00000371102 | ENST00000396858 | GNAS | chr20 | 57478846 | + | GAPDH | chr12 | 6647267 | + |
| Frame-shift | ENST00000371102 | ENST00000396859 | GNAS | chr20 | 57478846 | + | GAPDH | chr12 | 6647267 | + |
| In-frame | ENST00000265620 | ENST00000229239 | GNAS | chr20 | 57478846 | + | GAPDH | chr12 | 6647267 | + |
| In-frame | ENST00000265620 | ENST00000396861 | GNAS | chr20 | 57478846 | + | GAPDH | chr12 | 6647267 | + |
| In-frame | ENST00000306090 | ENST00000229239 | GNAS | chr20 | 57478846 | + | GAPDH | chr12 | 6647267 | + |
| In-frame | ENST00000306090 | ENST00000396861 | GNAS | chr20 | 57478846 | + | GAPDH | chr12 | 6647267 | + |
| In-frame | ENST00000354359 | ENST00000229239 | GNAS | chr20 | 57478846 | + | GAPDH | chr12 | 6647267 | + |
| In-frame | ENST00000354359 | ENST00000396861 | GNAS | chr20 | 57478846 | + | GAPDH | chr12 | 6647267 | + |
| In-frame | ENST00000371085 | ENST00000229239 | GNAS | chr20 | 57478846 | + | GAPDH | chr12 | 6647267 | + |
| In-frame | ENST00000371085 | ENST00000396861 | GNAS | chr20 | 57478846 | + | GAPDH | chr12 | 6647267 | + |
| In-frame | ENST00000371095 | ENST00000229239 | GNAS | chr20 | 57478846 | + | GAPDH | chr12 | 6647267 | + |
| In-frame | ENST00000371095 | ENST00000396861 | GNAS | chr20 | 57478846 | + | GAPDH | chr12 | 6647267 | + |
| In-frame | ENST00000371100 | ENST00000229239 | GNAS | chr20 | 57478846 | + | GAPDH | chr12 | 6647267 | + |
| In-frame | ENST00000371100 | ENST00000396861 | GNAS | chr20 | 57478846 | + | GAPDH | chr12 | 6647267 | + |
| In-frame | ENST00000371102 | ENST00000229239 | GNAS | chr20 | 57478846 | + | GAPDH | chr12 | 6647267 | + |
| In-frame | ENST00000371102 | ENST00000396861 | GNAS | chr20 | 57478846 | + | GAPDH | chr12 | 6647267 | + |
| intron-3CDS | ENST00000306120 | ENST00000229239 | GNAS | chr20 | 57478846 | + | GAPDH | chr12 | 6647267 | + |
| intron-3CDS | ENST00000306120 | ENST00000396856 | GNAS | chr20 | 57478846 | + | GAPDH | chr12 | 6647267 | + |
| intron-3CDS | ENST00000306120 | ENST00000396858 | GNAS | chr20 | 57478846 | + | GAPDH | chr12 | 6647267 | + |
| intron-3CDS | ENST00000306120 | ENST00000396859 | GNAS | chr20 | 57478846 | + | GAPDH | chr12 | 6647267 | + |
| intron-3CDS | ENST00000306120 | ENST00000396861 | GNAS | chr20 | 57478846 | + | GAPDH | chr12 | 6647267 | + |
| intron-3CDS | ENST00000371081 | ENST00000229239 | GNAS | chr20 | 57478846 | + | GAPDH | chr12 | 6647267 | + |
| intron-3CDS | ENST00000371081 | ENST00000396856 | GNAS | chr20 | 57478846 | + | GAPDH | chr12 | 6647267 | + |
| intron-3CDS | ENST00000371081 | ENST00000396858 | GNAS | chr20 | 57478846 | + | GAPDH | chr12 | 6647267 | + |
| intron-3CDS | ENST00000371081 | ENST00000396859 | GNAS | chr20 | 57478846 | + | GAPDH | chr12 | 6647267 | + |
| intron-3CDS | ENST00000371081 | ENST00000396861 | GNAS | chr20 | 57478846 | + | GAPDH | chr12 | 6647267 | + |
| intron-3CDS | ENST00000371098 | ENST00000229239 | GNAS | chr20 | 57478846 | + | GAPDH | chr12 | 6647267 | + |
| intron-3CDS | ENST00000371098 | ENST00000396856 | GNAS | chr20 | 57478846 | + | GAPDH | chr12 | 6647267 | + |
| intron-3CDS | ENST00000371098 | ENST00000396858 | GNAS | chr20 | 57478846 | + | GAPDH | chr12 | 6647267 | + |
| intron-3CDS | ENST00000371098 | ENST00000396859 | GNAS | chr20 | 57478846 | + | GAPDH | chr12 | 6647267 | + |
| intron-3CDS | ENST00000371098 | ENST00000396861 | GNAS | chr20 | 57478846 | + | GAPDH | chr12 | 6647267 | + |
| intron-3CDS | ENST00000371099 | ENST00000229239 | GNAS | chr20 | 57478846 | + | GAPDH | chr12 | 6647267 | + |
| intron-3CDS | ENST00000371099 | ENST00000396856 | GNAS | chr20 | 57478846 | + | GAPDH | chr12 | 6647267 | + |
| intron-3CDS | ENST00000371099 | ENST00000396858 | GNAS | chr20 | 57478846 | + | GAPDH | chr12 | 6647267 | + |
| intron-3CDS | ENST00000371099 | ENST00000396859 | GNAS | chr20 | 57478846 | + | GAPDH | chr12 | 6647267 | + |
| intron-3CDS | ENST00000371099 | ENST00000396861 | GNAS | chr20 | 57478846 | + | GAPDH | chr12 | 6647267 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000371100 | GNAS | chr20 | 57478846 | + | ENST00000229239 | GAPDH | chr12 | 6647267 | + | 3184 | 2913 | 531 | 2918 | 795 |
| ENST00000371100 | GNAS | chr20 | 57478846 | + | ENST00000396861 | GAPDH | chr12 | 6647267 | + | 3172 | 2913 | 531 | 2918 | 795 |
| ENST00000371102 | GNAS | chr20 | 57478846 | + | ENST00000229239 | GAPDH | chr12 | 6647267 | + | 2593 | 2322 | 3 | 2327 | 774 |
| ENST00000371102 | GNAS | chr20 | 57478846 | + | ENST00000396861 | GAPDH | chr12 | 6647267 | + | 2581 | 2322 | 3 | 2327 | 774 |
| ENST00000371095 | GNAS | chr20 | 57478846 | + | ENST00000229239 | GAPDH | chr12 | 6647267 | + | 1086 | 815 | 823 | 2 | 274 |
| ENST00000371095 | GNAS | chr20 | 57478846 | + | ENST00000396861 | GAPDH | chr12 | 6647267 | + | 1074 | 815 | 823 | 2 | 274 |
| ENST00000371085 | GNAS | chr20 | 57478846 | + | ENST00000229239 | GAPDH | chr12 | 6647267 | + | 1127 | 856 | 864 | 1 | 288 |
| ENST00000371085 | GNAS | chr20 | 57478846 | + | ENST00000396861 | GAPDH | chr12 | 6647267 | + | 1115 | 856 | 864 | 1 | 288 |
| ENST00000354359 | GNAS | chr20 | 57478846 | + | ENST00000229239 | GAPDH | chr12 | 6647267 | + | 1130 | 859 | 867 | 1 | 289 |
| ENST00000354359 | GNAS | chr20 | 57478846 | + | ENST00000396861 | GAPDH | chr12 | 6647267 | + | 1118 | 859 | 867 | 1 | 289 |
| ENST00000265620 | GNAS | chr20 | 57478846 | + | ENST00000229239 | GAPDH | chr12 | 6647267 | + | 1021 | 750 | 758 | 0 | 253 |
| ENST00000265620 | GNAS | chr20 | 57478846 | + | ENST00000396861 | GAPDH | chr12 | 6647267 | + | 1009 | 750 | 758 | 0 | 253 |
| ENST00000306090 | GNAS | chr20 | 57478846 | + | ENST00000229239 | GAPDH | chr12 | 6647267 | + | 803 | 532 | 540 | 1 | 180 |
| ENST00000306090 | GNAS | chr20 | 57478846 | + | ENST00000396861 | GAPDH | chr12 | 6647267 | + | 791 | 532 | 540 | 1 | 180 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000371100 | ENST00000229239 | GNAS | chr20 | 57478846 | + | GAPDH | chr12 | 6647267 | + | 0.01068128 | 0.9893188 |
| ENST00000371100 | ENST00000396861 | GNAS | chr20 | 57478846 | + | GAPDH | chr12 | 6647267 | + | 0.010635193 | 0.98936486 |
| ENST00000371102 | ENST00000229239 | GNAS | chr20 | 57478846 | + | GAPDH | chr12 | 6647267 | + | 0.016691785 | 0.9833082 |
| ENST00000371102 | ENST00000396861 | GNAS | chr20 | 57478846 | + | GAPDH | chr12 | 6647267 | + | 0.016581496 | 0.9834185 |
| ENST00000371095 | ENST00000229239 | GNAS | chr20 | 57478846 | + | GAPDH | chr12 | 6647267 | + | 0.008682908 | 0.99131715 |
| ENST00000371095 | ENST00000396861 | GNAS | chr20 | 57478846 | + | GAPDH | chr12 | 6647267 | + | 0.008667513 | 0.9913325 |
| ENST00000371085 | ENST00000229239 | GNAS | chr20 | 57478846 | + | GAPDH | chr12 | 6647267 | + | 0.005251807 | 0.9947482 |
| ENST00000371085 | ENST00000396861 | GNAS | chr20 | 57478846 | + | GAPDH | chr12 | 6647267 | + | 0.005287748 | 0.99471223 |
| ENST00000354359 | ENST00000229239 | GNAS | chr20 | 57478846 | + | GAPDH | chr12 | 6647267 | + | 0.004871563 | 0.9951284 |
| ENST00000354359 | ENST00000396861 | GNAS | chr20 | 57478846 | + | GAPDH | chr12 | 6647267 | + | 0.005117199 | 0.9948828 |
| ENST00000265620 | ENST00000229239 | GNAS | chr20 | 57478846 | + | GAPDH | chr12 | 6647267 | + | 0.008899255 | 0.9911007 |
| ENST00000265620 | ENST00000396861 | GNAS | chr20 | 57478846 | + | GAPDH | chr12 | 6647267 | + | 0.009380726 | 0.9906193 |
| ENST00000306090 | ENST00000229239 | GNAS | chr20 | 57478846 | + | GAPDH | chr12 | 6647267 | + | 0.010252884 | 0.98974717 |
| ENST00000306090 | ENST00000396861 | GNAS | chr20 | 57478846 | + | GAPDH | chr12 | 6647267 | + | 0.010419838 | 0.9895802 |
Top |
Fusion Genomic Features for GNAS-GAPDH |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| GNAS | chr20 | 57478846 | + | GAPDH | chr12 | 6647266 | + | 0.000806652 | 0.9991934 |
| GNAS | chr20 | 57478846 | + | GAPDH | chr12 | 6647266 | + | 0.000806652 | 0.9991934 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
 |
Top |
Fusion Protein Features for GNAS-GAPDH |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr20:57478846/chr12:6647267) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| GNAS | GAPDH |
| FUNCTION: Guanine nucleotide-binding proteins (G proteins) function as transducers in numerous signaling pathways controlled by G protein-coupled receptors (GPCRs) (PubMed:17110384). Signaling involves the activation of adenylyl cyclases, resulting in increased levels of the signaling molecule cAMP (PubMed:26206488, PubMed:8702665). GNAS functions downstream of several GPCRs, including beta-adrenergic receptors (PubMed:21488135). Stimulates the Ras signaling pathway via RAPGEF2 (PubMed:12391161). {ECO:0000269|PubMed:12391161, ECO:0000269|PubMed:17110384, ECO:0000269|PubMed:21488135, ECO:0000269|PubMed:26206488, ECO:0000269|PubMed:8702665}. | FUNCTION: Has both glyceraldehyde-3-phosphate dehydrogenase and nitrosylase activities, thereby playing a role in glycolysis and nuclear functions, respectively. Participates in nuclear events including transcription, RNA transport, DNA replication and apoptosis. Nuclear functions are probably due to the nitrosylase activity that mediates cysteine S-nitrosylation of nuclear target proteins such as SIRT1, HDAC2 and PRKDC. Modulates the organization and assembly of the cytoskeleton. Facilitates the CHP1-dependent microtubule and membrane associations through its ability to stimulate the binding of CHP1 to microtubules (By similarity). Glyceraldehyde-3-phosphate dehydrogenase is a key enzyme in glycolysis that catalyzes the first step of the pathway by converting D-glyceraldehyde 3-phosphate (G3P) into 3-phospho-D-glyceroyl phosphate. Component of the GAIT (gamma interferon-activated inhibitor of translation) complex which mediates interferon-gamma-induced transcript-selective translation inhibition in inflammation processes. Upon interferon-gamma treatment assembles into the GAIT complex which binds to stem loop-containing GAIT elements in the 3'-UTR of diverse inflammatory mRNAs (such as ceruplasmin) and suppresses their translation. {ECO:0000250, ECO:0000269|PubMed:11724794, ECO:0000269|PubMed:23071094, ECO:0000269|PubMed:3170585}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | GNAS | chr20:57478846 | chr12:6647267 | ENST00000371100 | + | 5 | 13 | 641_667 | 787 | 1038.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | GNAS | chr20:57478846 | chr12:6647267 | ENST00000371100 | + | 5 | 13 | 730_756 | 787 | 1038.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | GNAS | chr20:57478846 | chr12:6647267 | ENST00000371102 | + | 4 | 12 | 641_667 | 773 | 1024.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | GNAS | chr20:57478846 | chr12:6647267 | ENST00000371102 | + | 4 | 12 | 730_756 | 773 | 1024.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | GNAS | chr20:57478846 | chr12:6647267 | ENST00000313949 | + | 5 | 13 | 78_142 | 357 | 26.333333333333332 | Compositional bias | Glu-rich |
| Hgene | GNAS | chr20:57478846 | chr12:6647267 | ENST00000371075 | + | 5 | 13 | 78_142 | 358 | 36.333333333333336 | Compositional bias | Glu-rich |
| Hgene | GNAS | chr20:57478846 | chr12:6647267 | ENST00000371100 | + | 5 | 13 | 358_522 | 787 | 1038.0 | Compositional bias | Ala-rich |
| Hgene | GNAS | chr20:57478846 | chr12:6647267 | ENST00000371102 | + | 4 | 12 | 358_522 | 773 | 1024.0 | Compositional bias | Ala-rich |
| Hgene | GNAS | chr20:57478846 | chr12:6647267 | ENST00000265620 | + | 4 | 12 | 47_55 | 129 | 380.0 | Nucleotide binding | GTP |
| Hgene | GNAS | chr20:57478846 | chr12:6647267 | ENST00000306090 | + | 5 | 13 | 47_55 | 130 | 381.0 | Nucleotide binding | GTP |
| Hgene | GNAS | chr20:57478846 | chr12:6647267 | ENST00000354359 | + | 5 | 13 | 47_55 | 145 | 396.0 | Nucleotide binding | GTP |
| Hgene | GNAS | chr20:57478846 | chr12:6647267 | ENST00000371085 | + | 5 | 13 | 47_55 | 144 | 395.0 | Nucleotide binding | GTP |
| Hgene | GNAS | chr20:57478846 | chr12:6647267 | ENST00000371095 | + | 4 | 12 | 47_55 | 130 | 381.0 | Nucleotide binding | GTP |
| Hgene | GNAS | chr20:57478846 | chr12:6647267 | ENST00000371100 | + | 5 | 13 | 690_698 | 787 | 1038.0 | Nucleotide binding | GTP |
| Hgene | GNAS | chr20:57478846 | chr12:6647267 | ENST00000371102 | + | 4 | 12 | 690_698 | 773 | 1024.0 | Nucleotide binding | GTP |
| Hgene | GNAS | chr20:57478846 | chr12:6647267 | ENST00000265620 | + | 4 | 12 | 42_55 | 129 | 380.0 | Region | G1 motif |
| Hgene | GNAS | chr20:57478846 | chr12:6647267 | ENST00000306090 | + | 5 | 13 | 42_55 | 130 | 381.0 | Region | G1 motif |
| Hgene | GNAS | chr20:57478846 | chr12:6647267 | ENST00000354359 | + | 5 | 13 | 42_55 | 145 | 396.0 | Region | G1 motif |
| Hgene | GNAS | chr20:57478846 | chr12:6647267 | ENST00000371085 | + | 5 | 13 | 42_55 | 144 | 395.0 | Region | G1 motif |
| Hgene | GNAS | chr20:57478846 | chr12:6647267 | ENST00000371095 | + | 4 | 12 | 42_55 | 130 | 381.0 | Region | G1 motif |
| Hgene | GNAS | chr20:57478846 | chr12:6647267 | ENST00000371100 | + | 5 | 13 | 685_698 | 787 | 1038.0 | Region | G1 motif |
| Hgene | GNAS | chr20:57478846 | chr12:6647267 | ENST00000371102 | + | 4 | 12 | 685_698 | 773 | 1024.0 | Region | G1 motif |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | GNAS | chr20:57478846 | chr12:6647267 | ENST00000306120 | + | 1 | 1 | 238_476 | 0 | 626.0 | Compositional bias | Pro-rich |
| Hgene | GNAS | chr20:57478846 | chr12:6647267 | ENST00000371098 | + | 1 | 4 | 78_142 | 0 | 20.333333333333332 | Compositional bias | Glu-rich |
| Hgene | GNAS | chr20:57478846 | chr12:6647267 | ENST00000265620 | + | 4 | 12 | 39_394 | 129 | 380.0 | Domain | G-alpha |
| Hgene | GNAS | chr20:57478846 | chr12:6647267 | ENST00000306090 | + | 5 | 13 | 39_394 | 130 | 381.0 | Domain | G-alpha |
| Hgene | GNAS | chr20:57478846 | chr12:6647267 | ENST00000354359 | + | 5 | 13 | 39_394 | 145 | 396.0 | Domain | G-alpha |
| Hgene | GNAS | chr20:57478846 | chr12:6647267 | ENST00000371085 | + | 5 | 13 | 39_394 | 144 | 395.0 | Domain | G-alpha |
| Hgene | GNAS | chr20:57478846 | chr12:6647267 | ENST00000371095 | + | 4 | 12 | 39_394 | 130 | 381.0 | Domain | G-alpha |
| Hgene | GNAS | chr20:57478846 | chr12:6647267 | ENST00000371100 | + | 5 | 13 | 682_1037 | 787 | 1038.0 | Domain | G-alpha |
| Hgene | GNAS | chr20:57478846 | chr12:6647267 | ENST00000371102 | + | 4 | 12 | 682_1037 | 773 | 1024.0 | Domain | G-alpha |
| Hgene | GNAS | chr20:57478846 | chr12:6647267 | ENST00000265620 | + | 4 | 12 | 197_204 | 129 | 380.0 | Nucleotide binding | GTP |
| Hgene | GNAS | chr20:57478846 | chr12:6647267 | ENST00000265620 | + | 4 | 12 | 223_227 | 129 | 380.0 | Nucleotide binding | GTP |
| Hgene | GNAS | chr20:57478846 | chr12:6647267 | ENST00000265620 | + | 4 | 12 | 292_295 | 129 | 380.0 | Nucleotide binding | GTP |
| Hgene | GNAS | chr20:57478846 | chr12:6647267 | ENST00000306090 | + | 5 | 13 | 197_204 | 130 | 381.0 | Nucleotide binding | GTP |
| Hgene | GNAS | chr20:57478846 | chr12:6647267 | ENST00000306090 | + | 5 | 13 | 223_227 | 130 | 381.0 | Nucleotide binding | GTP |
| Hgene | GNAS | chr20:57478846 | chr12:6647267 | ENST00000306090 | + | 5 | 13 | 292_295 | 130 | 381.0 | Nucleotide binding | GTP |
| Hgene | GNAS | chr20:57478846 | chr12:6647267 | ENST00000354359 | + | 5 | 13 | 197_204 | 145 | 396.0 | Nucleotide binding | GTP |
| Hgene | GNAS | chr20:57478846 | chr12:6647267 | ENST00000354359 | + | 5 | 13 | 223_227 | 145 | 396.0 | Nucleotide binding | GTP |
| Hgene | GNAS | chr20:57478846 | chr12:6647267 | ENST00000354359 | + | 5 | 13 | 292_295 | 145 | 396.0 | Nucleotide binding | GTP |
| Hgene | GNAS | chr20:57478846 | chr12:6647267 | ENST00000371085 | + | 5 | 13 | 197_204 | 144 | 395.0 | Nucleotide binding | GTP |
| Hgene | GNAS | chr20:57478846 | chr12:6647267 | ENST00000371085 | + | 5 | 13 | 223_227 | 144 | 395.0 | Nucleotide binding | GTP |
| Hgene | GNAS | chr20:57478846 | chr12:6647267 | ENST00000371085 | + | 5 | 13 | 292_295 | 144 | 395.0 | Nucleotide binding | GTP |
| Hgene | GNAS | chr20:57478846 | chr12:6647267 | ENST00000371095 | + | 4 | 12 | 197_204 | 130 | 381.0 | Nucleotide binding | GTP |
| Hgene | GNAS | chr20:57478846 | chr12:6647267 | ENST00000371095 | + | 4 | 12 | 223_227 | 130 | 381.0 | Nucleotide binding | GTP |
| Hgene | GNAS | chr20:57478846 | chr12:6647267 | ENST00000371095 | + | 4 | 12 | 292_295 | 130 | 381.0 | Nucleotide binding | GTP |
| Hgene | GNAS | chr20:57478846 | chr12:6647267 | ENST00000371100 | + | 5 | 13 | 840_847 | 787 | 1038.0 | Nucleotide binding | GTP |
| Hgene | GNAS | chr20:57478846 | chr12:6647267 | ENST00000371100 | + | 5 | 13 | 866_870 | 787 | 1038.0 | Nucleotide binding | GTP |
| Hgene | GNAS | chr20:57478846 | chr12:6647267 | ENST00000371100 | + | 5 | 13 | 935_938 | 787 | 1038.0 | Nucleotide binding | GTP |
| Hgene | GNAS | chr20:57478846 | chr12:6647267 | ENST00000371102 | + | 4 | 12 | 840_847 | 773 | 1024.0 | Nucleotide binding | GTP |
| Hgene | GNAS | chr20:57478846 | chr12:6647267 | ENST00000371102 | + | 4 | 12 | 866_870 | 773 | 1024.0 | Nucleotide binding | GTP |
| Hgene | GNAS | chr20:57478846 | chr12:6647267 | ENST00000371102 | + | 4 | 12 | 935_938 | 773 | 1024.0 | Nucleotide binding | GTP |
| Hgene | GNAS | chr20:57478846 | chr12:6647267 | ENST00000265620 | + | 4 | 12 | 196_204 | 129 | 380.0 | Region | G2 motif |
| Hgene | GNAS | chr20:57478846 | chr12:6647267 | ENST00000265620 | + | 4 | 12 | 219_228 | 129 | 380.0 | Region | G3 motif |
| Hgene | GNAS | chr20:57478846 | chr12:6647267 | ENST00000265620 | + | 4 | 12 | 288_295 | 129 | 380.0 | Region | G4 motif |
| Hgene | GNAS | chr20:57478846 | chr12:6647267 | ENST00000265620 | + | 4 | 12 | 364_369 | 129 | 380.0 | Region | G5 motif |
| Hgene | GNAS | chr20:57478846 | chr12:6647267 | ENST00000306090 | + | 5 | 13 | 196_204 | 130 | 381.0 | Region | G2 motif |
| Hgene | GNAS | chr20:57478846 | chr12:6647267 | ENST00000306090 | + | 5 | 13 | 219_228 | 130 | 381.0 | Region | G3 motif |
| Hgene | GNAS | chr20:57478846 | chr12:6647267 | ENST00000306090 | + | 5 | 13 | 288_295 | 130 | 381.0 | Region | G4 motif |
| Hgene | GNAS | chr20:57478846 | chr12:6647267 | ENST00000306090 | + | 5 | 13 | 364_369 | 130 | 381.0 | Region | G5 motif |
| Hgene | GNAS | chr20:57478846 | chr12:6647267 | ENST00000354359 | + | 5 | 13 | 196_204 | 145 | 396.0 | Region | G2 motif |
| Hgene | GNAS | chr20:57478846 | chr12:6647267 | ENST00000354359 | + | 5 | 13 | 219_228 | 145 | 396.0 | Region | G3 motif |
| Hgene | GNAS | chr20:57478846 | chr12:6647267 | ENST00000354359 | + | 5 | 13 | 288_295 | 145 | 396.0 | Region | G4 motif |
| Hgene | GNAS | chr20:57478846 | chr12:6647267 | ENST00000354359 | + | 5 | 13 | 364_369 | 145 | 396.0 | Region | G5 motif |
| Hgene | GNAS | chr20:57478846 | chr12:6647267 | ENST00000371085 | + | 5 | 13 | 196_204 | 144 | 395.0 | Region | G2 motif |
| Hgene | GNAS | chr20:57478846 | chr12:6647267 | ENST00000371085 | + | 5 | 13 | 219_228 | 144 | 395.0 | Region | G3 motif |
| Hgene | GNAS | chr20:57478846 | chr12:6647267 | ENST00000371085 | + | 5 | 13 | 288_295 | 144 | 395.0 | Region | G4 motif |
| Hgene | GNAS | chr20:57478846 | chr12:6647267 | ENST00000371085 | + | 5 | 13 | 364_369 | 144 | 395.0 | Region | G5 motif |
| Hgene | GNAS | chr20:57478846 | chr12:6647267 | ENST00000371095 | + | 4 | 12 | 196_204 | 130 | 381.0 | Region | G2 motif |
| Hgene | GNAS | chr20:57478846 | chr12:6647267 | ENST00000371095 | + | 4 | 12 | 219_228 | 130 | 381.0 | Region | G3 motif |
| Hgene | GNAS | chr20:57478846 | chr12:6647267 | ENST00000371095 | + | 4 | 12 | 288_295 | 130 | 381.0 | Region | G4 motif |
| Hgene | GNAS | chr20:57478846 | chr12:6647267 | ENST00000371095 | + | 4 | 12 | 364_369 | 130 | 381.0 | Region | G5 motif |
| Hgene | GNAS | chr20:57478846 | chr12:6647267 | ENST00000371100 | + | 5 | 13 | 1007_1012 | 787 | 1038.0 | Region | G5 motif |
| Hgene | GNAS | chr20:57478846 | chr12:6647267 | ENST00000371100 | + | 5 | 13 | 839_847 | 787 | 1038.0 | Region | G2 motif |
| Hgene | GNAS | chr20:57478846 | chr12:6647267 | ENST00000371100 | + | 5 | 13 | 862_871 | 787 | 1038.0 | Region | G3 motif |
| Hgene | GNAS | chr20:57478846 | chr12:6647267 | ENST00000371100 | + | 5 | 13 | 931_938 | 787 | 1038.0 | Region | G4 motif |
| Hgene | GNAS | chr20:57478846 | chr12:6647267 | ENST00000371102 | + | 4 | 12 | 1007_1012 | 773 | 1024.0 | Region | G5 motif |
| Hgene | GNAS | chr20:57478846 | chr12:6647267 | ENST00000371102 | + | 4 | 12 | 839_847 | 773 | 1024.0 | Region | G2 motif |
| Hgene | GNAS | chr20:57478846 | chr12:6647267 | ENST00000371102 | + | 4 | 12 | 862_871 | 773 | 1024.0 | Region | G3 motif |
| Hgene | GNAS | chr20:57478846 | chr12:6647267 | ENST00000371102 | + | 4 | 12 | 931_938 | 773 | 1024.0 | Region | G4 motif |
| Tgene | GAPDH | chr20:57478846 | chr12:6647267 | ENST00000229239 | 7 | 9 | 245_250 | 312 | 336.0 | Motif | [IL]-x-C-x-x-[DE] motif | |
| Tgene | GAPDH | chr20:57478846 | chr12:6647267 | ENST00000396858 | 6 | 8 | 245_250 | 270 | 294.0 | Motif | [IL]-x-C-x-x-[DE] motif | |
| Tgene | GAPDH | chr20:57478846 | chr12:6647267 | ENST00000396859 | 6 | 8 | 245_250 | 312 | 336.0 | Motif | [IL]-x-C-x-x-[DE] motif | |
| Tgene | GAPDH | chr20:57478846 | chr12:6647267 | ENST00000396861 | 7 | 9 | 245_250 | 312 | 336.0 | Motif | [IL]-x-C-x-x-[DE] motif | |
| Tgene | GAPDH | chr20:57478846 | chr12:6647267 | ENST00000229239 | 7 | 9 | 13_14 | 312 | 336.0 | Nucleotide binding | NAD | |
| Tgene | GAPDH | chr20:57478846 | chr12:6647267 | ENST00000396858 | 6 | 8 | 13_14 | 270 | 294.0 | Nucleotide binding | NAD | |
| Tgene | GAPDH | chr20:57478846 | chr12:6647267 | ENST00000396859 | 6 | 8 | 13_14 | 312 | 336.0 | Nucleotide binding | NAD | |
| Tgene | GAPDH | chr20:57478846 | chr12:6647267 | ENST00000396861 | 7 | 9 | 13_14 | 312 | 336.0 | Nucleotide binding | NAD | |
| Tgene | GAPDH | chr20:57478846 | chr12:6647267 | ENST00000229239 | 7 | 9 | 151_153 | 312 | 336.0 | Region | Glyceraldehyde 3-phosphate binding | |
| Tgene | GAPDH | chr20:57478846 | chr12:6647267 | ENST00000229239 | 7 | 9 | 211_212 | 312 | 336.0 | Region | Glyceraldehyde 3-phosphate binding | |
| Tgene | GAPDH | chr20:57478846 | chr12:6647267 | ENST00000396858 | 6 | 8 | 151_153 | 270 | 294.0 | Region | Glyceraldehyde 3-phosphate binding | |
| Tgene | GAPDH | chr20:57478846 | chr12:6647267 | ENST00000396858 | 6 | 8 | 211_212 | 270 | 294.0 | Region | Glyceraldehyde 3-phosphate binding | |
| Tgene | GAPDH | chr20:57478846 | chr12:6647267 | ENST00000396859 | 6 | 8 | 151_153 | 312 | 336.0 | Region | Glyceraldehyde 3-phosphate binding | |
| Tgene | GAPDH | chr20:57478846 | chr12:6647267 | ENST00000396859 | 6 | 8 | 211_212 | 312 | 336.0 | Region | Glyceraldehyde 3-phosphate binding | |
| Tgene | GAPDH | chr20:57478846 | chr12:6647267 | ENST00000396861 | 7 | 9 | 151_153 | 312 | 336.0 | Region | Glyceraldehyde 3-phosphate binding | |
| Tgene | GAPDH | chr20:57478846 | chr12:6647267 | ENST00000396861 | 7 | 9 | 211_212 | 312 | 336.0 | Region | Glyceraldehyde 3-phosphate binding |
Top |
Fusion Gene Sequence for GNAS-GAPDH |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >33624_33624_1_GNAS-GAPDH_GNAS_chr20_57478846_ENST00000265620_GAPDH_chr12_6647267_ENST00000229239_length(transcript)=1021nt_BP=750nt GGGCGGCGGCGGGGGCCCGGCCGAGGCAATAAGAGCGGCGGCGGCGGCAGCGGCGGCAGCAGCTCCCGCAGCTCCTGCTCTGGTCCGCCT CGGCCCGGCGGCGGCCATCAGCCCCCTCGGCCTCGGCTCGAGGGGCGGGGAGCTGCGCGCGCCCCTCGGTCCGACCGACACCCTCCCCTT CCCGCCCGTCCGCGCGCCCCGCGGCCCGCGGCCCGCAGTCCGCCCCGCGCGCTCCTTGCCGAGGAGCCGAGCCCGCGCCCGGCCCGCCCG CCCGGCGCTGCCCCGGCCCTCCCGGCCCGCGTGAGGCCGCCCGCGCCCGCCGCCGCCGCAGCCCGGCCGCGCCCCGCCGCCGCCGCCGCC GCCATGGGCTGCCTCGGGAACAGTAAGACCGAGGACCAGCGCAACGAGGAGAAGGCGCAGCGTGAGGCCAACAAAAAGATCGAGAAGCAG CTGCAGAAGGACAAGCAGGTCTACCGGGCCACGCACCGCCTGCTGCTGCTGGGTGCTGGAGAATCTGGTAAAAGCACCATTGTGAAGCAG ATGAGGATCCTGCATGTTAATGGGTTTAATGGAGATGAGAAGGCAACCAAAGTGCAGGACATCAAAAACAACCTGAAAGAGGCGATTGAA ACCATTGTGGCCGCCATGAGCAACCTGGTGCCCCCCGTGGAGCTGGCCAACCCCGAGAACCAGTTCAGAGTGGACTACATCCTGAGTGTG ATGAACGTGCCTGACTTTGACTTCCCTCCCGTATGACAACGAATTTGGCTACAGCAACAGGGTGGTGGACCTCATGGCCCACATGGCCTC CAAGGAGTAAGACCCCTGGACCACCAGCCCCAGCAAGAGCACAAGAGGAAGAGAGAGACCCTCACTGCTGGGGAGTCCCTGCCACACTCA GTCCCCCACCACACTGAATCTCCCCTCCTCACAGTTGCCATGTAGACCCCTTGAAGAGGGGAGGGGCCTAGGGAGCCGCACCTTGTCATG >33624_33624_1_GNAS-GAPDH_GNAS_chr20_57478846_ENST00000265620_GAPDH_chr12_6647267_ENST00000229239_length(amino acids)=253AA_BP= MSYGREVKVRHVHHTQDVVHSELVLGVGQLHGGHQVAHGGHNGFNRLFQVVFDVLHFGCLLISIKPINMQDPHLLHNGAFTRFSSTQQQQ AVRGPVDLLVLLQLLLDLFVGLTLRLLLVALVLGLTVPEAAHGGGGGGGARPGCGGGGRGRPHAGREGRGSAGRAGRARARLLGKERAGR -------------------------------------------------------------- >33624_33624_2_GNAS-GAPDH_GNAS_chr20_57478846_ENST00000265620_GAPDH_chr12_6647267_ENST00000396861_length(transcript)=1009nt_BP=750nt GGGCGGCGGCGGGGGCCCGGCCGAGGCAATAAGAGCGGCGGCGGCGGCAGCGGCGGCAGCAGCTCCCGCAGCTCCTGCTCTGGTCCGCCT CGGCCCGGCGGCGGCCATCAGCCCCCTCGGCCTCGGCTCGAGGGGCGGGGAGCTGCGCGCGCCCCTCGGTCCGACCGACACCCTCCCCTT CCCGCCCGTCCGCGCGCCCCGCGGCCCGCGGCCCGCAGTCCGCCCCGCGCGCTCCTTGCCGAGGAGCCGAGCCCGCGCCCGGCCCGCCCG CCCGGCGCTGCCCCGGCCCTCCCGGCCCGCGTGAGGCCGCCCGCGCCCGCCGCCGCCGCAGCCCGGCCGCGCCCCGCCGCCGCCGCCGCC GCCATGGGCTGCCTCGGGAACAGTAAGACCGAGGACCAGCGCAACGAGGAGAAGGCGCAGCGTGAGGCCAACAAAAAGATCGAGAAGCAG CTGCAGAAGGACAAGCAGGTCTACCGGGCCACGCACCGCCTGCTGCTGCTGGGTGCTGGAGAATCTGGTAAAAGCACCATTGTGAAGCAG ATGAGGATCCTGCATGTTAATGGGTTTAATGGAGATGAGAAGGCAACCAAAGTGCAGGACATCAAAAACAACCTGAAAGAGGCGATTGAA ACCATTGTGGCCGCCATGAGCAACCTGGTGCCCCCCGTGGAGCTGGCCAACCCCGAGAACCAGTTCAGAGTGGACTACATCCTGAGTGTG ATGAACGTGCCTGACTTTGACTTCCCTCCCGTATGACAACGAATTTGGCTACAGCAACAGGGTGGTGGACCTCATGGCCCACATGGCCTC CAAGGAGTAAGACCCCTGGACCACCAGCCCCAGCAAGAGCACAAGAGGAAGAGAGAGACCCTCACTGCTGGGGAGTCCCTGCCACACTCA GTCCCCCACCACACTGAATCTCCCCTCCTCACAGTTGCCATGTAGACCCCTTGAAGAGGGGAGGGGCCTAGGGAGCCGCACCTTGTCATG >33624_33624_2_GNAS-GAPDH_GNAS_chr20_57478846_ENST00000265620_GAPDH_chr12_6647267_ENST00000396861_length(amino acids)=253AA_BP= MSYGREVKVRHVHHTQDVVHSELVLGVGQLHGGHQVAHGGHNGFNRLFQVVFDVLHFGCLLISIKPINMQDPHLLHNGAFTRFSSTQQQQ AVRGPVDLLVLLQLLLDLFVGLTLRLLLVALVLGLTVPEAAHGGGGGGGARPGCGGGGRGRPHAGREGRGSAGRAGRARARLLGKERAGR -------------------------------------------------------------- >33624_33624_3_GNAS-GAPDH_GNAS_chr20_57478846_ENST00000306090_GAPDH_chr12_6647267_ENST00000229239_length(transcript)=803nt_BP=532nt CAGCGGCGGCAGCAGCTCCCGCAGCTCCTGCTCTGGTCCGCCTCGGCCCGGCGGCGGCCATCAGCCCCCTCGGCCTCGGCTCGAGGGGCG GGGAGCTGCGCGCGCCCCTCGGTCCGACCGACACCCTCCCCTTCCCGCCCCCATGGGCTGCCTCGGGAACAGTAAGACCGAGGACCAGCG CAACGAGGAGAAGGCGCAGCGTGAGGCCAACAAAAAGATCGAGAAGCAGCTGCAGAAGGACAAGCAGGTCTACCGGGCCACGCACCGCCT GCTGCTGCTGGGTGCTGGAGAATCTGGTAAAAGCACCATTGTGAAGCAGATGAGGATCCTGCATGTTAATGGGTTTAATGGAGACAGTGA GAAGGCAACCAAAGTGCAGGACATCAAAAACAACCTGAAAGAGGCGATTGAAACCATTGTGGCCGCCATGAGCAACCTGGTGCCCCCCGT GGAGCTGGCCAACCCCGAGAACCAGTTCAGAGTGGACTACATCCTGAGTGTGATGAACGTGCCTGACTTTGACTTCCCTCCCGTATGACA ACGAATTTGGCTACAGCAACAGGGTGGTGGACCTCATGGCCCACATGGCCTCCAAGGAGTAAGACCCCTGGACCACCAGCCCCAGCAAGA GCACAAGAGGAAGAGAGAGACCCTCACTGCTGGGGAGTCCCTGCCACACTCAGTCCCCCACCACACTGAATCTCCCCTCCTCACAGTTGC >33624_33624_3_GNAS-GAPDH_GNAS_chr20_57478846_ENST00000306090_GAPDH_chr12_6647267_ENST00000229239_length(amino acids)=180AA_BP= MSYGREVKVRHVHHTQDVVHSELVLGVGQLHGGHQVAHGGHNGFNRLFQVVFDVLHFGCLLTVSIKPINMQDPHLLHNGAFTRFSSTQQQ QAVRGPVDLLVLLQLLLDLFVGLTLRLLLVALVLGLTVPEAAHGGGKGRVSVGPRGARSSPPLEPRPRGLMAAAGPRRTRAGAAGAAAAA -------------------------------------------------------------- >33624_33624_4_GNAS-GAPDH_GNAS_chr20_57478846_ENST00000306090_GAPDH_chr12_6647267_ENST00000396861_length(transcript)=791nt_BP=532nt CAGCGGCGGCAGCAGCTCCCGCAGCTCCTGCTCTGGTCCGCCTCGGCCCGGCGGCGGCCATCAGCCCCCTCGGCCTCGGCTCGAGGGGCG GGGAGCTGCGCGCGCCCCTCGGTCCGACCGACACCCTCCCCTTCCCGCCCCCATGGGCTGCCTCGGGAACAGTAAGACCGAGGACCAGCG CAACGAGGAGAAGGCGCAGCGTGAGGCCAACAAAAAGATCGAGAAGCAGCTGCAGAAGGACAAGCAGGTCTACCGGGCCACGCACCGCCT GCTGCTGCTGGGTGCTGGAGAATCTGGTAAAAGCACCATTGTGAAGCAGATGAGGATCCTGCATGTTAATGGGTTTAATGGAGACAGTGA GAAGGCAACCAAAGTGCAGGACATCAAAAACAACCTGAAAGAGGCGATTGAAACCATTGTGGCCGCCATGAGCAACCTGGTGCCCCCCGT GGAGCTGGCCAACCCCGAGAACCAGTTCAGAGTGGACTACATCCTGAGTGTGATGAACGTGCCTGACTTTGACTTCCCTCCCGTATGACA ACGAATTTGGCTACAGCAACAGGGTGGTGGACCTCATGGCCCACATGGCCTCCAAGGAGTAAGACCCCTGGACCACCAGCCCCAGCAAGA GCACAAGAGGAAGAGAGAGACCCTCACTGCTGGGGAGTCCCTGCCACACTCAGTCCCCCACCACACTGAATCTCCCCTCCTCACAGTTGC >33624_33624_4_GNAS-GAPDH_GNAS_chr20_57478846_ENST00000306090_GAPDH_chr12_6647267_ENST00000396861_length(amino acids)=180AA_BP= MSYGREVKVRHVHHTQDVVHSELVLGVGQLHGGHQVAHGGHNGFNRLFQVVFDVLHFGCLLTVSIKPINMQDPHLLHNGAFTRFSSTQQQ QAVRGPVDLLVLLQLLLDLFVGLTLRLLLVALVLGLTVPEAAHGGGKGRVSVGPRGARSSPPLEPRPRGLMAAAGPRRTRAGAAGAAAAA -------------------------------------------------------------- >33624_33624_5_GNAS-GAPDH_GNAS_chr20_57478846_ENST00000354359_GAPDH_chr12_6647267_ENST00000229239_length(transcript)=1130nt_BP=859nt TCGCCGCCGCCGCCTCCTCCTCCCCCCGCCTCCCCCCGCCCGCCGCGGCGCTTTTGGCTCGGGGCGGCGGCGGGGGCCCGGCCGAGGCAA TAAGAGCGGCGGCGGCGGCAGCGGCGGCAGCAGCTCCCGCAGCTCCTGCTCTGGTCCGCCTCGGCCCGGCGGCGGCCATCAGCCCCCTCG GCCTCGGCTCGAGGGGCGGGGAGCTGCGCGCGCCCCTCGGTCCGACCGACACCCTCCCCTTCCCGCCCGTCCGCGCGCCCCGCGGCCCGC GGCCCGCAGTCCGCCCCGCGCGCTCCTTGCCGAGGAGCCGAGCCCGCGCCCGGCCCGCCCGCCCGGCGCTGCCCCGGCCCTCCCGGCCCG CGTGAGGCCGCCCGCGCCCGCCGCCGCCGCAGCCCGGCCGCGCCCCGCCGCCGCCGCCGCCGCCATGGGCTGCCTCGGGAACAGTAAGAC CGAGGACCAGCGCAACGAGGAGAAGGCGCAGCGTGAGGCCAACAAAAAGATCGAGAAGCAGCTGCAGAAGGACAAGCAGGTCTACCGGGC CACGCACCGCCTGCTGCTGCTGGGTGCTGGAGAATCTGGTAAAAGCACCATTGTGAAGCAGATGAGGATCCTGCATGTTAATGGGTTTAA TGGAGAGGGCGGCGAAGAGGACCCGCAGGCTGCAAGGAGCAACAGCGATGGCAGTGAGAAGGCAACCAAAGTGCAGGACATCAAAAACAA CCTGAAAGAGGCGATTGAAACCATTGTGGCCGCCATGAGCAACCTGGTGCCCCCCGTGGAGCTGGCCAACCCCGAGAACCAGTTCAGAGT GGACTACATCCTGAGTGTGATGAACGTGCCTGACTTTGACTTCCCTCCCGTATGACAACGAATTTGGCTACAGCAACAGGGTGGTGGACC TCATGGCCCACATGGCCTCCAAGGAGTAAGACCCCTGGACCACCAGCCCCAGCAAGAGCACAAGAGGAAGAGAGAGACCCTCACTGCTGG GGAGTCCCTGCCACACTCAGTCCCCCACCACACTGAATCTCCCCTCCTCACAGTTGCCATGTAGACCCCTTGAAGAGGGGAGGGGCCTAG >33624_33624_5_GNAS-GAPDH_GNAS_chr20_57478846_ENST00000354359_GAPDH_chr12_6647267_ENST00000229239_length(amino acids)=289AA_BP= MSYGREVKVRHVHHTQDVVHSELVLGVGQLHGGHQVAHGGHNGFNRLFQVVFDVLHFGCLLTAIAVAPCSLRVLFAALSIKPINMQDPHL LHNGAFTRFSSTQQQQAVRGPVDLLVLLQLLLDLFVGLTLRLLLVALVLGLTVPEAAHGGGGGGGARPGCGGGGRGRPHAGREGRGSAGR AGRARARLLGKERAGRTAGRGPRGARTGGKGRVSVGPRGARSSPPLEPRPRGLMAAAGPRRTRAGAAGAAAAAAAAAALIASAGPPPPPR -------------------------------------------------------------- >33624_33624_6_GNAS-GAPDH_GNAS_chr20_57478846_ENST00000354359_GAPDH_chr12_6647267_ENST00000396861_length(transcript)=1118nt_BP=859nt TCGCCGCCGCCGCCTCCTCCTCCCCCCGCCTCCCCCCGCCCGCCGCGGCGCTTTTGGCTCGGGGCGGCGGCGGGGGCCCGGCCGAGGCAA TAAGAGCGGCGGCGGCGGCAGCGGCGGCAGCAGCTCCCGCAGCTCCTGCTCTGGTCCGCCTCGGCCCGGCGGCGGCCATCAGCCCCCTCG GCCTCGGCTCGAGGGGCGGGGAGCTGCGCGCGCCCCTCGGTCCGACCGACACCCTCCCCTTCCCGCCCGTCCGCGCGCCCCGCGGCCCGC GGCCCGCAGTCCGCCCCGCGCGCTCCTTGCCGAGGAGCCGAGCCCGCGCCCGGCCCGCCCGCCCGGCGCTGCCCCGGCCCTCCCGGCCCG CGTGAGGCCGCCCGCGCCCGCCGCCGCCGCAGCCCGGCCGCGCCCCGCCGCCGCCGCCGCCGCCATGGGCTGCCTCGGGAACAGTAAGAC CGAGGACCAGCGCAACGAGGAGAAGGCGCAGCGTGAGGCCAACAAAAAGATCGAGAAGCAGCTGCAGAAGGACAAGCAGGTCTACCGGGC CACGCACCGCCTGCTGCTGCTGGGTGCTGGAGAATCTGGTAAAAGCACCATTGTGAAGCAGATGAGGATCCTGCATGTTAATGGGTTTAA TGGAGAGGGCGGCGAAGAGGACCCGCAGGCTGCAAGGAGCAACAGCGATGGCAGTGAGAAGGCAACCAAAGTGCAGGACATCAAAAACAA CCTGAAAGAGGCGATTGAAACCATTGTGGCCGCCATGAGCAACCTGGTGCCCCCCGTGGAGCTGGCCAACCCCGAGAACCAGTTCAGAGT GGACTACATCCTGAGTGTGATGAACGTGCCTGACTTTGACTTCCCTCCCGTATGACAACGAATTTGGCTACAGCAACAGGGTGGTGGACC TCATGGCCCACATGGCCTCCAAGGAGTAAGACCCCTGGACCACCAGCCCCAGCAAGAGCACAAGAGGAAGAGAGAGACCCTCACTGCTGG GGAGTCCCTGCCACACTCAGTCCCCCACCACACTGAATCTCCCCTCCTCACAGTTGCCATGTAGACCCCTTGAAGAGGGGAGGGGCCTAG >33624_33624_6_GNAS-GAPDH_GNAS_chr20_57478846_ENST00000354359_GAPDH_chr12_6647267_ENST00000396861_length(amino acids)=289AA_BP= MSYGREVKVRHVHHTQDVVHSELVLGVGQLHGGHQVAHGGHNGFNRLFQVVFDVLHFGCLLTAIAVAPCSLRVLFAALSIKPINMQDPHL LHNGAFTRFSSTQQQQAVRGPVDLLVLLQLLLDLFVGLTLRLLLVALVLGLTVPEAAHGGGGGGGARPGCGGGGRGRPHAGREGRGSAGR AGRARARLLGKERAGRTAGRGPRGARTGGKGRVSVGPRGARSSPPLEPRPRGLMAAAGPRRTRAGAAGAAAAAAAAAALIASAGPPPPPR -------------------------------------------------------------- >33624_33624_7_GNAS-GAPDH_GNAS_chr20_57478846_ENST00000371085_GAPDH_chr12_6647267_ENST00000229239_length(transcript)=1127nt_BP=856nt TCGCCGCCGCCGCCTCCTCCTCCCCCCGCCTCCCCCCGCCCGCCGCGGCGCTTTTGGCTCGGGGCGGCGGCGGGGGCCCGGCCGAGGCAA TAAGAGCGGCGGCGGCGGCAGCGGCGGCAGCAGCTCCCGCAGCTCCTGCTCTGGTCCGCCTCGGCCCGGCGGCGGCCATCAGCCCCCTCG GCCTCGGCTCGAGGGGCGGGGAGCTGCGCGCGCCCCTCGGTCCGACCGACACCCTCCCCTTCCCGCCCGTCCGCGCGCCCCGCGGCCCGC GGCCCGCAGTCCGCCCCGCGCGCTCCTTGCCGAGGAGCCGAGCCCGCGCCCGGCCCGCCCGCCCGGCGCTGCCCCGGCCCTCCCGGCCCG CGTGAGGCCGCCCGCGCCCGCCGCCGCCGCAGCCCGGCCGCGCCCCGCCGCCGCCGCCGCCGCCATGGGCTGCCTCGGGAACAGTAAGAC CGAGGACCAGCGCAACGAGGAGAAGGCGCAGCGTGAGGCCAACAAAAAGATCGAGAAGCAGCTGCAGAAGGACAAGCAGGTCTACCGGGC CACGCACCGCCTGCTGCTGCTGGGTGCTGGAGAATCTGGTAAAAGCACCATTGTGAAGCAGATGAGGATCCTGCATGTTAATGGGTTTAA TGGAGAGGGCGGCGAAGAGGACCCGCAGGCTGCAAGGAGCAACAGCGATGGTGAGAAGGCAACCAAAGTGCAGGACATCAAAAACAACCT GAAAGAGGCGATTGAAACCATTGTGGCCGCCATGAGCAACCTGGTGCCCCCCGTGGAGCTGGCCAACCCCGAGAACCAGTTCAGAGTGGA CTACATCCTGAGTGTGATGAACGTGCCTGACTTTGACTTCCCTCCCGTATGACAACGAATTTGGCTACAGCAACAGGGTGGTGGACCTCA TGGCCCACATGGCCTCCAAGGAGTAAGACCCCTGGACCACCAGCCCCAGCAAGAGCACAAGAGGAAGAGAGAGACCCTCACTGCTGGGGA GTCCCTGCCACACTCAGTCCCCCACCACACTGAATCTCCCCTCCTCACAGTTGCCATGTAGACCCCTTGAAGAGGGGAGGGGCCTAGGGA >33624_33624_7_GNAS-GAPDH_GNAS_chr20_57478846_ENST00000371085_GAPDH_chr12_6647267_ENST00000229239_length(amino acids)=288AA_BP= MSYGREVKVRHVHHTQDVVHSELVLGVGQLHGGHQVAHGGHNGFNRLFQVVFDVLHFGCLLTIAVAPCSLRVLFAALSIKPINMQDPHLL HNGAFTRFSSTQQQQAVRGPVDLLVLLQLLLDLFVGLTLRLLLVALVLGLTVPEAAHGGGGGGGARPGCGGGGRGRPHAGREGRGSAGRA GRARARLLGKERAGRTAGRGPRGARTGGKGRVSVGPRGARSSPPLEPRPRGLMAAAGPRRTRAGAAGAAAAAAAAAALIASAGPPPPPRA -------------------------------------------------------------- >33624_33624_8_GNAS-GAPDH_GNAS_chr20_57478846_ENST00000371085_GAPDH_chr12_6647267_ENST00000396861_length(transcript)=1115nt_BP=856nt TCGCCGCCGCCGCCTCCTCCTCCCCCCGCCTCCCCCCGCCCGCCGCGGCGCTTTTGGCTCGGGGCGGCGGCGGGGGCCCGGCCGAGGCAA TAAGAGCGGCGGCGGCGGCAGCGGCGGCAGCAGCTCCCGCAGCTCCTGCTCTGGTCCGCCTCGGCCCGGCGGCGGCCATCAGCCCCCTCG GCCTCGGCTCGAGGGGCGGGGAGCTGCGCGCGCCCCTCGGTCCGACCGACACCCTCCCCTTCCCGCCCGTCCGCGCGCCCCGCGGCCCGC GGCCCGCAGTCCGCCCCGCGCGCTCCTTGCCGAGGAGCCGAGCCCGCGCCCGGCCCGCCCGCCCGGCGCTGCCCCGGCCCTCCCGGCCCG CGTGAGGCCGCCCGCGCCCGCCGCCGCCGCAGCCCGGCCGCGCCCCGCCGCCGCCGCCGCCGCCATGGGCTGCCTCGGGAACAGTAAGAC CGAGGACCAGCGCAACGAGGAGAAGGCGCAGCGTGAGGCCAACAAAAAGATCGAGAAGCAGCTGCAGAAGGACAAGCAGGTCTACCGGGC CACGCACCGCCTGCTGCTGCTGGGTGCTGGAGAATCTGGTAAAAGCACCATTGTGAAGCAGATGAGGATCCTGCATGTTAATGGGTTTAA TGGAGAGGGCGGCGAAGAGGACCCGCAGGCTGCAAGGAGCAACAGCGATGGTGAGAAGGCAACCAAAGTGCAGGACATCAAAAACAACCT GAAAGAGGCGATTGAAACCATTGTGGCCGCCATGAGCAACCTGGTGCCCCCCGTGGAGCTGGCCAACCCCGAGAACCAGTTCAGAGTGGA CTACATCCTGAGTGTGATGAACGTGCCTGACTTTGACTTCCCTCCCGTATGACAACGAATTTGGCTACAGCAACAGGGTGGTGGACCTCA TGGCCCACATGGCCTCCAAGGAGTAAGACCCCTGGACCACCAGCCCCAGCAAGAGCACAAGAGGAAGAGAGAGACCCTCACTGCTGGGGA GTCCCTGCCACACTCAGTCCCCCACCACACTGAATCTCCCCTCCTCACAGTTGCCATGTAGACCCCTTGAAGAGGGGAGGGGCCTAGGGA >33624_33624_8_GNAS-GAPDH_GNAS_chr20_57478846_ENST00000371085_GAPDH_chr12_6647267_ENST00000396861_length(amino acids)=288AA_BP= MSYGREVKVRHVHHTQDVVHSELVLGVGQLHGGHQVAHGGHNGFNRLFQVVFDVLHFGCLLTIAVAPCSLRVLFAALSIKPINMQDPHLL HNGAFTRFSSTQQQQAVRGPVDLLVLLQLLLDLFVGLTLRLLLVALVLGLTVPEAAHGGGGGGGARPGCGGGGRGRPHAGREGRGSAGRA GRARARLLGKERAGRTAGRGPRGARTGGKGRVSVGPRGARSSPPLEPRPRGLMAAAGPRRTRAGAAGAAAAAAAAAALIASAGPPPPPRA -------------------------------------------------------------- >33624_33624_9_GNAS-GAPDH_GNAS_chr20_57478846_ENST00000371095_GAPDH_chr12_6647267_ENST00000229239_length(transcript)=1086nt_BP=815nt CTCGCCGCCGCCGCCTCCTCCTCCCCCCGCCTCCCCCCGCCCGCCGCGGCGCTTTTGGCTCGGGGCGGCGGCGGGGGCCCGGCCGAGGCA ATAAGAGCGGCGGCGGCGGCAGCGGCGGCAGCAGCTCCCGCAGCTCCTGCTCTGGTCCGCCTCGGCCCGGCGGCGGCCATCAGCCCCCTC GGCCTCGGCTCGAGGGGCGGGGAGCTGCGCGCGCCCCTCGGTCCGACCGACACCCTCCCCTTCCCGCCCGTCCGCGCGCCCCGCGGCCCG CGGCCCGCAGTCCGCCCCGCGCGCTCCTTGCCGAGGAGCCGAGCCCGCGCCCGGCCCGCCCGCCCGGCGCTGCCCCGGCCCTCCCGGCCC GCGTGAGGCCGCCCGCGCCCGCCGCCGCCGCAGCCCGGCCGCGCCCCGCCGCCGCCGCCGCCGCCATGGGCTGCCTCGGGAACAGTAAGA CCGAGGACCAGCGCAACGAGGAGAAGGCGCAGCGTGAGGCCAACAAAAAGATCGAGAAGCAGCTGCAGAAGGACAAGCAGGTCTACCGGG CCACGCACCGCCTGCTGCTGCTGGGTGCTGGAGAATCTGGTAAAAGCACCATTGTGAAGCAGATGAGGATCCTGCATGTTAATGGGTTTA ATGGAGACAGTGAGAAGGCAACCAAAGTGCAGGACATCAAAAACAACCTGAAAGAGGCGATTGAAACCATTGTGGCCGCCATGAGCAACC TGGTGCCCCCCGTGGAGCTGGCCAACCCCGAGAACCAGTTCAGAGTGGACTACATCCTGAGTGTGATGAACGTGCCTGACTTTGACTTCC CTCCCGTATGACAACGAATTTGGCTACAGCAACAGGGTGGTGGACCTCATGGCCCACATGGCCTCCAAGGAGTAAGACCCCTGGACCACC AGCCCCAGCAAGAGCACAAGAGGAAGAGAGAGACCCTCACTGCTGGGGAGTCCCTGCCACACTCAGTCCCCCACCACACTGAATCTCCCC TCCTCACAGTTGCCATGTAGACCCCTTGAAGAGGGGAGGGGCCTAGGGAGCCGCACCTTGTCATGTACCATCAATAAAGTACCCTGTGCT >33624_33624_9_GNAS-GAPDH_GNAS_chr20_57478846_ENST00000371095_GAPDH_chr12_6647267_ENST00000229239_length(amino acids)=274AA_BP= MSYGREVKVRHVHHTQDVVHSELVLGVGQLHGGHQVAHGGHNGFNRLFQVVFDVLHFGCLLTVSIKPINMQDPHLLHNGAFTRFSSTQQQ QAVRGPVDLLVLLQLLLDLFVGLTLRLLLVALVLGLTVPEAAHGGGGGGGARPGCGGGGRGRPHAGREGRGSAGRAGRARARLLGKERAG RTAGRGPRGARTGGKGRVSVGPRGARSSPPLEPRPRGLMAAAGPRRTRAGAAGAAAAAAAAAALIASAGPPPPPRAKSAAAGGGRRGEEE -------------------------------------------------------------- >33624_33624_10_GNAS-GAPDH_GNAS_chr20_57478846_ENST00000371095_GAPDH_chr12_6647267_ENST00000396861_length(transcript)=1074nt_BP=815nt CTCGCCGCCGCCGCCTCCTCCTCCCCCCGCCTCCCCCCGCCCGCCGCGGCGCTTTTGGCTCGGGGCGGCGGCGGGGGCCCGGCCGAGGCA ATAAGAGCGGCGGCGGCGGCAGCGGCGGCAGCAGCTCCCGCAGCTCCTGCTCTGGTCCGCCTCGGCCCGGCGGCGGCCATCAGCCCCCTC GGCCTCGGCTCGAGGGGCGGGGAGCTGCGCGCGCCCCTCGGTCCGACCGACACCCTCCCCTTCCCGCCCGTCCGCGCGCCCCGCGGCCCG CGGCCCGCAGTCCGCCCCGCGCGCTCCTTGCCGAGGAGCCGAGCCCGCGCCCGGCCCGCCCGCCCGGCGCTGCCCCGGCCCTCCCGGCCC GCGTGAGGCCGCCCGCGCCCGCCGCCGCCGCAGCCCGGCCGCGCCCCGCCGCCGCCGCCGCCGCCATGGGCTGCCTCGGGAACAGTAAGA CCGAGGACCAGCGCAACGAGGAGAAGGCGCAGCGTGAGGCCAACAAAAAGATCGAGAAGCAGCTGCAGAAGGACAAGCAGGTCTACCGGG CCACGCACCGCCTGCTGCTGCTGGGTGCTGGAGAATCTGGTAAAAGCACCATTGTGAAGCAGATGAGGATCCTGCATGTTAATGGGTTTA ATGGAGACAGTGAGAAGGCAACCAAAGTGCAGGACATCAAAAACAACCTGAAAGAGGCGATTGAAACCATTGTGGCCGCCATGAGCAACC TGGTGCCCCCCGTGGAGCTGGCCAACCCCGAGAACCAGTTCAGAGTGGACTACATCCTGAGTGTGATGAACGTGCCTGACTTTGACTTCC CTCCCGTATGACAACGAATTTGGCTACAGCAACAGGGTGGTGGACCTCATGGCCCACATGGCCTCCAAGGAGTAAGACCCCTGGACCACC AGCCCCAGCAAGAGCACAAGAGGAAGAGAGAGACCCTCACTGCTGGGGAGTCCCTGCCACACTCAGTCCCCCACCACACTGAATCTCCCC >33624_33624_10_GNAS-GAPDH_GNAS_chr20_57478846_ENST00000371095_GAPDH_chr12_6647267_ENST00000396861_length(amino acids)=274AA_BP= MSYGREVKVRHVHHTQDVVHSELVLGVGQLHGGHQVAHGGHNGFNRLFQVVFDVLHFGCLLTVSIKPINMQDPHLLHNGAFTRFSSTQQQ QAVRGPVDLLVLLQLLLDLFVGLTLRLLLVALVLGLTVPEAAHGGGGGGGARPGCGGGGRGRPHAGREGRGSAGRAGRARARLLGKERAG RTAGRGPRGARTGGKGRVSVGPRGARSSPPLEPRPRGLMAAAGPRRTRAGAAGAAAAAAAAAALIASAGPPPPPRAKSAAAGGGRRGEEE -------------------------------------------------------------- >33624_33624_11_GNAS-GAPDH_GNAS_chr20_57478846_ENST00000371100_GAPDH_chr12_6647267_ENST00000229239_length(transcript)=3184nt_BP=2913nt AAAGACTCCAAATCAGTCAGGGAGAGGAGTGGAAGGAGCCAAAACGTCCCTGCGAGGCCTGCGCTCACCAGCCCCCTTACCCAAAGTCCC GGGCCCCTGGCGCCAGGAGCAGGCTCCTGGAGCCGGAGCCAGCTCGGCACTGGAACCGGCGTCCTCTGGTGGCAGAGAGAGAGCGCTACT GGCGATTTTCGGACCGAATCGGCACGCTCGTCAGATCCAAGCAGGCGGGACTGGCCTGGAGCAAAAAGAAAGAGAGAGGAGGGCGTAAGG ATAGACCAAGGAAGAGGGGCTGGGGGGCAGCCTGGGGGCATGAAAAGTGGCCAGGAAGGAGCCAAGACTCCACCAGCAACAATTGAGTTG CTTCAGCCTCAGTCTAGGGTTCCTTCCAGGCCTTGAACCCCCCAACCTCACAAGGGTTGGAAAGTGAGGCCGGTGAACTTTCCAGCTGGT ACTTTGATTTTAAAATAATAATAATAATTTTTTCACCCTAGTTCGGTTGGGTGCTCCATCTTACGGAGCCCCAAACTTATTTTGAGAGGC CGCCACCGTGTTATGGGCGTGCGCAACTGCCTCTACGGCAATAATATGTCAGGACAACGCGATATCCCCCCTGAAATCGGGGAACAGCCC GAGCAACCACCTTTGGAGGCCCCAGGGGCAGCTGCCCCCGGTGCTGGGCCTAGCCCAGCCGAAGAGATGGAGACCGAACCGCCTCACAAC GAGCCCATCCCCGTCGAGAATGATGGCGAGGCCTGTGGACCCCCAGAGGTCTCCAGACCCAACTTTCAGGTCCTCAACCCGGCATTCAGG GAAGCTGGAGCCCATGGAAGCTACAGCCCACCTCCTGAGGAAGCAATGCCCTTCGAGGCTGAACAGCCCAGCTTGGGAGGCTTCTGGCCT ACACTGGAGCAGCCTGGATTCCCCAGTGGGGTCCATGCAGGCCTTGAGGCCTTCGGCCCAGCACTCATGGAGCCCGGAGCCTTCAGTGGT GCCAGACCAGGCCTGGGAGGATACAGCCCTCCACCAGAAGAAGCTATGCCCTTTGAGTTTGACCAGCCTGCCCAGAGAGGCTGCAGTCAA CTTCTCTTACAGGTCCCAGACCTTGCTCCAGGAGGCCCAGGTGCTGCAGGGGTCCCCGGAGCTCCTCCCGAGGAGCCCCAAGCCCTCAGG CCTGCAAAGGCTGGCTCCAGAGGAGGCTACAGCCCTCCCCCTGAGGAGACTATGCCATTTGAGCTTGATGGAGAAGGATTTGGGGACGAC AGCCCACCCCCGGGGCTTTCCCGAGTTATCGCACAAGTCGACGGCAGCAGCCAGTTCGCGGCAGTCGCGGCCTCGAGTGCGGTCCGCCTC ACTCCCGCCGCGAACGCGCCTCCCCTCTGGGTCCCAGGCGCCATCGGCAGCCCATCCCAAGAGGCTGTCAGACCTCCTTCTAACTTCACG GGCAGCAGCCCCTGGATGGAGATCTCCGGACCCCCGTTCGAGATTGGCAGCGCCCCCGCTGGGGTCGACGACACTCCCGTCAACATGGAC AGCCCCCCAATCGCGCTTGACGGCCCGCCCATCAAGGTCTCCGGAGCCCCAGATAAGAGAGAGCGAGCAGAGAGACCCCCAGTTGAGGAG GAAGCAGCAGAGATGGAAGGAGCCGCTGATGCCGCGGAGGGAGGAAAAGTACCCTCTCCGGGGTACGGATCCCCTGCCGCCGGGGCAGCC TCAGCGGATACCGCTGCCAGGGCAGCCCCTGCAGCCCCAGCCGATCCTGACTCCGGGGCAACCCCAGAAGATCCCGACTCCGGGACAGCA CCAGCCGATCCTGACTCCGGGGCATTCGCAGCCGATCCCGACTCCGGGGCAGCCCCTGCCGCCCCAGCCGATCCCGACTCCGGGGCGGCC CCTGACGCCCCAGCCGATCCCGACTCCGGGGCGGCCCCTGACGCCCCAGCCGATCCAGATGCCGGGGCGGCCCCTGAGGCTCCCGCCGCC CCTGCGGCTGCTGAGACCCGGGCAGCCCATGTCGCCCCAGCTGCGCCAGACGCAGGGGCTCCCACTGCCCCAGCCGCTTCTGCCACCCGG GCAGCCCAAGTCCGCCGGGCGGCCTCTGCAGCCCCTGCCTCCGGGGCCAGACGCAAGATCCATCTCAGACCCCCCAGCCCCGAGATCCAG GCTGCCGATCCGCCTACTCCGCGGCCTACTCGCGCGTCTGCCTGGCGGGGCAAGTCCGAGAGCAGCCGCGGCCGCCGCGTGTACTACGAT GAAGGGGTGGCCAGCAGCGACGATGACTCCAGCGGAGACGAGTCCGACGATGGGACCTCCGGATGCCTCCGCTGGTTTCAGCATCGGCGA AATCGCCGCCGCCGAAAGCCCCAGCGCAACTTACTCCGCAACTTTCTCGTGCAAGCCTTCGGGGGCTGCTTCGGTCGATCTGAGAGTCCC CAGCCCAAAGCCTCGCGCTCTCTCAAGGTCAAGAAGGTACCCCTGGCGGAGAAGCGCAGACAGATGCGCAAAGAAGCCCTGGAGAAGCGG GCCCAGAAGCGCGCAGAGAAGAAACGCAGTAAGCTCATCGACAAACAACTCCAGGACGAAAAGATGGGCTACATGTGTACGCACCGCCTG CTGCTTCTAGGTGCTGGAGAATCTGGTAAAAGCACCATTGTGAAGCAGATGAGGATCCTGCATGTTAATGGGTTTAATGGAGAGGGCGGC GAAGAGGACCCGCAGGCTGCAAGGAGCAACAGCGATGGTGAGAAGGCAACCAAAGTGCAGGACATCAAAAACAACCTGAAAGAGGCGATT GAAACCATTGTGGCCGCCATGAGCAACCTGGTGCCCCCCGTGGAGCTGGCCAACCCCGAGAACCAGTTCAGAGTGGACTACATCCTGAGT GTGATGAACGTGCCTGACTTTGACTTCCCTCCCGTATGACAACGAATTTGGCTACAGCAACAGGGTGGTGGACCTCATGGCCCACATGGC CTCCAAGGAGTAAGACCCCTGGACCACCAGCCCCAGCAAGAGCACAAGAGGAAGAGAGAGACCCTCACTGCTGGGGAGTCCCTGCCACAC TCAGTCCCCCACCACACTGAATCTCCCCTCCTCACAGTTGCCATGTAGACCCCTTGAAGAGGGGAGGGGCCTAGGGAGCCGCACCTTGTC >33624_33624_11_GNAS-GAPDH_GNAS_chr20_57478846_ENST00000371100_GAPDH_chr12_6647267_ENST00000229239_length(amino acids)=795AA_BP= MRGRHRVMGVRNCLYGNNMSGQRDIPPEIGEQPEQPPLEAPGAAAPGAGPSPAEEMETEPPHNEPIPVENDGEACGPPEVSRPNFQVLNP AFREAGAHGSYSPPPEEAMPFEAEQPSLGGFWPTLEQPGFPSGVHAGLEAFGPALMEPGAFSGARPGLGGYSPPPEEAMPFEFDQPAQRG CSQLLLQVPDLAPGGPGAAGVPGAPPEEPQALRPAKAGSRGGYSPPPEETMPFELDGEGFGDDSPPPGLSRVIAQVDGSSQFAAVAASSA VRLTPAANAPPLWVPGAIGSPSQEAVRPPSNFTGSSPWMEISGPPFEIGSAPAGVDDTPVNMDSPPIALDGPPIKVSGAPDKRERAERPP VEEEAAEMEGAADAAEGGKVPSPGYGSPAAGAASADTAARAAPAAPADPDSGATPEDPDSGTAPADPDSGAFAADPDSGAAPAAPADPDS GAAPDAPADPDSGAAPDAPADPDAGAAPEAPAAPAAAETRAAHVAPAAPDAGAPTAPAASATRAAQVRRAASAAPASGARRKIHLRPPSP EIQAADPPTPRPTRASAWRGKSESSRGRRVYYDEGVASSDDDSSGDESDDGTSGCLRWFQHRRNRRRRKPQRNLLRNFLVQAFGGCFGRS ESPQPKASRSLKVKKVPLAEKRRQMRKEALEKRAQKRAEKKRSKLIDKQLQDEKMGYMCTHRLLLLGAGESGKSTIVKQMRILHVNGFNG -------------------------------------------------------------- >33624_33624_12_GNAS-GAPDH_GNAS_chr20_57478846_ENST00000371100_GAPDH_chr12_6647267_ENST00000396861_length(transcript)=3172nt_BP=2913nt AAAGACTCCAAATCAGTCAGGGAGAGGAGTGGAAGGAGCCAAAACGTCCCTGCGAGGCCTGCGCTCACCAGCCCCCTTACCCAAAGTCCC GGGCCCCTGGCGCCAGGAGCAGGCTCCTGGAGCCGGAGCCAGCTCGGCACTGGAACCGGCGTCCTCTGGTGGCAGAGAGAGAGCGCTACT GGCGATTTTCGGACCGAATCGGCACGCTCGTCAGATCCAAGCAGGCGGGACTGGCCTGGAGCAAAAAGAAAGAGAGAGGAGGGCGTAAGG ATAGACCAAGGAAGAGGGGCTGGGGGGCAGCCTGGGGGCATGAAAAGTGGCCAGGAAGGAGCCAAGACTCCACCAGCAACAATTGAGTTG CTTCAGCCTCAGTCTAGGGTTCCTTCCAGGCCTTGAACCCCCCAACCTCACAAGGGTTGGAAAGTGAGGCCGGTGAACTTTCCAGCTGGT ACTTTGATTTTAAAATAATAATAATAATTTTTTCACCCTAGTTCGGTTGGGTGCTCCATCTTACGGAGCCCCAAACTTATTTTGAGAGGC CGCCACCGTGTTATGGGCGTGCGCAACTGCCTCTACGGCAATAATATGTCAGGACAACGCGATATCCCCCCTGAAATCGGGGAACAGCCC GAGCAACCACCTTTGGAGGCCCCAGGGGCAGCTGCCCCCGGTGCTGGGCCTAGCCCAGCCGAAGAGATGGAGACCGAACCGCCTCACAAC GAGCCCATCCCCGTCGAGAATGATGGCGAGGCCTGTGGACCCCCAGAGGTCTCCAGACCCAACTTTCAGGTCCTCAACCCGGCATTCAGG GAAGCTGGAGCCCATGGAAGCTACAGCCCACCTCCTGAGGAAGCAATGCCCTTCGAGGCTGAACAGCCCAGCTTGGGAGGCTTCTGGCCT ACACTGGAGCAGCCTGGATTCCCCAGTGGGGTCCATGCAGGCCTTGAGGCCTTCGGCCCAGCACTCATGGAGCCCGGAGCCTTCAGTGGT GCCAGACCAGGCCTGGGAGGATACAGCCCTCCACCAGAAGAAGCTATGCCCTTTGAGTTTGACCAGCCTGCCCAGAGAGGCTGCAGTCAA CTTCTCTTACAGGTCCCAGACCTTGCTCCAGGAGGCCCAGGTGCTGCAGGGGTCCCCGGAGCTCCTCCCGAGGAGCCCCAAGCCCTCAGG CCTGCAAAGGCTGGCTCCAGAGGAGGCTACAGCCCTCCCCCTGAGGAGACTATGCCATTTGAGCTTGATGGAGAAGGATTTGGGGACGAC AGCCCACCCCCGGGGCTTTCCCGAGTTATCGCACAAGTCGACGGCAGCAGCCAGTTCGCGGCAGTCGCGGCCTCGAGTGCGGTCCGCCTC ACTCCCGCCGCGAACGCGCCTCCCCTCTGGGTCCCAGGCGCCATCGGCAGCCCATCCCAAGAGGCTGTCAGACCTCCTTCTAACTTCACG GGCAGCAGCCCCTGGATGGAGATCTCCGGACCCCCGTTCGAGATTGGCAGCGCCCCCGCTGGGGTCGACGACACTCCCGTCAACATGGAC AGCCCCCCAATCGCGCTTGACGGCCCGCCCATCAAGGTCTCCGGAGCCCCAGATAAGAGAGAGCGAGCAGAGAGACCCCCAGTTGAGGAG GAAGCAGCAGAGATGGAAGGAGCCGCTGATGCCGCGGAGGGAGGAAAAGTACCCTCTCCGGGGTACGGATCCCCTGCCGCCGGGGCAGCC TCAGCGGATACCGCTGCCAGGGCAGCCCCTGCAGCCCCAGCCGATCCTGACTCCGGGGCAACCCCAGAAGATCCCGACTCCGGGACAGCA CCAGCCGATCCTGACTCCGGGGCATTCGCAGCCGATCCCGACTCCGGGGCAGCCCCTGCCGCCCCAGCCGATCCCGACTCCGGGGCGGCC CCTGACGCCCCAGCCGATCCCGACTCCGGGGCGGCCCCTGACGCCCCAGCCGATCCAGATGCCGGGGCGGCCCCTGAGGCTCCCGCCGCC CCTGCGGCTGCTGAGACCCGGGCAGCCCATGTCGCCCCAGCTGCGCCAGACGCAGGGGCTCCCACTGCCCCAGCCGCTTCTGCCACCCGG GCAGCCCAAGTCCGCCGGGCGGCCTCTGCAGCCCCTGCCTCCGGGGCCAGACGCAAGATCCATCTCAGACCCCCCAGCCCCGAGATCCAG GCTGCCGATCCGCCTACTCCGCGGCCTACTCGCGCGTCTGCCTGGCGGGGCAAGTCCGAGAGCAGCCGCGGCCGCCGCGTGTACTACGAT GAAGGGGTGGCCAGCAGCGACGATGACTCCAGCGGAGACGAGTCCGACGATGGGACCTCCGGATGCCTCCGCTGGTTTCAGCATCGGCGA AATCGCCGCCGCCGAAAGCCCCAGCGCAACTTACTCCGCAACTTTCTCGTGCAAGCCTTCGGGGGCTGCTTCGGTCGATCTGAGAGTCCC CAGCCCAAAGCCTCGCGCTCTCTCAAGGTCAAGAAGGTACCCCTGGCGGAGAAGCGCAGACAGATGCGCAAAGAAGCCCTGGAGAAGCGG GCCCAGAAGCGCGCAGAGAAGAAACGCAGTAAGCTCATCGACAAACAACTCCAGGACGAAAAGATGGGCTACATGTGTACGCACCGCCTG CTGCTTCTAGGTGCTGGAGAATCTGGTAAAAGCACCATTGTGAAGCAGATGAGGATCCTGCATGTTAATGGGTTTAATGGAGAGGGCGGC GAAGAGGACCCGCAGGCTGCAAGGAGCAACAGCGATGGTGAGAAGGCAACCAAAGTGCAGGACATCAAAAACAACCTGAAAGAGGCGATT GAAACCATTGTGGCCGCCATGAGCAACCTGGTGCCCCCCGTGGAGCTGGCCAACCCCGAGAACCAGTTCAGAGTGGACTACATCCTGAGT GTGATGAACGTGCCTGACTTTGACTTCCCTCCCGTATGACAACGAATTTGGCTACAGCAACAGGGTGGTGGACCTCATGGCCCACATGGC CTCCAAGGAGTAAGACCCCTGGACCACCAGCCCCAGCAAGAGCACAAGAGGAAGAGAGAGACCCTCACTGCTGGGGAGTCCCTGCCACAC TCAGTCCCCCACCACACTGAATCTCCCCTCCTCACAGTTGCCATGTAGACCCCTTGAAGAGGGGAGGGGCCTAGGGAGCCGCACCTTGTC >33624_33624_12_GNAS-GAPDH_GNAS_chr20_57478846_ENST00000371100_GAPDH_chr12_6647267_ENST00000396861_length(amino acids)=795AA_BP= MRGRHRVMGVRNCLYGNNMSGQRDIPPEIGEQPEQPPLEAPGAAAPGAGPSPAEEMETEPPHNEPIPVENDGEACGPPEVSRPNFQVLNP AFREAGAHGSYSPPPEEAMPFEAEQPSLGGFWPTLEQPGFPSGVHAGLEAFGPALMEPGAFSGARPGLGGYSPPPEEAMPFEFDQPAQRG CSQLLLQVPDLAPGGPGAAGVPGAPPEEPQALRPAKAGSRGGYSPPPEETMPFELDGEGFGDDSPPPGLSRVIAQVDGSSQFAAVAASSA VRLTPAANAPPLWVPGAIGSPSQEAVRPPSNFTGSSPWMEISGPPFEIGSAPAGVDDTPVNMDSPPIALDGPPIKVSGAPDKRERAERPP VEEEAAEMEGAADAAEGGKVPSPGYGSPAAGAASADTAARAAPAAPADPDSGATPEDPDSGTAPADPDSGAFAADPDSGAAPAAPADPDS GAAPDAPADPDSGAAPDAPADPDAGAAPEAPAAPAAAETRAAHVAPAAPDAGAPTAPAASATRAAQVRRAASAAPASGARRKIHLRPPSP EIQAADPPTPRPTRASAWRGKSESSRGRRVYYDEGVASSDDDSSGDESDDGTSGCLRWFQHRRNRRRRKPQRNLLRNFLVQAFGGCFGRS ESPQPKASRSLKVKKVPLAEKRRQMRKEALEKRAQKRAEKKRSKLIDKQLQDEKMGYMCTHRLLLLGAGESGKSTIVKQMRILHVNGFNG -------------------------------------------------------------- >33624_33624_13_GNAS-GAPDH_GNAS_chr20_57478846_ENST00000371102_GAPDH_chr12_6647267_ENST00000229239_length(transcript)=2593nt_BP=2322nt GTTATGGGCGTGCGCAACTGCCTCTACGGCAATAATATGTCAGGACAACGCGATATCCCCCCTGAAATCGGGGAACAGCCCGAGCAACCA CCTTTGGAGGCCCCAGGGGCAGCTGCCCCCGGTGCTGGGCCTAGCCCAGCCGAAGAGATGGAGACCGAACCGCCTCACAACGAGCCCATC CCCGTCGAGAATGATGGCGAGGCCTGTGGACCCCCAGAGGTCTCCAGACCCAACTTTCAGGTCCTCAACCCGGCATTCAGGGAAGCTGGA GCCCATGGAAGCTACAGCCCACCTCCTGAGGAAGCAATGCCCTTCGAGGCTGAACAGCCCAGCTTGGGAGGCTTCTGGCCTACACTGGAG CAGCCTGGATTCCCCAGTGGGGTCCATGCAGGCCTTGAGGCCTTCGGCCCAGCACTCATGGAGCCCGGAGCCTTCAGTGGTGCCAGACCA GGCCTGGGAGGATACAGCCCTCCACCAGAAGAAGCTATGCCCTTTGAGTTTGACCAGCCTGCCCAGAGAGGCTGCAGTCAACTTCTCTTA CAGGTCCCAGACCTTGCTCCAGGAGGCCCAGGTGCTGCAGGGGTCCCCGGAGCTCCTCCCGAGGAGCCCCAAGCCCTCAGGCCTGCAAAG GCTGGCTCCAGAGGAGGCTACAGCCCTCCCCCTGAGGAGACTATGCCATTTGAGCTTGATGGAGAAGGATTTGGGGACGACAGCCCACCC CCGGGGCTTTCCCGAGTTATCGCACAAGTCGACGGCAGCAGCCAGTTCGCGGCAGTCGCGGCCTCGAGTGCGGTCCGCCTCACTCCCGCC GCGAACGCGCCTCCCCTCTGGGTCCCAGGCGCCATCGGCAGCCCATCCCAAGAGGCTGTCAGACCTCCTTCTAACTTCACGGGCAGCAGC CCCTGGATGGAGATCTCCGGACCCCCGTTCGAGATTGGCAGCGCCCCCGCTGGGGTCGACGACACTCCCGTCAACATGGACAGCCCCCCA ATCGCGCTTGACGGCCCGCCCATCAAGGTCTCCGGAGCCCCAGATAAGAGAGAGCGAGCAGAGAGACCCCCAGTTGAGGAGGAAGCAGCA GAGATGGAAGGAGCCGCTGATGCCGCGGAGGGAGGAAAAGTACCCTCTCCGGGGTACGGATCCCCTGCCGCCGGGGCAGCCTCAGCGGAT ACCGCTGCCAGGGCAGCCCCTGCAGCCCCAGCCGATCCTGACTCCGGGGCAACCCCAGAAGATCCCGACTCCGGGACAGCACCAGCCGAT CCTGACTCCGGGGCATTCGCAGCCGATCCCGACTCCGGGGCAGCCCCTGCCGCCCCAGCCGATCCCGACTCCGGGGCGGCCCCTGACGCC CCAGCCGATCCCGACTCCGGGGCGGCCCCTGACGCCCCAGCCGATCCAGATGCCGGGGCGGCCCCTGAGGCTCCCGCCGCCCCTGCGGCT GCTGAGACCCGGGCAGCCCATGTCGCCCCAGCTGCGCCAGACGCAGGGGCTCCCACTGCCCCAGCCGCTTCTGCCACCCGGGCAGCCCAA GTCCGCCGGGCGGCCTCTGCAGCCCCTGCCTCCGGGGCCAGACGCAAGATCCATCTCAGACCCCCCAGCCCCGAGATCCAGGCTGCCGAT CCGCCTACTCCGCGGCCTACTCGCGCGTCTGCCTGGCGGGGCAAGTCCGAGAGCAGCCGCGGCCGCCGCGTGTACTACGATGAAGGGGTG GCCAGCAGCGACGATGACTCCAGCGGAGACGAGTCCGACGATGGGACCTCCGGATGCCTCCGCTGGTTTCAGCATCGGCGAAATCGCCGC CGCCGAAAGCCCCAGCGCAACTTACTCCGCAACTTTCTCGTGCAAGCCTTCGGGGGCTGCTTCGGTCGATCTGAGAGTCCCCAGCCCAAA GCCTCGCGCTCTCTCAAGGTCAAGAAGGTACCCCTGGCGGAGAAGCGCAGACAGATGCGCAAAGAAGCCCTGGAGAAGCGGGCCCAGAAG CGCGCAGAGAAGAAACGCAGTAAGCTCATCGACAAACAACTCCAGGACGAAAAGATGGGCTACATGTGTACGCACCGCCTGCTGCTTCTA GGTGCTGGAGAATCTGGTAAAAGCACCATTGTGAAGCAGATGAGGATCCTGCATGTTAATGGGTTTAATGGAGACAGTGAGAAGGCAACC AAAGTGCAGGACATCAAAAACAACCTGAAAGAGGCGATTGAAACCATTGTGGCCGCCATGAGCAACCTGGTGCCCCCCGTGGAGCTGGCC AACCCCGAGAACCAGTTCAGAGTGGACTACATCCTGAGTGTGATGAACGTGCCTGACTTTGACTTCCCTCCCGTATGACAACGAATTTGG CTACAGCAACAGGGTGGTGGACCTCATGGCCCACATGGCCTCCAAGGAGTAAGACCCCTGGACCACCAGCCCCAGCAAGAGCACAAGAGG AAGAGAGAGACCCTCACTGCTGGGGAGTCCCTGCCACACTCAGTCCCCCACCACACTGAATCTCCCCTCCTCACAGTTGCCATGTAGACC >33624_33624_13_GNAS-GAPDH_GNAS_chr20_57478846_ENST00000371102_GAPDH_chr12_6647267_ENST00000229239_length(amino acids)=774AA_BP= MGVRNCLYGNNMSGQRDIPPEIGEQPEQPPLEAPGAAAPGAGPSPAEEMETEPPHNEPIPVENDGEACGPPEVSRPNFQVLNPAFREAGA HGSYSPPPEEAMPFEAEQPSLGGFWPTLEQPGFPSGVHAGLEAFGPALMEPGAFSGARPGLGGYSPPPEEAMPFEFDQPAQRGCSQLLLQ VPDLAPGGPGAAGVPGAPPEEPQALRPAKAGSRGGYSPPPEETMPFELDGEGFGDDSPPPGLSRVIAQVDGSSQFAAVAASSAVRLTPAA NAPPLWVPGAIGSPSQEAVRPPSNFTGSSPWMEISGPPFEIGSAPAGVDDTPVNMDSPPIALDGPPIKVSGAPDKRERAERPPVEEEAAE MEGAADAAEGGKVPSPGYGSPAAGAASADTAARAAPAAPADPDSGATPEDPDSGTAPADPDSGAFAADPDSGAAPAAPADPDSGAAPDAP ADPDSGAAPDAPADPDAGAAPEAPAAPAAAETRAAHVAPAAPDAGAPTAPAASATRAAQVRRAASAAPASGARRKIHLRPPSPEIQAADP PTPRPTRASAWRGKSESSRGRRVYYDEGVASSDDDSSGDESDDGTSGCLRWFQHRRNRRRRKPQRNLLRNFLVQAFGGCFGRSESPQPKA SRSLKVKKVPLAEKRRQMRKEALEKRAQKRAEKKRSKLIDKQLQDEKMGYMCTHRLLLLGAGESGKSTIVKQMRILHVNGFNGDSEKATK -------------------------------------------------------------- >33624_33624_14_GNAS-GAPDH_GNAS_chr20_57478846_ENST00000371102_GAPDH_chr12_6647267_ENST00000396861_length(transcript)=2581nt_BP=2322nt GTTATGGGCGTGCGCAACTGCCTCTACGGCAATAATATGTCAGGACAACGCGATATCCCCCCTGAAATCGGGGAACAGCCCGAGCAACCA CCTTTGGAGGCCCCAGGGGCAGCTGCCCCCGGTGCTGGGCCTAGCCCAGCCGAAGAGATGGAGACCGAACCGCCTCACAACGAGCCCATC CCCGTCGAGAATGATGGCGAGGCCTGTGGACCCCCAGAGGTCTCCAGACCCAACTTTCAGGTCCTCAACCCGGCATTCAGGGAAGCTGGA GCCCATGGAAGCTACAGCCCACCTCCTGAGGAAGCAATGCCCTTCGAGGCTGAACAGCCCAGCTTGGGAGGCTTCTGGCCTACACTGGAG CAGCCTGGATTCCCCAGTGGGGTCCATGCAGGCCTTGAGGCCTTCGGCCCAGCACTCATGGAGCCCGGAGCCTTCAGTGGTGCCAGACCA GGCCTGGGAGGATACAGCCCTCCACCAGAAGAAGCTATGCCCTTTGAGTTTGACCAGCCTGCCCAGAGAGGCTGCAGTCAACTTCTCTTA CAGGTCCCAGACCTTGCTCCAGGAGGCCCAGGTGCTGCAGGGGTCCCCGGAGCTCCTCCCGAGGAGCCCCAAGCCCTCAGGCCTGCAAAG GCTGGCTCCAGAGGAGGCTACAGCCCTCCCCCTGAGGAGACTATGCCATTTGAGCTTGATGGAGAAGGATTTGGGGACGACAGCCCACCC CCGGGGCTTTCCCGAGTTATCGCACAAGTCGACGGCAGCAGCCAGTTCGCGGCAGTCGCGGCCTCGAGTGCGGTCCGCCTCACTCCCGCC GCGAACGCGCCTCCCCTCTGGGTCCCAGGCGCCATCGGCAGCCCATCCCAAGAGGCTGTCAGACCTCCTTCTAACTTCACGGGCAGCAGC CCCTGGATGGAGATCTCCGGACCCCCGTTCGAGATTGGCAGCGCCCCCGCTGGGGTCGACGACACTCCCGTCAACATGGACAGCCCCCCA ATCGCGCTTGACGGCCCGCCCATCAAGGTCTCCGGAGCCCCAGATAAGAGAGAGCGAGCAGAGAGACCCCCAGTTGAGGAGGAAGCAGCA GAGATGGAAGGAGCCGCTGATGCCGCGGAGGGAGGAAAAGTACCCTCTCCGGGGTACGGATCCCCTGCCGCCGGGGCAGCCTCAGCGGAT ACCGCTGCCAGGGCAGCCCCTGCAGCCCCAGCCGATCCTGACTCCGGGGCAACCCCAGAAGATCCCGACTCCGGGACAGCACCAGCCGAT CCTGACTCCGGGGCATTCGCAGCCGATCCCGACTCCGGGGCAGCCCCTGCCGCCCCAGCCGATCCCGACTCCGGGGCGGCCCCTGACGCC CCAGCCGATCCCGACTCCGGGGCGGCCCCTGACGCCCCAGCCGATCCAGATGCCGGGGCGGCCCCTGAGGCTCCCGCCGCCCCTGCGGCT GCTGAGACCCGGGCAGCCCATGTCGCCCCAGCTGCGCCAGACGCAGGGGCTCCCACTGCCCCAGCCGCTTCTGCCACCCGGGCAGCCCAA GTCCGCCGGGCGGCCTCTGCAGCCCCTGCCTCCGGGGCCAGACGCAAGATCCATCTCAGACCCCCCAGCCCCGAGATCCAGGCTGCCGAT CCGCCTACTCCGCGGCCTACTCGCGCGTCTGCCTGGCGGGGCAAGTCCGAGAGCAGCCGCGGCCGCCGCGTGTACTACGATGAAGGGGTG GCCAGCAGCGACGATGACTCCAGCGGAGACGAGTCCGACGATGGGACCTCCGGATGCCTCCGCTGGTTTCAGCATCGGCGAAATCGCCGC CGCCGAAAGCCCCAGCGCAACTTACTCCGCAACTTTCTCGTGCAAGCCTTCGGGGGCTGCTTCGGTCGATCTGAGAGTCCCCAGCCCAAA GCCTCGCGCTCTCTCAAGGTCAAGAAGGTACCCCTGGCGGAGAAGCGCAGACAGATGCGCAAAGAAGCCCTGGAGAAGCGGGCCCAGAAG CGCGCAGAGAAGAAACGCAGTAAGCTCATCGACAAACAACTCCAGGACGAAAAGATGGGCTACATGTGTACGCACCGCCTGCTGCTTCTA GGTGCTGGAGAATCTGGTAAAAGCACCATTGTGAAGCAGATGAGGATCCTGCATGTTAATGGGTTTAATGGAGACAGTGAGAAGGCAACC AAAGTGCAGGACATCAAAAACAACCTGAAAGAGGCGATTGAAACCATTGTGGCCGCCATGAGCAACCTGGTGCCCCCCGTGGAGCTGGCC AACCCCGAGAACCAGTTCAGAGTGGACTACATCCTGAGTGTGATGAACGTGCCTGACTTTGACTTCCCTCCCGTATGACAACGAATTTGG CTACAGCAACAGGGTGGTGGACCTCATGGCCCACATGGCCTCCAAGGAGTAAGACCCCTGGACCACCAGCCCCAGCAAGAGCACAAGAGG AAGAGAGAGACCCTCACTGCTGGGGAGTCCCTGCCACACTCAGTCCCCCACCACACTGAATCTCCCCTCCTCACAGTTGCCATGTAGACC >33624_33624_14_GNAS-GAPDH_GNAS_chr20_57478846_ENST00000371102_GAPDH_chr12_6647267_ENST00000396861_length(amino acids)=774AA_BP= MGVRNCLYGNNMSGQRDIPPEIGEQPEQPPLEAPGAAAPGAGPSPAEEMETEPPHNEPIPVENDGEACGPPEVSRPNFQVLNPAFREAGA HGSYSPPPEEAMPFEAEQPSLGGFWPTLEQPGFPSGVHAGLEAFGPALMEPGAFSGARPGLGGYSPPPEEAMPFEFDQPAQRGCSQLLLQ VPDLAPGGPGAAGVPGAPPEEPQALRPAKAGSRGGYSPPPEETMPFELDGEGFGDDSPPPGLSRVIAQVDGSSQFAAVAASSAVRLTPAA NAPPLWVPGAIGSPSQEAVRPPSNFTGSSPWMEISGPPFEIGSAPAGVDDTPVNMDSPPIALDGPPIKVSGAPDKRERAERPPVEEEAAE MEGAADAAEGGKVPSPGYGSPAAGAASADTAARAAPAAPADPDSGATPEDPDSGTAPADPDSGAFAADPDSGAAPAAPADPDSGAAPDAP ADPDSGAAPDAPADPDAGAAPEAPAAPAAAETRAAHVAPAAPDAGAPTAPAASATRAAQVRRAASAAPASGARRKIHLRPPSPEIQAADP PTPRPTRASAWRGKSESSRGRRVYYDEGVASSDDDSSGDESDDGTSGCLRWFQHRRNRRRRKPQRNLLRNFLVQAFGGCFGRSESPQPKA SRSLKVKKVPLAEKRRQMRKEALEKRAQKRAEKKRSKLIDKQLQDEKMGYMCTHRLLLLGAGESGKSTIVKQMRILHVNGFNGDSEKATK -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for GNAS-GAPDH |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
| Tgene | GAPDH | chr20:57478846 | chr12:6647267 | ENST00000229239 | 7 | 9 | 2_148 | 312.6666666666667 | 336.0 | WARS1 | |
| Tgene | GAPDH | chr20:57478846 | chr12:6647267 | ENST00000396858 | 6 | 8 | 2_148 | 270.6666666666667 | 294.0 | WARS1 | |
| Tgene | GAPDH | chr20:57478846 | chr12:6647267 | ENST00000396859 | 6 | 8 | 2_148 | 312.6666666666667 | 336.0 | WARS1 | |
| Tgene | GAPDH | chr20:57478846 | chr12:6647267 | ENST00000396861 | 7 | 9 | 2_148 | 312.6666666666667 | 336.0 | WARS1 |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for GNAS-GAPDH |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for GNAS-GAPDH |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
