
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:AKAP8-BRD4 (FusionGDB2 ID:3365) |
Fusion Gene Summary for AKAP8-BRD4 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: AKAP8-BRD4 | Fusion gene ID: 3365 | Hgene | Tgene | Gene symbol | AKAP8 | BRD4 | Gene ID | 10270 | 23476 |
| Gene name | A-kinase anchoring protein 8 | bromodomain containing 4 | |
| Synonyms | AKAP 95|AKAP-8|AKAP-95|AKAP95 | CAP|HUNK1|HUNKI|MCAP | |
| Cytomap | 19p13.12 | 19p13.12 | |
| Type of gene | protein-coding | protein-coding | |
| Description | A-kinase anchor protein 8A kinase (PRKA) anchor protein 8A-kinase anchor protein, 95kDa | bromodomain-containing protein 4chromosome-associated proteinmitotic chromosome-associated protein | |
| Modification date | 20200313 | 20200329 | |
| UniProtAcc | O43823 | O60885 | |
| Ensembl transtripts involved in fusion gene | ENST00000269701, | ENST00000602230, ENST00000263377, ENST00000360016, ENST00000371835, | |
| Fusion gene scores | * DoF score | 13 X 14 X 9=1638 | 15 X 19 X 13=3705 |
| # samples | 17 | 28 | |
| ** MAII score | log2(17/1638*10)=-3.26832870550331 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(28/3705*10)=-3.72597481024823 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: AKAP8 [Title/Abstract] AND BRD4 [Title/Abstract] AND fusion [Title/Abstract] | ||
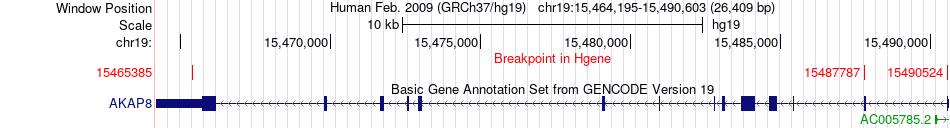
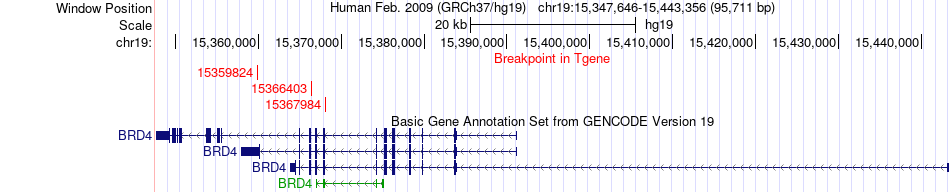
| Most frequent breakpoint | AKAP8(15465385)-BRD4(15359824), # samples:1 AKAP8(15487787)-BRD4(15367984), # samples:1 AKAP8(15490524)-BRD4(15367984), # samples:1 AKAP8(15487787)-BRD4(15366403), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | AKAP8-BRD4 seems lost the major protein functional domain in Hgene partner, which is a essential gene due to the frame-shifted ORF. AKAP8-BRD4 seems lost the major protein functional domain in Hgene partner, which is a transcription factor due to the frame-shifted ORF. AKAP8-BRD4 seems lost the major protein functional domain in Tgene partner, which is a CGC due to the frame-shifted ORF. AKAP8-BRD4 seems lost the major protein functional domain in Tgene partner, which is a epigenetic factor due to the frame-shifted ORF. AKAP8-BRD4 seems lost the major protein functional domain in Tgene partner, which is a IUPHAR drug target due to the frame-shifted ORF. AKAP8-BRD4 seems lost the major protein functional domain in Tgene partner, which is a kinase due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Tgene | BRD4 | GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter | 19103749|23086925 |
| Tgene | BRD4 | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling | 19103749 |
| Tgene | BRD4 | GO:0045944 | positive regulation of transcription by RNA polymerase II | 23086925|23317504|24360279 |
| Tgene | BRD4 | GO:0050727 | regulation of inflammatory response | 19103749 |
| Tgene | BRD4 | GO:1901407 | regulation of phosphorylation of RNA polymerase II C-terminal domain | 23086925 |
 Fusion gene breakpoints across AKAP8 (5'-gene) Fusion gene breakpoints across AKAP8 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
 |
 Fusion gene breakpoints across BRD4 (3'-gene) Fusion gene breakpoints across BRD4 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | BLCA | TCGA-GV-A3QI-01A | AKAP8 | chr19 | 15465385 | - | BRD4 | chr19 | 15359824 | - |
| ChimerDB4 | BLCA | TCGA-ZF-AA4V-01A | AKAP8 | chr19 | 15487787 | - | BRD4 | chr19 | 15367984 | - |
| ChimerDB4 | BLCA | TCGA-ZF-AA4V-01A | AKAP8 | chr19 | 15490524 | - | BRD4 | chr19 | 15367984 | - |
| ChimerDB4 | OV | TCGA-04-1338-01A | AKAP8 | chr19 | 15487787 | - | BRD4 | chr19 | 15366403 | - |
Top |
Fusion Gene ORF analysis for AKAP8-BRD4 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-5UTR | ENST00000269701 | ENST00000602230 | AKAP8 | chr19 | 15487787 | - | BRD4 | chr19 | 15367984 | - |
| 5CDS-5UTR | ENST00000269701 | ENST00000602230 | AKAP8 | chr19 | 15490524 | - | BRD4 | chr19 | 15367984 | - |
| 5CDS-intron | ENST00000269701 | ENST00000602230 | AKAP8 | chr19 | 15487787 | - | BRD4 | chr19 | 15366403 | - |
| Frame-shift | ENST00000269701 | ENST00000263377 | AKAP8 | chr19 | 15487787 | - | BRD4 | chr19 | 15367984 | - |
| Frame-shift | ENST00000269701 | ENST00000263377 | AKAP8 | chr19 | 15487787 | - | BRD4 | chr19 | 15366403 | - |
| Frame-shift | ENST00000269701 | ENST00000360016 | AKAP8 | chr19 | 15487787 | - | BRD4 | chr19 | 15367984 | - |
| Frame-shift | ENST00000269701 | ENST00000360016 | AKAP8 | chr19 | 15487787 | - | BRD4 | chr19 | 15366403 | - |
| Frame-shift | ENST00000269701 | ENST00000371835 | AKAP8 | chr19 | 15487787 | - | BRD4 | chr19 | 15367984 | - |
| Frame-shift | ENST00000269701 | ENST00000371835 | AKAP8 | chr19 | 15487787 | - | BRD4 | chr19 | 15366403 | - |
| In-frame | ENST00000269701 | ENST00000263377 | AKAP8 | chr19 | 15490524 | - | BRD4 | chr19 | 15367984 | - |
| In-frame | ENST00000269701 | ENST00000360016 | AKAP8 | chr19 | 15490524 | - | BRD4 | chr19 | 15367984 | - |
| In-frame | ENST00000269701 | ENST00000371835 | AKAP8 | chr19 | 15490524 | - | BRD4 | chr19 | 15367984 | - |
| intron-3UTR | ENST00000269701 | ENST00000371835 | AKAP8 | chr19 | 15465385 | - | BRD4 | chr19 | 15359824 | - |
| intron-intron | ENST00000269701 | ENST00000263377 | AKAP8 | chr19 | 15465385 | - | BRD4 | chr19 | 15359824 | - |
| intron-intron | ENST00000269701 | ENST00000360016 | AKAP8 | chr19 | 15465385 | - | BRD4 | chr19 | 15359824 | - |
| intron-intron | ENST00000269701 | ENST00000602230 | AKAP8 | chr19 | 15465385 | - | BRD4 | chr19 | 15359824 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000269701 | AKAP8 | chr19 | 15490524 | - | ENST00000263377 | BRD4 | chr19 | 15367984 | - | 4369 | 80 | 14 | 2827 | 937 |
| ENST00000269701 | AKAP8 | chr19 | 15490524 | - | ENST00000371835 | BRD4 | chr19 | 15367984 | - | 3131 | 80 | 14 | 907 | 297 |
| ENST00000269701 | AKAP8 | chr19 | 15490524 | - | ENST00000360016 | BRD4 | chr19 | 15367984 | - | 1689 | 80 | 14 | 1123 | 369 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000269701 | ENST00000263377 | AKAP8 | chr19 | 15490524 | - | BRD4 | chr19 | 15367984 | - | 0.010930412 | 0.98906964 |
| ENST00000269701 | ENST00000371835 | AKAP8 | chr19 | 15490524 | - | BRD4 | chr19 | 15367984 | - | 0.00372581 | 0.99627423 |
| ENST00000269701 | ENST00000360016 | AKAP8 | chr19 | 15490524 | - | BRD4 | chr19 | 15367984 | - | 0.007903014 | 0.992097 |
Top |
Fusion Genomic Features for AKAP8-BRD4 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
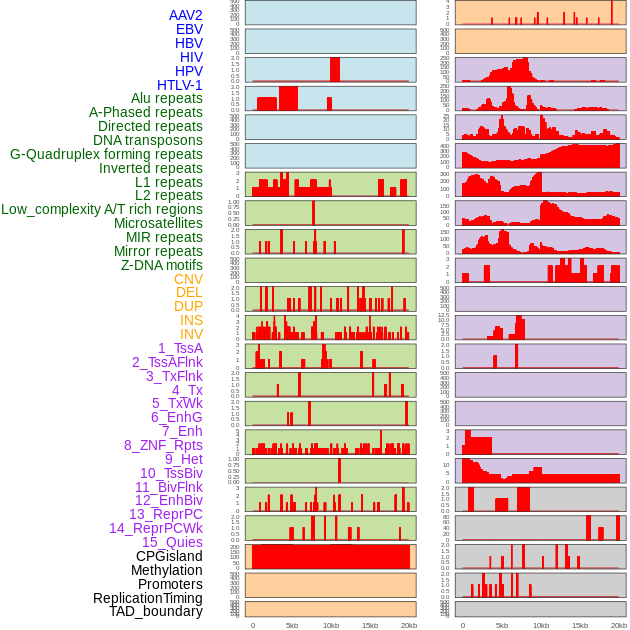
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for AKAP8-BRD4 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr19:15465385/chr19:15359824) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| AKAP8 | BRD4 |
| FUNCTION: Anchoring protein that mediates the subcellular compartmentation of cAMP-dependent protein kinase (PKA type II) (PubMed:9473338). Acts as an anchor for a PKA-signaling complex onto mitotic chromosomes, which is required for maintenance of chromosomes in a condensed form throughout mitosis. Recruits condensin complex subunit NCAPD2 to chromosomes required for chromatin condensation; the function appears to be independent from PKA-anchoring (PubMed:10601332, PubMed:10791967, PubMed:11964380). May help to deliver cyclin D/E to CDK4 to facilitate cell cycle progression (PubMed:14641107). Required for cell cycle G2/M transition and histone deacetylation during mitosis. In mitotic cells recruits HDAC3 to the vicinity of chromatin leading to deacetylation and subsequent phosphorylation at 'Ser-10' of histone H3; in this function may act redundantly with AKAP8L (PubMed:16980585). Involved in nuclear retention of RPS6KA1 upon ERK activation thus inducing cell proliferation (PubMed:22130794). May be involved in regulation of DNA replication by acting as scaffold for MCM2 (PubMed:12740381). Enhances HMT activity of the KMT2 family MLL4/WBP7 complex and is involved in transcriptional regulation. In a teratocarcinoma cell line is involved in retinoic acid-mediated induction of developmental genes implicating H3 'Lys-4' methylation (PubMed:23995757). May be involved in recruitment of active CASP3 to the nucleus in apoptotic cells (PubMed:16227597). May act as a carrier protein of GJA1 for its transport to the nucleus (PubMed:26880274). May play a repressive role in the regulation of rDNA transcription. Preferentially binds GC-rich DNA in vitro. In cells, associates with ribosomal RNA (rRNA) chromatin, preferentially with rRNA promoter and transcribed regions (PubMed:26683827). Involved in modulation of Toll-like receptor signaling. Required for the cAMP-dependent suppression of TNF-alpha in early stages of LPS-induced macrophage activation; the function probably implicates targeting of PKA to NFKB1 (By similarity). {ECO:0000250|UniProtKB:Q63014, ECO:0000250|UniProtKB:Q9DBR0, ECO:0000269|PubMed:10601332, ECO:0000269|PubMed:10791967, ECO:0000269|PubMed:11964380, ECO:0000269|PubMed:16980585, ECO:0000269|PubMed:22130794, ECO:0000269|PubMed:26683827, ECO:0000269|PubMed:26880274, ECO:0000305|PubMed:14641107, ECO:0000305|PubMed:9473338}. | FUNCTION: Chromatin reader protein that recognizes and binds acetylated histones and plays a key role in transmission of epigenetic memory across cell divisions and transcription regulation. Remains associated with acetylated chromatin throughout the entire cell cycle and provides epigenetic memory for postmitotic G1 gene transcription by preserving acetylated chromatin status and maintaining high-order chromatin structure (PubMed:23589332, PubMed:23317504, PubMed:22334664). During interphase, plays a key role in regulating the transcription of signal-inducible genes by associating with the P-TEFb complex and recruiting it to promoters. Also recruits P-TEFb complex to distal enhancers, so called anti-pause enhancers in collaboration with JMJD6. BRD4 and JMJD6 are required to form the transcriptionally active P-TEFb complex by displacing negative regulators such as HEXIM1 and 7SKsnRNA complex from P-TEFb, thereby transforming it into an active form that can then phosphorylate the C-terminal domain (CTD) of RNA polymerase II (PubMed:23589332, PubMed:19596240, PubMed:16109377, PubMed:16109376, PubMed:24360279). Promotes phosphorylation of 'Ser-2' of the C-terminal domain (CTD) of RNA polymerase II (PubMed:23086925). According to a report, directly acts as an atypical protein kinase and mediates phosphorylation of 'Ser-2' of the C-terminal domain (CTD) of RNA polymerase II; these data however need additional evidences in vivo (PubMed:22509028). In addition to acetylated histones, also recognizes and binds acetylated RELA, leading to further recruitment of the P-TEFb complex and subsequent activation of NF-kappa-B (PubMed:19103749). Also acts as a regulator of p53/TP53-mediated transcription: following phosphorylation by CK2, recruited to p53/TP53 specific target promoters (PubMed:23317504). {ECO:0000269|PubMed:16109376, ECO:0000269|PubMed:16109377, ECO:0000269|PubMed:19103749, ECO:0000269|PubMed:19596240, ECO:0000269|PubMed:22334664, ECO:0000269|PubMed:22509028, ECO:0000269|PubMed:23086925, ECO:0000269|PubMed:23317504, ECO:0000269|PubMed:23589332, ECO:0000269|PubMed:24360279}.; FUNCTION: [Isoform B]: Acts as a chromatin insulator in the DNA damage response pathway. Inhibits DNA damage response signaling by recruiting the condensin-2 complex to acetylated histones, leading to chromatin structure remodeling, insulating the region from DNA damage response by limiting spreading of histone H2AX/H2A.x phosphorylation. {ECO:0000269|PubMed:23728299}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | BRD4 | chr19:15490524 | chr19:15367984 | ENST00000263377 | 6 | 20 | 1011_1014 | 447 | 1363.0 | Compositional bias | Note=Poly-Pro | |
| Tgene | BRD4 | chr19:15490524 | chr19:15367984 | ENST00000263377 | 6 | 20 | 1028_1033 | 447 | 1363.0 | Compositional bias | Note=Poly-Pro | |
| Tgene | BRD4 | chr19:15490524 | chr19:15367984 | ENST00000263377 | 6 | 20 | 1283_1300 | 447 | 1363.0 | Compositional bias | Note=Poly-Gln | |
| Tgene | BRD4 | chr19:15490524 | chr19:15367984 | ENST00000263377 | 6 | 20 | 1301_1308 | 447 | 1363.0 | Compositional bias | Note=Poly-Ala | |
| Tgene | BRD4 | chr19:15490524 | chr19:15367984 | ENST00000263377 | 6 | 20 | 1335_1338 | 447 | 1363.0 | Compositional bias | Note=Poly-Arg | |
| Tgene | BRD4 | chr19:15490524 | chr19:15367984 | ENST00000263377 | 6 | 20 | 535_594 | 447 | 1363.0 | Compositional bias | Note=Lys-rich | |
| Tgene | BRD4 | chr19:15490524 | chr19:15367984 | ENST00000263377 | 6 | 20 | 692_717 | 447 | 1363.0 | Compositional bias | Note=Ser-rich | |
| Tgene | BRD4 | chr19:15490524 | chr19:15367984 | ENST00000263377 | 6 | 20 | 703_714 | 447 | 1363.0 | Compositional bias | Note=Poly-Ser | |
| Tgene | BRD4 | chr19:15490524 | chr19:15367984 | ENST00000263377 | 6 | 20 | 738_743 | 447 | 1363.0 | Compositional bias | Note=Poly-His | |
| Tgene | BRD4 | chr19:15490524 | chr19:15367984 | ENST00000263377 | 6 | 20 | 757_761 | 447 | 1363.0 | Compositional bias | Note=Poly-Pro | |
| Tgene | BRD4 | chr19:15490524 | chr19:15367984 | ENST00000263377 | 6 | 20 | 764_770 | 447 | 1363.0 | Compositional bias | Note=Poly-Pro | |
| Tgene | BRD4 | chr19:15490524 | chr19:15367984 | ENST00000263377 | 6 | 20 | 771_775 | 447 | 1363.0 | Compositional bias | Note=Poly-Gln | |
| Tgene | BRD4 | chr19:15490524 | chr19:15367984 | ENST00000263377 | 6 | 20 | 776_783 | 447 | 1363.0 | Compositional bias | Note=Poly-Pro | |
| Tgene | BRD4 | chr19:15490524 | chr19:15367984 | ENST00000263377 | 6 | 20 | 954_964 | 447 | 1363.0 | Compositional bias | Note=Poly-Pro | |
| Tgene | BRD4 | chr19:15490524 | chr19:15367984 | ENST00000263377 | 6 | 20 | 974_986 | 447 | 1363.0 | Compositional bias | Note=Poly-Pro | |
| Tgene | BRD4 | chr19:15490524 | chr19:15367984 | ENST00000360016 | 6 | 12 | 1011_1014 | 447 | 795.0 | Compositional bias | Note=Poly-Pro | |
| Tgene | BRD4 | chr19:15490524 | chr19:15367984 | ENST00000360016 | 6 | 12 | 1028_1033 | 447 | 795.0 | Compositional bias | Note=Poly-Pro | |
| Tgene | BRD4 | chr19:15490524 | chr19:15367984 | ENST00000360016 | 6 | 12 | 1283_1300 | 447 | 795.0 | Compositional bias | Note=Poly-Gln | |
| Tgene | BRD4 | chr19:15490524 | chr19:15367984 | ENST00000360016 | 6 | 12 | 1301_1308 | 447 | 795.0 | Compositional bias | Note=Poly-Ala | |
| Tgene | BRD4 | chr19:15490524 | chr19:15367984 | ENST00000360016 | 6 | 12 | 1335_1338 | 447 | 795.0 | Compositional bias | Note=Poly-Arg | |
| Tgene | BRD4 | chr19:15490524 | chr19:15367984 | ENST00000360016 | 6 | 12 | 535_594 | 447 | 795.0 | Compositional bias | Note=Lys-rich | |
| Tgene | BRD4 | chr19:15490524 | chr19:15367984 | ENST00000360016 | 6 | 12 | 692_717 | 447 | 795.0 | Compositional bias | Note=Ser-rich | |
| Tgene | BRD4 | chr19:15490524 | chr19:15367984 | ENST00000360016 | 6 | 12 | 703_714 | 447 | 795.0 | Compositional bias | Note=Poly-Ser | |
| Tgene | BRD4 | chr19:15490524 | chr19:15367984 | ENST00000360016 | 6 | 12 | 738_743 | 447 | 795.0 | Compositional bias | Note=Poly-His | |
| Tgene | BRD4 | chr19:15490524 | chr19:15367984 | ENST00000360016 | 6 | 12 | 757_761 | 447 | 795.0 | Compositional bias | Note=Poly-Pro | |
| Tgene | BRD4 | chr19:15490524 | chr19:15367984 | ENST00000360016 | 6 | 12 | 764_770 | 447 | 795.0 | Compositional bias | Note=Poly-Pro | |
| Tgene | BRD4 | chr19:15490524 | chr19:15367984 | ENST00000360016 | 6 | 12 | 771_775 | 447 | 795.0 | Compositional bias | Note=Poly-Gln | |
| Tgene | BRD4 | chr19:15490524 | chr19:15367984 | ENST00000360016 | 6 | 12 | 776_783 | 447 | 795.0 | Compositional bias | Note=Poly-Pro | |
| Tgene | BRD4 | chr19:15490524 | chr19:15367984 | ENST00000360016 | 6 | 12 | 954_964 | 447 | 795.0 | Compositional bias | Note=Poly-Pro | |
| Tgene | BRD4 | chr19:15490524 | chr19:15367984 | ENST00000360016 | 6 | 12 | 974_986 | 447 | 795.0 | Compositional bias | Note=Poly-Pro | |
| Tgene | BRD4 | chr19:15490524 | chr19:15367984 | ENST00000371835 | 6 | 12 | 1011_1014 | 447 | 723.0 | Compositional bias | Note=Poly-Pro | |
| Tgene | BRD4 | chr19:15490524 | chr19:15367984 | ENST00000371835 | 6 | 12 | 1028_1033 | 447 | 723.0 | Compositional bias | Note=Poly-Pro | |
| Tgene | BRD4 | chr19:15490524 | chr19:15367984 | ENST00000371835 | 6 | 12 | 1283_1300 | 447 | 723.0 | Compositional bias | Note=Poly-Gln | |
| Tgene | BRD4 | chr19:15490524 | chr19:15367984 | ENST00000371835 | 6 | 12 | 1301_1308 | 447 | 723.0 | Compositional bias | Note=Poly-Ala | |
| Tgene | BRD4 | chr19:15490524 | chr19:15367984 | ENST00000371835 | 6 | 12 | 1335_1338 | 447 | 723.0 | Compositional bias | Note=Poly-Arg | |
| Tgene | BRD4 | chr19:15490524 | chr19:15367984 | ENST00000371835 | 6 | 12 | 535_594 | 447 | 723.0 | Compositional bias | Note=Lys-rich | |
| Tgene | BRD4 | chr19:15490524 | chr19:15367984 | ENST00000371835 | 6 | 12 | 692_717 | 447 | 723.0 | Compositional bias | Note=Ser-rich | |
| Tgene | BRD4 | chr19:15490524 | chr19:15367984 | ENST00000371835 | 6 | 12 | 703_714 | 447 | 723.0 | Compositional bias | Note=Poly-Ser | |
| Tgene | BRD4 | chr19:15490524 | chr19:15367984 | ENST00000371835 | 6 | 12 | 738_743 | 447 | 723.0 | Compositional bias | Note=Poly-His | |
| Tgene | BRD4 | chr19:15490524 | chr19:15367984 | ENST00000371835 | 6 | 12 | 757_761 | 447 | 723.0 | Compositional bias | Note=Poly-Pro | |
| Tgene | BRD4 | chr19:15490524 | chr19:15367984 | ENST00000371835 | 6 | 12 | 764_770 | 447 | 723.0 | Compositional bias | Note=Poly-Pro | |
| Tgene | BRD4 | chr19:15490524 | chr19:15367984 | ENST00000371835 | 6 | 12 | 771_775 | 447 | 723.0 | Compositional bias | Note=Poly-Gln | |
| Tgene | BRD4 | chr19:15490524 | chr19:15367984 | ENST00000371835 | 6 | 12 | 776_783 | 447 | 723.0 | Compositional bias | Note=Poly-Pro | |
| Tgene | BRD4 | chr19:15490524 | chr19:15367984 | ENST00000371835 | 6 | 12 | 954_964 | 447 | 723.0 | Compositional bias | Note=Poly-Pro | |
| Tgene | BRD4 | chr19:15490524 | chr19:15367984 | ENST00000371835 | 6 | 12 | 974_986 | 447 | 723.0 | Compositional bias | Note=Poly-Pro | |
| Tgene | BRD4 | chr19:15490524 | chr19:15367984 | ENST00000263377 | 6 | 20 | 600_682 | 447 | 1363.0 | Domain | NET | |
| Tgene | BRD4 | chr19:15490524 | chr19:15367984 | ENST00000360016 | 6 | 12 | 600_682 | 447 | 795.0 | Domain | NET | |
| Tgene | BRD4 | chr19:15490524 | chr19:15367984 | ENST00000371835 | 6 | 12 | 600_682 | 447 | 723.0 | Domain | NET | |
| Tgene | BRD4 | chr19:15490524 | chr19:15367984 | ENST00000263377 | 6 | 20 | 1047_1362 | 447 | 1363.0 | Region | Note=C-terminal (CTD) region | |
| Tgene | BRD4 | chr19:15490524 | chr19:15367984 | ENST00000263377 | 6 | 20 | 484_503 | 447 | 1363.0 | Region | Note=NPS region | |
| Tgene | BRD4 | chr19:15490524 | chr19:15367984 | ENST00000263377 | 6 | 20 | 524_579 | 447 | 1363.0 | Region | Note=BID region | |
| Tgene | BRD4 | chr19:15490524 | chr19:15367984 | ENST00000360016 | 6 | 12 | 1047_1362 | 447 | 795.0 | Region | Note=C-terminal (CTD) region | |
| Tgene | BRD4 | chr19:15490524 | chr19:15367984 | ENST00000360016 | 6 | 12 | 484_503 | 447 | 795.0 | Region | Note=NPS region | |
| Tgene | BRD4 | chr19:15490524 | chr19:15367984 | ENST00000360016 | 6 | 12 | 524_579 | 447 | 795.0 | Region | Note=BID region | |
| Tgene | BRD4 | chr19:15490524 | chr19:15367984 | ENST00000371835 | 6 | 12 | 1047_1362 | 447 | 723.0 | Region | Note=C-terminal (CTD) region | |
| Tgene | BRD4 | chr19:15490524 | chr19:15367984 | ENST00000371835 | 6 | 12 | 484_503 | 447 | 723.0 | Region | Note=NPS region | |
| Tgene | BRD4 | chr19:15490524 | chr19:15367984 | ENST00000371835 | 6 | 12 | 524_579 | 447 | 723.0 | Region | Note=BID region |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | AKAP8 | chr19:15490524 | chr19:15367984 | ENST00000269701 | - | 1 | 14 | 107_118 | 6 | 693.0 | Compositional bias | Note=Poly-Gly |
| Hgene | AKAP8 | chr19:15490524 | chr19:15367984 | ENST00000269701 | - | 1 | 14 | 289_306 | 6 | 693.0 | Motif | Bipartite nuclear localization signal |
| Hgene | AKAP8 | chr19:15490524 | chr19:15367984 | ENST00000269701 | - | 1 | 14 | 387_450 | 6 | 693.0 | Region | Involved in chromatin-binding |
| Hgene | AKAP8 | chr19:15490524 | chr19:15367984 | ENST00000269701 | - | 1 | 14 | 525_569 | 6 | 693.0 | Region | Involved in condensin complex recruitment |
| Hgene | AKAP8 | chr19:15490524 | chr19:15367984 | ENST00000269701 | - | 1 | 14 | 572_589 | 6 | 693.0 | Region | RII-binding |
| Hgene | AKAP8 | chr19:15490524 | chr19:15367984 | ENST00000269701 | - | 1 | 14 | 392_414 | 6 | 693.0 | Zinc finger | C2H2 AKAP95-type 1 |
| Hgene | AKAP8 | chr19:15490524 | chr19:15367984 | ENST00000269701 | - | 1 | 14 | 481_504 | 6 | 693.0 | Zinc finger | C2H2 AKAP95-type 2 |
| Tgene | BRD4 | chr19:15490524 | chr19:15367984 | ENST00000263377 | 6 | 20 | 368_440 | 447 | 1363.0 | Domain | Bromo 2 | |
| Tgene | BRD4 | chr19:15490524 | chr19:15367984 | ENST00000263377 | 6 | 20 | 75_147 | 447 | 1363.0 | Domain | Bromo 1 | |
| Tgene | BRD4 | chr19:15490524 | chr19:15367984 | ENST00000360016 | 6 | 12 | 368_440 | 447 | 795.0 | Domain | Bromo 2 | |
| Tgene | BRD4 | chr19:15490524 | chr19:15367984 | ENST00000360016 | 6 | 12 | 75_147 | 447 | 795.0 | Domain | Bromo 1 | |
| Tgene | BRD4 | chr19:15490524 | chr19:15367984 | ENST00000371835 | 6 | 12 | 368_440 | 447 | 723.0 | Domain | Bromo 2 | |
| Tgene | BRD4 | chr19:15490524 | chr19:15367984 | ENST00000371835 | 6 | 12 | 75_147 | 447 | 723.0 | Domain | Bromo 1 |
Top |
Fusion Gene Sequence for AKAP8-BRD4 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >3365_3365_1_AKAP8-BRD4_AKAP8_chr19_15490524_ENST00000269701_BRD4_chr19_15367984_ENST00000263377_length(transcript)=4369nt_BP=80nt TGAACGCATGCGTGCTGTGGTCGCCTAGTAAACGGGGCTGCTGGTGGGCCGCGTCGAAGACATGGACCAGGGCTACGGAGGATGTGTTCG AAATGCGCTTTGCCAAGATGCCGGACGAGCCTGAGGAGCCAGTGGTGGCCGTGTCCTCCCCGGCAGTGCCCCCTCCCACCAAGGTTGTGG CCCCGCCCTCATCCAGCGACAGCAGCAGCGATAGCTCCTCGGACAGTGACAGTTCGACTGATGACTCTGAGGAGGAGCGAGCCCAGCGGC TGGCTGAGCTCCAGGAGCAGCTCAAAGCCGTGCACGAGCAGCTTGCAGCCCTCTCTCAGCCCCAGCAGAACAAACCAAAGAAAAAGGAGA AAGACAAGAAGGAAAAGAAAAAAGAAAAGCACAAAAGGAAAGAGGAAGTGGAAGAGAATAAAAAAAGCAAAGCCAAGGAACCTCCTCCTA AAAAGACGAAGAAAAATAATAGCAGCAACAGCAATGTGAGCAAGAAGGAGCCAGCGCCCATGAAGAGCAAGCCCCCTCCCACGTATGAGT CGGAGGAAGAGGACAAGTGCAAGCCTATGTCCTATGAGGAGAAGCGGCAGCTCAGCTTGGACATCAACAAGCTCCCCGGCGAGAAGCTGG GCCGCGTGGTGCACATCATCCAGTCACGGGAGCCCTCCCTGAAGAATTCCAACCCCGACGAGATTGAAATCGACTTTGAGACCCTGAAGC CGTCCACACTGCGTGAGCTGGAGCGCTATGTCACCTCCTGTTTGCGGAAGAAAAGGAAACCTCAAGCTGAGAAAGTTGATGTGATTGCCG GCTCCTCCAAGATGAAGGGCTTCTCGTCCTCAGAGTCGGAGAGCTCCAGTGAGTCCAGCTCCTCTGACAGCGAAGACTCCGAAACAGAGA TGGCTCCGAAGTCAAAAAAGAAGGGGCACCCCGGGAGGGAGCAGAAGAAGCACCATCATCACCACCATCAGCAGATGCAGCAGGCCCCGG CTCCTGTGCCCCAGCAGCCGCCCCCGCCTCCCCAGCAGCCCCCACCGCCTCCACCTCCGCAGCAGCAACAGCAGCCGCCACCCCCGCCTC CCCCACCCTCCATGCCGCAGCAGGCAGCCCCGGCGATGAAGTCCTCGCCCCCACCCTTCATTGCCACCCAGGTGCCCGTCCTGGAGCCCC AGCTCCCAGGCAGCGTCTTTGACCCCATCGGCCACTTCACCCAGCCCATCCTGCACCTGCCGCAGCCTGAGCTGCCCCCTCACCTGCCCC AGCCGCCTGAGCACAGCACTCCACCCCATCTCAACCAGCACGCAGTGGTCTCTCCTCCAGCTTTGCACAACGCACTACCCCAGCAGCCAT CACGGCCCAGCAACCGAGCCGCTGCCCTGCCTCCCAAGCCCGCCCGGCCCCCAGCCGTGTCACCAGCCTTGACCCAAACACCCCTGCTCC CACAGCCCCCCATGGCCCAACCCCCCCAAGTGCTGCTGGAGGATGAAGAGCCACCTGCCCCACCCCTCACCTCCATGCAGATGCAGCTGT ACCTGCAGCAGCTGCAGAAGGTGCAGCCCCCTACGCCGCTACTCCCTTCCGTGAAGGTGCAGTCCCAGCCCCCACCCCCCCTGCCGCCCC CACCCCACCCCTCTGTGCAGCAGCAGCTGCAGCAGCAGCCGCCACCACCCCCACCACCCCAGCCCCAGCCTCCACCCCAGCAGCAGCATC AGCCCCCTCCACGGCCCGTGCACTTGCAGCCCATGCAGTTTTCCACCCACATCCAACAGCCCCCGCCACCCCAGGGCCAGCAGCCCCCCC ATCCGCCCCCAGGCCAGCAGCCACCCCCGCCGCAGCCTGCCAAGCCTCAGCAAGTCATCCAGCACCACCATTCACCCCGGCACCACAAGT CGGACCCCTACTCAACCGGTCACCTCCGCGAAGCCCCCTCCCCGCTTATGATACATTCCCCCCAGATGTCACAGTTCCAGAGCCTGACCC ACCAGTCTCCACCCCAGCAAAACGTCCAGCCTAAGAAACAGGAGCTGCGTGCTGCCTCCGTGGTCCAGCCCCAGCCCCTCGTGGTGGTGA AGGAGGAGAAGATCCACTCACCCATCATCCGCAGCGAGCCCTTCAGCCCCTCGCTGCGGCCGGAGCCCCCCAAGCACCCGGAGAGCATCA AGGCCCCCGTCCACCTGCCCCAGCGGCCGGAAATGAAGCCTGTGGATGTCGGGAGGCCTGTGATCCGGCCCCCAGAGCAGAACGCACCGC CACCAGGGGCCCCTGACAAGGACAAACAGAAACAGGAGCCGAAGACTCCAGTTGCGCCCAAAAAGGACCTGAAAATCAAGAACATGGGCT CCTGGGCCAGCCTAGTGCAGAAGCATCCGACCACCCCCTCCTCCACAGCCAAGTCATCCAGCGACAGCTTCGAGCAGTTCCGCCGCGCCG CTCGGGAGAAAGAGGAGCGTGAGAAGGCCCTGAAGGCTCAGGCCGAGCACGCTGAGAAGGAGAAGGAGCGGCTGCGGCAGGAGCGCATGA GGAGCCGAGAGGACGAGGATGCGCTGGAGCAGGCCCGGCGGGCCCATGAGGAGGCACGTCGGCGCCAGGAGCAGCAGCAGCAGCAGCGCC AGGAGCAACAGCAGCAGCAGCAACAGCAAGCAGCTGCGGTGGCTGCCGCCGCCACCCCACAGGCCCAGAGCTCCCAGCCCCAGTCCATGC TGGACCAGCAGAGGGAGTTGGCCCGGAAGCGGGAGCAGGAGCGAAGACGCCGGGAAGCCATGGCAGCTACCATTGACATGAATTTCCAGA GTGATCTATTGTCAATATTTGAAGAAAATCTTTTCTGAGCGCACCTAGGTGGCTTCTGACTTTGATTTTCTGGCAAAACATTGACTTTCC ATAGTGTTAGGGGCGGTGGTGGAGGTGGGATCAGCGGCCAGGGGATGCCTCAGGGCCTGGCCCTCCTGCATGCTATGCCCGGGGCAGGCC TGACGGGCAGCTGAGGATTGCAGAGCCTGTCTGCCTTACGGCCAGTCGGACAGACGTCCCGCCACCCACCACCCCTCACAGGACGTCCGC TCAGCACACGCCTTGTTACGAGCAAGTGCCGGCTGGACCCAAGCCCTGCATCCCCACATGCGGGGCAGAGGCCCTTCTCTCCGCCAAATG TCTACACAGTATACACAGGACATCGTTGCTGCCGCCGTGACTGGTTTTCTGTCCCCAAGAACGTGACGTTCGTGATGTCCTGCCCGCCGG GAGTCTTTCCCCACACCCCAGCCATCGCCGCCCGCTCCCAGGAGGCCAGGGCAGGCCTGCGTGGGCTGGAGGCGGGCGAGGCCGGCCCAC CCCCTCGCTGGCACTGACTTTGCCTTGAACAGACCCCCCGACCCTCCCCCACAAGCCTTTAATTGAGAGCCGCTCTCTGTAAGTGTTTGC TTGTGCAAAAGGGAATAGTGCCGTGGAGGTGTGTGTGTCCATGGCATCCGGAGCGAGGCGACTGTCCTGCGTGGGTAGCCCTCGGCCGGG GAGTGAGGCCACCAACCAAAGTCAGTTCCTTCCCACCTGTGTTTCTGTTTCGTTTTTTTTTTTCTTTTTTTTCTATATATATTTTTTGTT GAATTCTATTTTATTTTTAATTCTCTCTTCTCCTCCAGACACAATGGCACTGCTTATCTCCGAAATGGTGTGATCGTCTCCTCATTGAGC AGCGGCTGCCACCGCGCTGTGGGTAGTGTGTGACCGTGGCTGTACTGTATAGTGAACATAGTTGGCATATCTTTGTTTGAAGTTTGTTGG TGACTCCACCAAACTGGTGTGAAAAAAGAAAAAAGCTCAAAAAAATCCACAAAAAGACAAAACACACAAAAAAAATCCTGCCTATATTTT ACTCAGTTTCAAACTTTATTAGTCTATTTTTAATTATAAAACCAGAAAGCTACAATTTCTTTTCTTTCCCCTCCACCCCCCCCCCCCCCA CCCATTTGTTGGCTTTTTTGTTTTTTAATGTCAGATCTGTTGAGTTGGTTTTTTTGGTTTTGGTTTTTGTTTTTGTTTTTGTTTTTTACT GAGAAAGGAAGGGCCAAGGGATGAGGTGGGAACCGGGCCCTGGGGGCGCCACAGACTAAGGCAGAGACTCCCCTACCTGGCGCCCAGCCC CAACCAGCTGGCCGCTCCTGCCCATGCTTTTTTTTTTTTTTTTTTTTAATTTTTATAATTGGAGCCCCTGGTGAGGTTACGCGTGCCATG AGAACCCACTCTACACCACGACGCTGGTGCCTCAGTGTTGGCCAAACTCTGGAGTCACTGACTGGTTTGACTTTCATACGGTGAATATGC >3365_3365_1_AKAP8-BRD4_AKAP8_chr19_15490524_ENST00000269701_BRD4_chr19_15367984_ENST00000263377_length(amino acids)=937AA_BP=547 MWSPSKRGCWWAASKTWTRATEDVFEMRFAKMPDEPEEPVVAVSSPAVPPPTKVVAPPSSSDSSSDSSSDSDSSTDDSEEERAQRLAELQ EQLKAVHEQLAALSQPQQNKPKKKEKDKKEKKKEKHKRKEEVEENKKSKAKEPPPKKTKKNNSSNSNVSKKEPAPMKSKPPPTYESEEED KCKPMSYEEKRQLSLDINKLPGEKLGRVVHIIQSREPSLKNSNPDEIEIDFETLKPSTLRELERYVTSCLRKKRKPQAEKVDVIAGSSKM KGFSSSESESSSESSSSDSEDSETEMAPKSKKKGHPGREQKKHHHHHHQQMQQAPAPVPQQPPPPPQQPPPPPPPQQQQQPPPPPPPPSM PQQAAPAMKSSPPPFIATQVPVLEPQLPGSVFDPIGHFTQPILHLPQPELPPHLPQPPEHSTPPHLNQHAVVSPPALHNALPQQPSRPSN RAAALPPKPARPPAVSPALTQTPLLPQPPMAQPPQVLLEDEEPPAPPLTSMQMQLYLQQLQKVQPPTPLLPSVKVQSQPPPPLPPPPHPS VQQQLQQQPPPPPPPQPQPPPQQQHQPPPRPVHLQPMQFSTHIQQPPPPQGQQPPHPPPGQQPPPPQPAKPQQVIQHHHSPRHHKSDPYS TGHLREAPSPLMIHSPQMSQFQSLTHQSPPQQNVQPKKQELRAASVVQPQPLVVVKEEKIHSPIIRSEPFSPSLRPEPPKHPESIKAPVH LPQRPEMKPVDVGRPVIRPPEQNAPPPGAPDKDKQKQEPKTPVAPKKDLKIKNMGSWASLVQKHPTTPSSTAKSSSDSFEQFRRAAREKE EREKALKAQAEHAEKEKERLRQERMRSREDEDALEQARRAHEEARRRQEQQQQQRQEQQQQQQQQAAAVAAAATPQAQSSQPQSMLDQQR -------------------------------------------------------------- >3365_3365_2_AKAP8-BRD4_AKAP8_chr19_15490524_ENST00000269701_BRD4_chr19_15367984_ENST00000360016_length(transcript)=1689nt_BP=80nt TGAACGCATGCGTGCTGTGGTCGCCTAGTAAACGGGGCTGCTGGTGGGCCGCGTCGAAGACATGGACCAGGGCTACGGAGGATGTGTTCG AAATGCGCTTTGCCAAGATGCCGGACGAGCCTGAGGAGCCAGTGGTGGCCGTGTCCTCCCCGGCAGTGCCCCCTCCCACCAAGGTTGTGG CCCCGCCCTCATCCAGCGACAGCAGCAGCGATAGCTCCTCGGACAGTGACAGTTCGACTGATGACTCTGAGGAGGAGCGAGCCCAGCGGC TGGCTGAGCTCCAGGAGCAGCTCAAAGCCGTGCACGAGCAGCTTGCAGCCCTCTCTCAGCCCCAGCAGAACAAACCAAAGAAAAAGGAGA AAGACAAGAAGGAAAAGAAAAAAGAAAAGCACAAAAGGAAAGAGGAAGTGGAAGAGAATAAAAAAAGCAAAGCCAAGGAACCTCCTCCTA AAAAGACGAAGAAAAATAATAGCAGCAACAGCAATGTGAGCAAGAAGGAGCCAGCGCCCATGAAGAGCAAGCCCCCTCCCACGTATGAGT CGGAGGAAGAGGACAAGTGCAAGCCTATGTCCTATGAGGAGAAGCGGCAGCTCAGCTTGGACATCAACAAGCTCCCCGGCGAGAAGCTGG GCCGCGTGGTGCACATCATCCAGTCACGGGAGCCCTCCCTGAAGAATTCCAACCCCGACGAGATTGAAATCGACTTTGAGACCCTGAAGC CGTCCACACTGCGTGAGCTGGAGCGCTATGTCACCTCCTGTTTGCGGAAGAAAAGGAAACCTCAAGCTGAGAAAGTTGATGTGATTGCCG GCTCCTCCAAGATGAAGGGCTTCTCGTCCTCAGAGTCGGAGAGCTCCAGTGAGTCCAGCTCCTCTGACAGCGAAGACTCCGAAACAGCTT TCTGCACCAGTGGAGACTTCGTGTCCCCAGGGCCTTCCCCGTATCACAGTCACGTGCAGTGCGGCCGCTTCAGGGAGATGCTTCGCTGGT TTCTGGTGGATGTGGAGCAGACTGCAGCTGGCCAGCCGCATCGCCAGTCTGCTGCTGGCCCTGCCATCACCTGGGCCCCAGCCATTGCCT ACCCCTCCCCAGAGTGTGCTCGTTGCTGTGTTGGCTGCTCCTGAATCTGCCCTAACTCCACACGCACCTGGACTTGCGTGTCCCTCCTGC AGTTCTAACTAACAGTCCCTTCTTTTCAAGCCCCGTGGGTCTGCACGTTGGACCCTGGGTTCCCCATTAGAGCCCACCTTCTGAGCAGCA GCCTCAGTGGGAGGTGGAGGCAGGTAGTGATGCTGGGTGCCAGGTGGGAGTGGAGAGGGGACTGCTCTCCTCCAAGTGTGCACTTTCCTC ATTATTCTCAGGGGCGCAGATGCTCAGGCTGGCGCAGGGGAGAGGCTAGGGAGGGAGCCATGGTGCCCAGAAGGCCTGGCGACCAGCCCC TGCTGAGAGATGGAGCTAACATCCTGTGTTTACGGCCAACGGGGTTGCCGCTAGGCTGGTGCAGCTGTCAGTGAGCTGGCGTGCTGCAGA CCACCTGAGAGCTGGCCCTAGGGTCTCAGGCAGACTGGGGAGTGGGGATCCACAGTGGGAAACCTGTGTTTTGGCAGTAGACTCCTGCAT >3365_3365_2_AKAP8-BRD4_AKAP8_chr19_15490524_ENST00000269701_BRD4_chr19_15367984_ENST00000360016_length(amino acids)=369AA_BP=1 MWSPSKRGCWWAASKTWTRATEDVFEMRFAKMPDEPEEPVVAVSSPAVPPPTKVVAPPSSSDSSSDSSSDSDSSTDDSEEERAQRLAELQ EQLKAVHEQLAALSQPQQNKPKKKEKDKKEKKKEKHKRKEEVEENKKSKAKEPPPKKTKKNNSSNSNVSKKEPAPMKSKPPPTYESEEED KCKPMSYEEKRQLSLDINKLPGEKLGRVVHIIQSREPSLKNSNPDEIEIDFETLKPSTLRELERYVTSCLRKKRKPQAEKVDVIAGSSKM KGFSSSESESSSESSSSDSEDSETAFCTSGDFVSPGPSPYHSHVQCGRFREMLRWFLVDVEQTAAGQPHRQSAAGPAITWAPAIAYPSPE -------------------------------------------------------------- >3365_3365_3_AKAP8-BRD4_AKAP8_chr19_15490524_ENST00000269701_BRD4_chr19_15367984_ENST00000371835_length(transcript)=3131nt_BP=80nt TGAACGCATGCGTGCTGTGGTCGCCTAGTAAACGGGGCTGCTGGTGGGCCGCGTCGAAGACATGGACCAGGGCTACGGAGGATGTGTTCG AAATGCGCTTTGCCAAGATGCCGGACGAGCCTGAGGAGCCAGTGGTGGCCGTGTCCTCCCCGGCAGTGCCCCCTCCCACCAAGGTTGTGG CCCCGCCCTCATCCAGCGACAGCAGCAGCGATAGCTCCTCGGACAGTGACAGTTCGACTGATGACTCTGAGGAGGAGCGAGCCCAGCGGC TGGCTGAGCTCCAGGAGCAGCTCAAAGCCGTGCACGAGCAGCTTGCAGCCCTCTCTCAGCCCCAGCAGAACAAACCAAAGAAAAAGGAGA AAGACAAGAAGGAAAAGAAAAAAGAAAAGCACAAAAGGAAAGAGGAAGTGGAAGAGAATAAAAAAAGCAAAGCCAAGGAACCTCCTCCTA AAAAGACGAAGAAAAATAATAGCAGCAACAGCAATGTGAGCAAGAAGGAGCCAGCGCCCATGAAGAGCAAGCCCCCTCCCACGTATGAGT CGGAGGAAGAGGACAAGTGCAAGCCTATGTCCTATGAGGAGAAGCGGCAGCTCAGCTTGGACATCAACAAGCTCCCCGGCGAGAAGCTGG GCCGCGTGGTGCACATCATCCAGTCACGGGAGCCCTCCCTGAAGAATTCCAACCCCGACGAGATTGAAATCGACTTTGAGACCCTGAAGC CGTCCACACTGCGTGAGCTGGAGCGCTATGTCACCTCCTGTTTGCGGAAGAAAAGGAAACCTCAAGCTGAGAAAGTTGATGTGATTGCCG GCTCCTCCAAGATGAAGGGCTTCTCGTCCTCAGAGTCGGAGAGCTCCAGTGAGTCCAGCTCCTCTGACAGCGAAGACTCCGAAACAGGTC CTGCCTAATCATTGGACACGGACTCTTAATAAAACGGTCTTCAGTTCCAGATTCCTTCCCAGCAAGCTATAGCTTAAGTCCATTTTCTTC CGTGAAAGGGACAGGACTCCATCAAGTTATGGAATTCCTCAGAGCCCTGGGCCTGTCCCCCGGGGTGGATTAGTCATGTCCAGCAGCACA CGCCTAGTCCCGCCTTCGGGAAGGCTGCCTGCCTGGCCAGCCGCCCAGGCCTCTCTGTGTAAAGACTGCCTGGCTGTCCTGCCCAGCCTT CCTGGTTCTCTGGGGTCCTCTGGGTGGGTGGCATCTCCTGGAGGGTGATGACAATCCCCAACACATGCATTCATGTGGTGCTACTCTGTG TGCAAAGCCAGACCCCAAGTATGTTTTCTCTCTTTGTCCCATCCCTCTTTTTCTGGGACTTTGGACCCTAACTACTTCCCTCCTGAACCT TGCAGTGACATCAGTCCAGGAGAGCTCTCGTTCAGTGTGCGGAAGAACACTCTGACCTCTAGAGCTGTCCTAGATAAGGAGTGGGAGCTT TAGAGGCAAGGCCTCTAGACCCTGGAAGGCTCAGTGAGGCTCTTCCCACAGCATGCTTCTCACTGGTGCCCTGTAAGGCTCGAGCCACCG CTGACTCTGAGCCTTTTGGAGTCTTTCCTCCTTCGTCTCCATTGTTCCCGTGCATTTCCAAAAGCTTAAGTTGCCTGGTGGGCATTTCCC CAGTTTCTTTGGCCTCCGTCTTCTCAAGTCACATAGGGAAAGTACCTCCTGGAACCAGGCTGCAGTATGCAGGACCTGCCAGGCAGGCAC TGGTGAAGGGCCTTGGGCCTATCATCCCCCCAACCCCACCTCACCCCACCCGCCTCCTCTAGTGGGGTGAGTCTGGGCTGGTGGACCAGA GAGGGTGTCACAGACCCTCAGGGACTGCCCCATGGACACCTCTGACTGGTGTTAACAGTGTGAACATTTTCCCCGTCTTCAGTCCCTTAG AATGACGACAGCCCCTGGGGTTGGGGCAGGCGAGTGTGGCCACATCATCCAAGCCCTCCCAGAGACACAAATAGGCTTTTTTGCTCTAAA AATAAATACCAGCCCTTTTTTGGTCACAAATCCAGCATCTCAGCAGAAAACTGCCTGACATGAAAAGTCCCCTGAGGAACTGCATCTGCG TTTCAGGGGCTTTTCATTTTTTCTCCTTTTTTAAAGTGTAGATTGTGGGTGCTTCCTAGAGGCCTGCCTTCTTCTGGAACTGGAAGTGGG CTATCACCATGGGCAAGCCCTTGGGTGCAGGCTCCCCACCTGCCTGGGAACTCTGGCAGCTCTCCTCAGCTCCTTGGGCTTGAGCAGCTG CAACTGCCCCAGATTTGCTGTGGAAGCAGGGGCTAGCCCTGGCCTCACCAGGGCCTCCCGGGGCCCTGCATTGATGCTCAGGAGTTCCTG GGCTGCTCTTGATCCTTTCTGGGCATCCAGCTTCCAGTTAAGCTCTGTTTGCCAAACAAACTATTCTCAGCTGCCCTTTGGCCTGCGCCT GATGTGTTCCTGTTGCAGTCCCGCCTGCCTGAGACAGGAGCAGGCAGGAGAGCCTTCATGCCCAGATTCCCACAGGACAATTGGGGAGCT GCTGGCATTGTCTTTCTGGGAAGATTCTGCTTTCTTGGACCAAATGGCAGCCTGATTACCAGTGTCGGGCCTGCATGCTGCCCCCGACAC ACGCACGCACGCGCACACACGTGTGCACATGGGCCATAGCCACAAGCCAGCTCTCCTCCAGGGTCCTTTCAACCTCGCTGTCCAGGGACC CTGTCCTTCTTGCCCGTGGGGCTTCCATCTGGCAGAGAACGTTCAGGGCTTGTTGAACTTGAAAGCTCATTAGACTTAAGCTGTCACCTG TGCTTGGTGCCCCAGGAACAGCCAGAGAGGACAGTGCCCACTCACTTCTTGTTGGCAGCCTCCTGTGCAGGAAGTGCCAGCCGGGCCTCG ACGCACCAGCTGGCTGTGGGTCCTGAGGAGGGGCGGGAGGCGGCCGCTCAGTGCAGATGGGGACTCCTCTCCTCTGCCCTGACCTTACCC TCCATTACCTCCTTCACTGGAGTGGGGCTGGGGGGTGGGTGGAATCAGTGTTTTAATCGGATTTTTAAAAAACATTTTATTTCTTTGTAC >3365_3365_3_AKAP8-BRD4_AKAP8_chr19_15490524_ENST00000269701_BRD4_chr19_15367984_ENST00000371835_length(amino acids)=297AA_BP=1 MWSPSKRGCWWAASKTWTRATEDVFEMRFAKMPDEPEEPVVAVSSPAVPPPTKVVAPPSSSDSSSDSSSDSDSSTDDSEEERAQRLAELQ EQLKAVHEQLAALSQPQQNKPKKKEKDKKEKKKEKHKRKEEVEENKKSKAKEPPPKKTKKNNSSNSNVSKKEPAPMKSKPPPTYESEEED KCKPMSYEEKRQLSLDINKLPGEKLGRVVHIIQSREPSLKNSNPDEIEIDFETLKPSTLRELERYVTSCLRKKRKPQAEKVDVIAGSSKM -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for AKAP8-BRD4 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
| Hgene | AKAP8 | chr19:15490524 | chr19:15367984 | ENST00000269701 | - | 1 | 14 | 109_201 | 6.333333333333333 | 693.0 | DDX5 |
| Hgene | AKAP8 | chr19:15490524 | chr19:15367984 | ENST00000269701 | - | 1 | 14 | 1_210 | 6.333333333333333 | 693.0 | DPY30 |
| Hgene | AKAP8 | chr19:15490524 | chr19:15367984 | ENST00000269701 | - | 1 | 14 | 1_195 | 6.333333333333333 | 693.0 | MCM2 |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for AKAP8-BRD4 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for AKAP8-BRD4 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
