
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:GPATCH8-NPLOC4 (FusionGDB2 ID:34124) |
Fusion Gene Summary for GPATCH8-NPLOC4 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: GPATCH8-NPLOC4 | Fusion gene ID: 34124 | Hgene | Tgene | Gene symbol | GPATCH8 | NPLOC4 | Gene ID | 23131 | 55666 |
| Gene name | G-patch domain containing 8 | NPL4 homolog, ubiquitin recognition factor | |
| Synonyms | GPATC8|KIAA0553 | NPL4 | |
| Cytomap | 17q21.31 | 17q25.3 | |
| Type of gene | protein-coding | protein-coding | |
| Description | G patch domain-containing protein 8 | nuclear protein localization protein 4 homologNPLOC4 ubiquitin recognition factornuclear protein localization 4 homolog | |
| Modification date | 20200320 | 20200313 | |
| UniProtAcc | Q9UKJ3 | Q8TAT6 | |
| Ensembl transtripts involved in fusion gene | ENST00000434000, ENST00000591680, ENST00000586265, ENST00000588554, ENST00000592154, | ENST00000539314, ENST00000572760, ENST00000573876, ENST00000574344, ENST00000331134, ENST00000374747, | |
| Fusion gene scores | * DoF score | 22 X 14 X 12=3696 | 15 X 9 X 10=1350 |
| # samples | 27 | 18 | |
| ** MAII score | log2(27/3696*10)=-3.77493344436523 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(18/1350*10)=-2.90689059560852 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: GPATCH8 [Title/Abstract] AND NPLOC4 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | GPATCH8(42512433)-NPLOC4(79596831), # samples:3 | ||
| Anticipated loss of major functional domain due to fusion event. | GPATCH8-NPLOC4 seems lost the major protein functional domain in Hgene partner, which is a essential gene due to the frame-shifted ORF. GPATCH8-NPLOC4 seems lost the major protein functional domain in Tgene partner, which is a essential gene due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
 Fusion gene breakpoints across GPATCH8 (5'-gene) Fusion gene breakpoints across GPATCH8 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
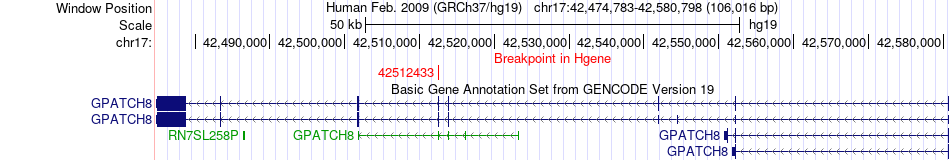 |
 Fusion gene breakpoints across NPLOC4 (3'-gene) Fusion gene breakpoints across NPLOC4 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
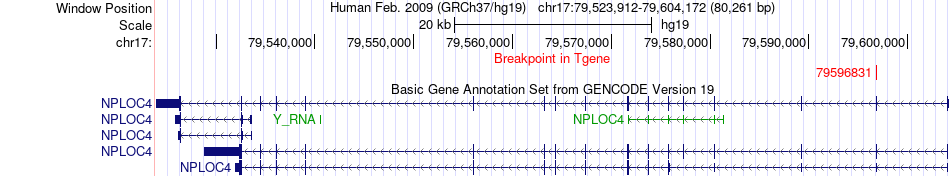 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | BRCA | TCGA-A8-A07B-01A | GPATCH8 | chr17 | 42512433 | - | NPLOC4 | chr17 | 79596831 | - |
Top |
Fusion Gene ORF analysis for GPATCH8-NPLOC4 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-5UTR | ENST00000434000 | ENST00000539314 | GPATCH8 | chr17 | 42512433 | - | NPLOC4 | chr17 | 79596831 | - |
| 5CDS-5UTR | ENST00000591680 | ENST00000539314 | GPATCH8 | chr17 | 42512433 | - | NPLOC4 | chr17 | 79596831 | - |
| 5CDS-intron | ENST00000434000 | ENST00000572760 | GPATCH8 | chr17 | 42512433 | - | NPLOC4 | chr17 | 79596831 | - |
| 5CDS-intron | ENST00000434000 | ENST00000573876 | GPATCH8 | chr17 | 42512433 | - | NPLOC4 | chr17 | 79596831 | - |
| 5CDS-intron | ENST00000434000 | ENST00000574344 | GPATCH8 | chr17 | 42512433 | - | NPLOC4 | chr17 | 79596831 | - |
| 5CDS-intron | ENST00000591680 | ENST00000572760 | GPATCH8 | chr17 | 42512433 | - | NPLOC4 | chr17 | 79596831 | - |
| 5CDS-intron | ENST00000591680 | ENST00000573876 | GPATCH8 | chr17 | 42512433 | - | NPLOC4 | chr17 | 79596831 | - |
| 5CDS-intron | ENST00000591680 | ENST00000574344 | GPATCH8 | chr17 | 42512433 | - | NPLOC4 | chr17 | 79596831 | - |
| 5UTR-3CDS | ENST00000586265 | ENST00000331134 | GPATCH8 | chr17 | 42512433 | - | NPLOC4 | chr17 | 79596831 | - |
| 5UTR-3CDS | ENST00000586265 | ENST00000374747 | GPATCH8 | chr17 | 42512433 | - | NPLOC4 | chr17 | 79596831 | - |
| 5UTR-5UTR | ENST00000586265 | ENST00000539314 | GPATCH8 | chr17 | 42512433 | - | NPLOC4 | chr17 | 79596831 | - |
| 5UTR-intron | ENST00000586265 | ENST00000572760 | GPATCH8 | chr17 | 42512433 | - | NPLOC4 | chr17 | 79596831 | - |
| 5UTR-intron | ENST00000586265 | ENST00000573876 | GPATCH8 | chr17 | 42512433 | - | NPLOC4 | chr17 | 79596831 | - |
| 5UTR-intron | ENST00000586265 | ENST00000574344 | GPATCH8 | chr17 | 42512433 | - | NPLOC4 | chr17 | 79596831 | - |
| Frame-shift | ENST00000434000 | ENST00000331134 | GPATCH8 | chr17 | 42512433 | - | NPLOC4 | chr17 | 79596831 | - |
| Frame-shift | ENST00000434000 | ENST00000374747 | GPATCH8 | chr17 | 42512433 | - | NPLOC4 | chr17 | 79596831 | - |
| In-frame | ENST00000591680 | ENST00000331134 | GPATCH8 | chr17 | 42512433 | - | NPLOC4 | chr17 | 79596831 | - |
| In-frame | ENST00000591680 | ENST00000374747 | GPATCH8 | chr17 | 42512433 | - | NPLOC4 | chr17 | 79596831 | - |
| intron-3CDS | ENST00000588554 | ENST00000331134 | GPATCH8 | chr17 | 42512433 | - | NPLOC4 | chr17 | 79596831 | - |
| intron-3CDS | ENST00000588554 | ENST00000374747 | GPATCH8 | chr17 | 42512433 | - | NPLOC4 | chr17 | 79596831 | - |
| intron-3CDS | ENST00000592154 | ENST00000331134 | GPATCH8 | chr17 | 42512433 | - | NPLOC4 | chr17 | 79596831 | - |
| intron-3CDS | ENST00000592154 | ENST00000374747 | GPATCH8 | chr17 | 42512433 | - | NPLOC4 | chr17 | 79596831 | - |
| intron-5UTR | ENST00000588554 | ENST00000539314 | GPATCH8 | chr17 | 42512433 | - | NPLOC4 | chr17 | 79596831 | - |
| intron-5UTR | ENST00000592154 | ENST00000539314 | GPATCH8 | chr17 | 42512433 | - | NPLOC4 | chr17 | 79596831 | - |
| intron-intron | ENST00000588554 | ENST00000572760 | GPATCH8 | chr17 | 42512433 | - | NPLOC4 | chr17 | 79596831 | - |
| intron-intron | ENST00000588554 | ENST00000573876 | GPATCH8 | chr17 | 42512433 | - | NPLOC4 | chr17 | 79596831 | - |
| intron-intron | ENST00000588554 | ENST00000574344 | GPATCH8 | chr17 | 42512433 | - | NPLOC4 | chr17 | 79596831 | - |
| intron-intron | ENST00000592154 | ENST00000572760 | GPATCH8 | chr17 | 42512433 | - | NPLOC4 | chr17 | 79596831 | - |
| intron-intron | ENST00000592154 | ENST00000573876 | GPATCH8 | chr17 | 42512433 | - | NPLOC4 | chr17 | 79596831 | - |
| intron-intron | ENST00000592154 | ENST00000574344 | GPATCH8 | chr17 | 42512433 | - | NPLOC4 | chr17 | 79596831 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000591680 | GPATCH8 | chr17 | 42512433 | - | ENST00000331134 | NPLOC4 | chr17 | 79596831 | - | 4563 | 379 | 31 | 2190 | 719 |
| ENST00000591680 | GPATCH8 | chr17 | 42512433 | - | ENST00000374747 | NPLOC4 | chr17 | 79596831 | - | 5772 | 379 | 31 | 2217 | 728 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000591680 | ENST00000331134 | GPATCH8 | chr17 | 42512433 | - | NPLOC4 | chr17 | 79596831 | - | 0.001668645 | 0.99833137 |
| ENST00000591680 | ENST00000374747 | GPATCH8 | chr17 | 42512433 | - | NPLOC4 | chr17 | 79596831 | - | 0.000896337 | 0.9991036 |
Top |
Fusion Genomic Features for GPATCH8-NPLOC4 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
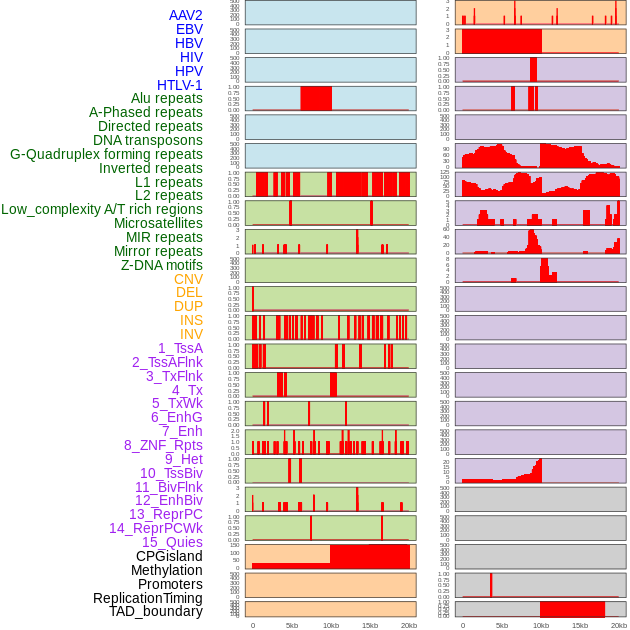 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for GPATCH8-NPLOC4 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr17:42512433/chr17:79596831) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| GPATCH8 | NPLOC4 |
| FUNCTION: The ternary complex containing UFD1, VCP and NPLOC4 binds ubiquitinated proteins and is necessary for the export of misfolded proteins from the ER to the cytoplasm, where they are degraded by the proteasome. The NPLOC4-UFD1-VCP complex regulates spindle disassembly at the end of mitosis and is necessary for the formation of a closed nuclear envelope (By similarity). Acts as a negative regulator of type I interferon production via the complex formed with VCP and UFD1, which binds to DDX58/RIG-I and recruits RNF125 to promote ubiquitination and degradation of DDX58/RIG-I (PubMed:26471729). {ECO:0000250|UniProtKB:Q9ES54, ECO:0000269|PubMed:26471729}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | GPATCH8 | chr17:42512433 | chr17:79596831 | ENST00000591680 | - | 5 | 8 | 40_86 | 116 | 1503.0 | Domain | G-patch |
| Tgene | NPLOC4 | chr17:42512433 | chr17:79596831 | ENST00000331134 | 0 | 17 | 226_363 | 5 | 609.0 | Domain | MPN | |
| Tgene | NPLOC4 | chr17:42512433 | chr17:79596831 | ENST00000374747 | 0 | 16 | 226_363 | 5 | 618.0 | Domain | MPN | |
| Tgene | NPLOC4 | chr17:42512433 | chr17:79596831 | ENST00000331134 | 0 | 17 | 580_608 | 5 | 609.0 | Zinc finger | RanBP2-type | |
| Tgene | NPLOC4 | chr17:42512433 | chr17:79596831 | ENST00000374747 | 0 | 16 | 580_608 | 5 | 618.0 | Zinc finger | RanBP2-type |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | GPATCH8 | chr17:42512433 | chr17:79596831 | ENST00000434000 | - | 6 | 9 | 89_124 | 38 | 1425.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | GPATCH8 | chr17:42512433 | chr17:79596831 | ENST00000591680 | - | 5 | 8 | 89_124 | 116 | 1503.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | GPATCH8 | chr17:42512433 | chr17:79596831 | ENST00000434000 | - | 6 | 9 | 1384_1390 | 38 | 1425.0 | Compositional bias | Note=Poly-Ala |
| Hgene | GPATCH8 | chr17:42512433 | chr17:79596831 | ENST00000434000 | - | 6 | 9 | 1472_1478 | 38 | 1425.0 | Compositional bias | Note=Poly-Ala |
| Hgene | GPATCH8 | chr17:42512433 | chr17:79596831 | ENST00000434000 | - | 6 | 9 | 665_720 | 38 | 1425.0 | Compositional bias | Note=Lys-rich |
| Hgene | GPATCH8 | chr17:42512433 | chr17:79596831 | ENST00000434000 | - | 6 | 9 | 745_749 | 38 | 1425.0 | Compositional bias | Note=Poly-Arg |
| Hgene | GPATCH8 | chr17:42512433 | chr17:79596831 | ENST00000434000 | - | 6 | 9 | 772_775 | 38 | 1425.0 | Compositional bias | Note=Poly-Gly |
| Hgene | GPATCH8 | chr17:42512433 | chr17:79596831 | ENST00000434000 | - | 6 | 9 | 804_1009 | 38 | 1425.0 | Compositional bias | Note=Ser-rich |
| Hgene | GPATCH8 | chr17:42512433 | chr17:79596831 | ENST00000591680 | - | 5 | 8 | 1384_1390 | 116 | 1503.0 | Compositional bias | Note=Poly-Ala |
| Hgene | GPATCH8 | chr17:42512433 | chr17:79596831 | ENST00000591680 | - | 5 | 8 | 1472_1478 | 116 | 1503.0 | Compositional bias | Note=Poly-Ala |
| Hgene | GPATCH8 | chr17:42512433 | chr17:79596831 | ENST00000591680 | - | 5 | 8 | 665_720 | 116 | 1503.0 | Compositional bias | Note=Lys-rich |
| Hgene | GPATCH8 | chr17:42512433 | chr17:79596831 | ENST00000591680 | - | 5 | 8 | 745_749 | 116 | 1503.0 | Compositional bias | Note=Poly-Arg |
| Hgene | GPATCH8 | chr17:42512433 | chr17:79596831 | ENST00000591680 | - | 5 | 8 | 772_775 | 116 | 1503.0 | Compositional bias | Note=Poly-Gly |
| Hgene | GPATCH8 | chr17:42512433 | chr17:79596831 | ENST00000591680 | - | 5 | 8 | 804_1009 | 116 | 1503.0 | Compositional bias | Note=Ser-rich |
| Hgene | GPATCH8 | chr17:42512433 | chr17:79596831 | ENST00000434000 | - | 6 | 9 | 40_86 | 38 | 1425.0 | Domain | G-patch |
| Hgene | GPATCH8 | chr17:42512433 | chr17:79596831 | ENST00000434000 | - | 6 | 9 | 136_160 | 38 | 1425.0 | Zinc finger | C2H2-type |
| Hgene | GPATCH8 | chr17:42512433 | chr17:79596831 | ENST00000591680 | - | 5 | 8 | 136_160 | 116 | 1503.0 | Zinc finger | C2H2-type |
Top |
Fusion Gene Sequence for GPATCH8-NPLOC4 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >34124_34124_1_GPATCH8-NPLOC4_GPATCH8_chr17_42512433_ENST00000591680_NPLOC4_chr17_79596831_ENST00000331134_length(transcript)=4563nt_BP=379nt GCGTCGTCCTGAGAGGAGTGAAGGCGGCAAAATGGCGGACCGCTTCTCCCGCTTCAACGAAGACCGAGACTTTCAGGGTAATCACTTTGA TCAGTATGAGGAAGGACACTTGGAAATTGAACAAGCGTCACTTGACAAGCCTATAGAATCGGATAATATTGGACACCGCTTACTCCAGAA ACATGGGTGGAAGCTGGGCCAGGGATTGGGAAAATCTCTTCAGGGGAGAACAGATCCCATTCCAATCGTTGTCAAGTATGATGTCATGGG CATGGGTCGCATGGAAATGGAGCTTGATTATGCTGAAGATGCTACCGAACGGCGCCGTGTCCTAGAAGTAGAAAAAGAAGACACAGAAGA GCTGAGACAAAAGTACAAGATAATTCGTGTCCAGTCCCCGGATGGAGTGAAGCGGATCACAGCAACAAAGAGAGAAACAGCAGCAACATT TTTGAAAAAGGTTGCAAAGGAGTTTGGCTTCCAAAATAATGGCTTCTCGGTTTACATCAATAGAAACAAGACCGGAGAGATAACAGCCTC CTCCAACAAATCCCTCAACTTGCTAAAAATCAAGCATGGCGATTTGTTGTTCCTGTTTCCCTCGAGCCTTGCTGGGCCCTCATCTGAAAT GGAGACGTCAGTTCCACCGGGCTTCAAAGTCTTTGGCGCTCCCAACGTGGTGGAGGATGAGATTGATCAGTACCTCAGCAAACAGGACGG GAAGATTTACAGAAGCCGAGACCCACAGCTATGCCGCCACGGCCCTTTGGGGAAATGCGTGCACTGCGTCCCTCTAGAGCCATTCGATGA GGACTATCTAAACCATCTCGAGCCTCCCGTGAAGCACATGTCCTTCCACGCCTACATCCGGAAGCTGACTGGAGGGGCTGACAAGGGGAA GTTTGTTGCCCTGGAGAACATCAGCTGCAAGATTAAGTCAGGGTGCGAGGGGCACCTCCCGTGGCCGAATGGCATCTGTACTAAGTGCCA GCCGAGCGCCATCACGCTGAACAGACAGAAGTACAGGCATGTGGACAATATCATGTTTGAGAATCACACCGTCGCTGACCGCTTTCTTGA CTTCTGGAGAAAGACAGGGAACCAGCATTTTGGGTACTTATACGGACGGTACACGGAGCACAAAGACATTCCCCTTGGCATCAGGGCTGA AGTGGCTGCGATTTATGAGCCACCTCAGATTGGTACACAGAACAGCTTGGAGCTTCTTGAGGATCCAAAAGCTGAAGTGGTCGATGAAAT TGCTGCCAAACTTGGCCTGCGGAAGGTTGGCTGGATATTTACAGACCTCGTCTCAGAAGATACCCGAAAGGGTACCGTCCGCTACAGTCG AAATAAGGACACCTATTTCCTAAGTTCAGAAGAGTGCATCACTGCAGGAGACTTCCAGAACAAGCATCCCAACATGTGCCGGCTCTCTCC AGACGGACATTTTGGATCCAAGTTTGTTACTGCAGTGGCTACAGGTGGTCCTGACAACCAAGTCCACTTTGAAGGGTACCAGGTGTCCAA TCAGTGTATGGCACTGGTCCGTGATGAGTGTTTGCTGCCATGCAAGGACGCCCCGGAGCTTGGCTACGCCAAGGAGTCTAGCAGTGAGCA GTACGTGCCTGATGTGTTTTATAAGGACGTAGACAAGTTTGGCAACGAGATCACCCAGCTGGCCCGGCCCCTGCCTGTGGAGTATCTCAT CATAGACATCACAACAACTTTCCCCAAGGATCCAGTTTACACTTTTTCTATTTCGCAAAATCCATTTCCTATTGAAAACCGGGATGTATT GGGTGAGACACAGGACTTCCATAGCTTGGCCACCTATTTGTCTCAGAATACCTCATCTGTGTTCTTGGATACCATCTCAGATTTCCACCT CTTGCTGTTCCTGGTCACCAATGAAGTTATGCCTCTGCAGGACAGCATCAGCTTGCTGCTGGAGGCCGTGCGGACCAGAAATGAGGAGCT CGCCCAGACATGGAAGAGGTCTGAGCAGTGGGCCACCATCGAGCAGCTGTGCAGCACAGTTGGCGGGCAGCTCCCAGGTCTCCATGAGTA CGGCGCCGTCGGGGGCTCCACACACACGGCCACTGCAGCCATGTGGGCCTGTCAGCACTGCACGTTCATGAACCAGCCAGGCACAGGCCA CTGCGAGATGTGCAGCCTCCCCAGGACCTAGGGCGCCTGCCCTCTGCTGGCTAGGACCGGGCCCAGCCCAGCCCTTCCTGAAGCCAGAAG CGTTGCTGAGTGTGTTCCCTGTAACTGCCCCATAGTGGGCAGCCCTGGAGGAACAAGGGGCTGGCTGTCCTGGGCTCCCTGACCCACTGA AGCTTCTCAGCACGTTCCTCCCTGGAGAGCGGGCGCCACGGCTGGTATTCTGCAGGCTGAATGCAGTCTCCAGACTGGAAACGCAGAGCG GCTCCTCACGCCTAATCCTGTTGACAAGTCCCCCGCCGTGTTGGAAAGACCTCTCGCCTCTACGTGGCACCTGGAATTGGGGCGCACAGG GAGGGGCCGATGCTGCCACCACCCAGCCTTCTCTTTATTTTCAGTGCTTTTTGTTTGGCCTCCCTGCCCCCATGTTTTTCTGGCTGGGAG CTCTTGGTTACCCCCTTGTCTGACTGCTTGGTGGCAGGCACCCTGCCTGTGAGGGCCACTTTCCTCTTCTAGTAGCTGTTGTCTGGTGGG GACGCAGTGGCCCTGGACACGGCTCCCCTCTGCAGGTCTGCAGGTCGGTTTGCTGCCTGCCCTCCTCCTCTCACCCGATGTCCAGGTGGG ATTTTAAAGTCTGCATTGGTTATAATAACAGTTATCAGTAATTCCTGCCCAGAAGACTTTTATTTATTTTTTTTTAAGATAAAAACTGCA CAAAAGGGGAGTGAGAGAGACTAGTTTCCACATCCTTCCCTCCTTTAGTGAAGCCCCCGAGGTTGTGTCCAGGGTGATGAGTGTGGACGG GGGCACCAGTCAGTTCTCCCTTGAAGTAAACCTCAGTGCCTGAGACTTTTCTACCAAGCCACACAGCTGCAGCAACTGCAGATACTGCGG GCTGAGCAGGAGCAGTGGTGGCGCCTGCCCTGAGGCTGCCCTGCGATGGCCTGGGTGGGAGAGCTGGGTCACCGGTGCCGATGCTCTGGC CCTCCCCAATACGCTGCTTCGTCCCACTGCACCGCCTGGCTGAGGGCGTTAGGGGCTGTGCCTCTTGTGAGGGCCATTTGGGACCTCCCT GGGGCACTGCACAATTGATAGTGTACCAATAGAGGGAGACTGGGCGATCTGGAACAGCACGTGGTGGGGTCCTGCTTGTGTGCTCTGCGT TCCTCTGTGGCGTGGCCAGGCCGGGGCATGGCTCTTACCCGGGGAGTGGTGGGCATCTCGATGCTTCTTTGCCTTAATGATGGCCACATC TGGGCTGCTCTGCACCCACGGGAGAGGCTGGCCCAGCTGCAGACTGCTTAGGGACTTCTGTGTCCATCCTGGGGGGTAAGCCCACGTGAC CCACATTCTTGGCACTATGAACAGAGAACATTTGCCTGTTGGCTTCTGAAGTGGTCAGGGCCATGGCTGACACCTCCAGGTCCGCCTGGC ATGGGACACCAAGTGGAAGGCCCAAGCAGCTCATCTGCTCTTGGGACCAGGGGCCAGTTGGGTTGGGTCTGGTCACGGCAGAGCTATTGT GGAGGGTCAGGAAGGGTGGAGAGGAGCTGGGTTGAAGCGGACTGCTGCGGATGCAACTCCCAGCTTGCCCACCGCGGGCTGTCTGCTCTC CCTCCTAGCAGCTGTCACACTGAAGTTTTGTCCTCTGCTGTCTCCTCTGGTCCTGAGATGAGCTGTGAGCCTAGGTGGCCAAGGCTTCCT GCATTGCTTCCCTGTGAGTCCAAGGCCTTCCCCCACCACTGGGCAGAGGCTGGACAGCACGGACTTCTAGAGAGAGCCGCGTTGCCAGTT CCTCTCCCACTCGCTCGTCCTTATCCACCACGCTATTATAGTTTCCGTTGTCCTCCACCAGCATTTCCCTTACTCTGAAGTTCCGGCATT CACATCATTCATGTTTTCTTTTGTCTTTTAGCTAAAGGAAAAGCATTGGCGATTTGTCTGATTCTGGTTTTGAGTTACTCTTTGTTCAGT AATGCACTTTATTTTATTGTCCAAAGAGAGTCAGAGCTAAGCATACAGGCTTGGGGGTGAGCCCTGCTGTGAGAGTTCAGGCCCTGGGAG GCTCAGCCACCTCCTCTTGTGGGAAGGAGGTCTCAGCCCCACCTCGCATCTTCACCTGCCCTTGGTGTGGACACACCCTCTCATGCTACC AGCACCATAATCCAGTGGGGGTGACTGGGTGCACACCTGCCCAGGTGAACACAGCGGCTGCCAGTCTCCTGGTCCCGAGAGGAGGTGGGG CCTGGCCCTGGCTCCCTCCAACCAGCTGCTCCTGGGACACAGGTGCTCCTGCTTCGGCTCTGTTTCGGCTCACAGGTGTGCATCACTGGG >34124_34124_1_GPATCH8-NPLOC4_GPATCH8_chr17_42512433_ENST00000591680_NPLOC4_chr17_79596831_ENST00000331134_length(amino acids)=719AA_BP=116 MADRFSRFNEDRDFQGNHFDQYEEGHLEIEQASLDKPIESDNIGHRLLQKHGWKLGQGLGKSLQGRTDPIPIVVKYDVMGMGRMEMELDY AEDATERRRVLEVEKEDTEELRQKYKIIRVQSPDGVKRITATKRETAATFLKKVAKEFGFQNNGFSVYINRNKTGEITASSNKSLNLLKI KHGDLLFLFPSSLAGPSSEMETSVPPGFKVFGAPNVVEDEIDQYLSKQDGKIYRSRDPQLCRHGPLGKCVHCVPLEPFDEDYLNHLEPPV KHMSFHAYIRKLTGGADKGKFVALENISCKIKSGCEGHLPWPNGICTKCQPSAITLNRQKYRHVDNIMFENHTVADRFLDFWRKTGNQHF GYLYGRYTEHKDIPLGIRAEVAAIYEPPQIGTQNSLELLEDPKAEVVDEIAAKLGLRKVGWIFTDLVSEDTRKGTVRYSRNKDTYFLSSE ECITAGDFQNKHPNMCRLSPDGHFGSKFVTAVATGGPDNQVHFEGYQVSNQCMALVRDECLLPCKDAPELGYAKESSSEQYVPDVFYKDV DKFGNEITQLARPLPVEYLIIDITTTFPKDPVYTFSISQNPFPIENRDVLGETQDFHSLATYLSQNTSSVFLDTISDFHLLLFLVTNEVM -------------------------------------------------------------- >34124_34124_2_GPATCH8-NPLOC4_GPATCH8_chr17_42512433_ENST00000591680_NPLOC4_chr17_79596831_ENST00000374747_length(transcript)=5772nt_BP=379nt GCGTCGTCCTGAGAGGAGTGAAGGCGGCAAAATGGCGGACCGCTTCTCCCGCTTCAACGAAGACCGAGACTTTCAGGGTAATCACTTTGA TCAGTATGAGGAAGGACACTTGGAAATTGAACAAGCGTCACTTGACAAGCCTATAGAATCGGATAATATTGGACACCGCTTACTCCAGAA ACATGGGTGGAAGCTGGGCCAGGGATTGGGAAAATCTCTTCAGGGGAGAACAGATCCCATTCCAATCGTTGTCAAGTATGATGTCATGGG CATGGGTCGCATGGAAATGGAGCTTGATTATGCTGAAGATGCTACCGAACGGCGCCGTGTCCTAGAAGTAGAAAAAGAAGACACAGAAGA GCTGAGACAAAAGTACAAGATAATTCGTGTCCAGTCCCCGGATGGAGTGAAGCGGATCACAGCAACAAAGAGAGAAACAGCAGCAACATT TTTGAAAAAGGTTGCAAAGGAGTTTGGCTTCCAAAATAATGGCTTCTCGGTTTACATCAATAGAAACAAGACCGGAGAGATAACAGCCTC CTCCAACAAATCCCTCAACTTGCTAAAAATCAAGCATGGCGATTTGTTGTTCCTGTTTCCCTCGAGCCTTGCTGGGCCCTCATCTGAAAT GGAGACGTCAGTTCCACCGGGCTTCAAAGTCTTTGGCGCTCCCAACGTGGTGGAGGATGAGATTGATCAGTACCTCAGCAAACAGGACGG GAAGATTTACAGAAGCCGAGACCCACAGCTATGCCGCCACGGCCCTTTGGGGAAATGCGTGCACTGCGTCCCTCTAGAGCCATTCGATGA GGACTATCTAAACCATCTCGAGCCTCCCGTGAAGCACATGTCCTTCCACGCCTACATCCGGAAGCTGACTGGAGGGGCTGACAAGGGGAA GTTTGTTGCCCTGGAGAACATCAGCTGCAAGATTAAGTCAGGGTGCGAGGGGCACCTCCCGTGGCCGAATGGCATCTGTACTAAGTGCCA GCCGAGCGCCATCACGCTGAACAGACAGAAGTACAGGCATGTGGACAATATCATGTTTGAGAATCACACCGTCGCTGACCGCTTTCTTGA CTTCTGGAGAAAGACAGGGAACCAGCATTTTGGGTACTTATACGGACGGTACACGGAGCACAAAGACATTCCCCTTGGCATCAGGGCTGA AGTGGCTGCGATTTATGAGCCACCTCAGATTGGTACACAGAACAGCTTGGAGCTTCTTGAGGATCCAAAAGCTGAAGTGGTCGATGAAAT TGCTGCCAAACTTGGCCTGCGGAAGGTTGGCTGGATATTTACAGACCTCGTCTCAGAAGATACCCGAAAGGGTACCGTCCGCTACAGTCG AAATAAGGACACCTATTTCCTAAGTTCAGAAGAGTGCATCACTGCAGGAGACTTCCAGAACAAGCATCCCAACATGTGCCGGCTCTCTCC AGACGGACATTTTGGATCCAAGTTTGTTACTGCAGTGGCTACAGGTGGTCCTGACAACCAAGTCCACTTTGAAGGGTACCAGGTGTCCAA TCAGTGTATGGCACTGGTCCGTGATGAGTGTTTGCTGCCATGCAAGGACGCCCCGGAGCTTGGCTACGCCAAGGAGTCTAGCAGTGAGCA GTACGTGCCTGATGTGTTTTATAAGGACGTAGACAAGTTTGGCAACGAGATCACCCAGCTGGCCCGGCCCCTGCCTGTGGAGTATCTCAT CATAGACATCACAACAACTTTCCCCAAGGATCCAGTTTACACTTTTTCTATTTCGCAAAATCCATTTCCTATTGAAAACCGGGATGTATT GGGTGAGACACAGGACTTCCATAGCTTGGCCACCTATTTGTCTCAGAATACCTCATCTGTGTTCTTGGATACCATCTCAGATTTCCACCT CTTGCTGTTCCTGGTCACCAATGAAGTTATGCCTCTGCAGGACAGCATCAGCTTGCTGCTGGAGGCCGTGCGGACCAGAAATGAGGAGCT CGCCCAGACATGGAAGAGGTCTGAGCAGTGGGCCACCATCGAGCAGCTGTGCAGTGAGTACCCCCACCCCCTGCCCCGGCACCCCGTGGC TGGGGCCGGAGAGCAGCCCACTCTGCACAGCTCCCCACTGCCGGTGGTGCCCTGGATTCCGCACCCAGCTGCATCCTGGCAGGTGCCATC GGCAATGCAACGAGTAGAGACTCGGCCTCCCTGCCAGGCACGTGGCAGGCTCCGGTGATGGAGCAGACATGTTGTCCAGGCCGTACGTCT TAGGTACACAGCCCTGTAAATTTTATAAACCTGTACATCAGAGATACTAATCTCCAGGCATCCTAGGAGCACTTTGAACATCATTGTCTT AAGAGAATCAGAAACTTTATGGAAGCGCCGGAAGCGTGGGACCTTTTGGTCTCCTTGAGAATTTCCCTTTAGTGCTTAAAGCTGTAAGGA GGAGAAGCCTGCAGCATCCATGTTGTTGCTGGTGGACACTACGGAACGGACCGCAGGCATTGCGTGTCTTCCGGACCCTCAGTCACCAGG TCCTGAACGATTCCCCACTCGTGAGAACCTTCTGTCAGCATTGCTGCAGGGACTCCTCACGCACCGTGGTGCCTCGTGCAGCCCGAGTGC CCTGAGTCCCTGCAGCTGAGAGCACCAGAGTGCCCGGGAGCTACGGCCCTGGACAAATGCTAGAATCCTTCTGTTAACACAGCCTTCAAA AGACACATCAAACACCTTTATACCATTTCCTGTTTTTTTTTTTTTTTTTGAGACGGAGTTTGGCTCTTGTCACCCAGGCTGGAGTGTGGT GGCGTGATCTTGCCTCACTGCAACCTCCACCTCCTGGGTTCAAACGATTCTCCTGCCTCAGCCTCCTGAGTAGCTGGGACTACAGGCATG TGCCACCATGCCTGGATAATTTTTGTATTCTTAGTAGAGCCAGGGTTTCACCATGTTAGCCAGGCTGGTCTCGAACTCCTGACCTTGTGA TCTGCTTGCCTCTGCCTCCCAGAGTGTTGGGATTACAGGTGTGAGCCACCACGCCCAGCCGAGGTTTTCTTTTCACACAATTCTTTGTTT ATTATTTTTATTTTTGTAGAGACAGGGTCTCACTCTATTGCCGAGGCTGGAGTGCAGTTGCACAATCGTAACTCACTGCAGCCTCAAACT CCAGGCTCAAGTGATCTTTCTGCTTCAGCCTGCTGAGTAACTGGGACCACAGGCTTGCAACACCACACCCAGCTAATTCTTGGGGCTTTG TTGTTGCTTTTTATTTGTTTGTTTTTTAAAAATAATTTTAACTTATTTTAGATTCACAGGGTACATGTGCAGGTTTGTCACCTGGGTATA TTGCCTGATGCTGAGGTTGGGGTATGATTGATCACATCACCCAGGTACTACACATAGTACCTAATAGTTTTTCAGCCCGCCCTGCTCTTT GTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTTTTGAGCTGGAGCCTCACCCTGTCTCCCAGGCTGGAGTGCAGTGGCGCAATCTTAG CTCACTGCAAACTCTGCCTCCTGGGTTGAAGCAATTCCCCTCCCTCAGCCTCCCGAGTAGCTGGGATTACAGATGTGTACCACCACACCT GGCGTTGCCATCTTTATGTACGTGAGTACCCATGTTTAGCTCCCACTTATAAATGAAAACATGTAGTATTTGGTTTGCTGTTCCTGCATT AGTTTGCTTAGAATAATGGCCTTCAACTGCATCCATGTTGCTGCAAAGGACATGATTTCATTCCTTTTTATGGCTGCAAAATTTTTTAAT ATTTTTGTATCAATGAGGTCTCACTGTGTTGCCCAGACTGGTCTCAAACTTTGGCCTCAAGCATTCCTTGCACCTCAGCCTCACAAAGTC CTGAGATTATACCTGGGAGCCACTGCACCCAGCCCTTTGATGCAGTTCTAACAATGACACCAGTCTTTTTTTTTTTTTTCTTTTTTGTTT CCCTTTTAGAGACAGGGTTTCACTCTGTTGCCTAGGCTGGAGTGCAGTGGAGTGATCATAGCTCACTACAGCCTTCAATTCCTGAGCTCA AGCGATCCTCCTGCATCAGCTTTCGGAGTACCTAGGAGTATAAACATGAGCCACTACACCCAACTAATGTTTTTGGTTGGTGTTTTTTTT TTTTCTTTAAGAGATAGGGTTGGCCGGGCCCGGTGGCTGGCTCACACCTGTAATCCCAGCACTTTGGGAGGCTGAGGCAGGTGGATCACA ACGTCAAGGGATTGAGACCATCCTGGCCAACATGGTGAAACCCCATTTCTGTTAAAACTACAAAAATTAGCTGGGCATGGTGGCGAGCGC CTGTAATCCCAGCTACTCAGGAGGCTGAGGCAGGAGAATCACTTGACCCAGGAAGCGTAGGTTGTAGTGAGCTGAGATCGTGCCACTGTA CTCCAGCCTGGGCGACAGAGCAAGACTCTGTCTCAAAAAAAAAAAAAAGATGTGGTCTTGCCATGTTGCCCAGGCTGGTCTCAAACTCCT AGGCTCAAGTGATCTTCCTGCCTTGGTTCCCAAAGTGCTGAGATTACAGCTGTGAGCCACTGCACCTGACCTTTTTTTTTTCCCCCCTCC TTTTTTTTTTGAGACAGGTTCTCACTCTGTCACCCAGGCTGAAGTGCAGTGGCATTTGAGGTCAGTCTCACCTTGACCTCCCCAGGCTCA AACAATCCTCCCGCCTCAGCCTCCTGAGCAGCTGGGACTACAAGCATGTGCCACCATCCCTGGCTGATTTTTTAAATTTGTTGTAGAGAT GAGGTCTCACTATGTTGCCCAGGCTGGTCTTGAACTCTTGGGCCCAAGCGATCCTCCCGCCTCAGTGCTGAGATTGCAGTGGTAAGCCAC CATGTCCAGCCAACTCTTTAATAATAGGCAGTTCTGACTGGTGTAAGTTCTGACTGGTATCTCACTGTGGTTTTAATTTGCATCTCTGAT GATTAGTGATGTTGGGCATTTTTTCATGGGTTTTTTGTTTGTTTGTTTTGAGACAAAGTCTTGTTCTGTTGCCCAGGCTGGAGTGCAGTA GTGTGATCTCAGCTCACTGCAGCCTCCACCTCCCGGGTTCAAGCCATTGTCCAGCCTCTGCCTCCTAAGTAGCTGGAATTACAGGTGGCT GCCACCACACCCAGCTAATTTTTTTAGTTTTAGTAGAGACAGGGTTTCTCCATGTTGGCCAGGCTGGTCTCAAACTCCTGACCTCAAGTG ATCCACCCACCTCGGCTTCCCAAAGTGCTGGGATTATAGGTGTGAGCCACCGCGCCCGGCCCTCATGGGTTGTTTTGTTTTGTTTTGAGA CGGAATCTCACCCTGTCACCCAGGCTGCAGTGCAATCTCATGATCTCCACTTACTGCAACCTCCACGTCCTGGGTTCAAGTGATACTCCT GCCTCAGCCTCCCGAGTAGCTGGGACTACAGGCGTGCACCCACGCCTGGCTAATTTTTATATTTTTAGTAGAGATGGGGTTTCACCATGT TGGTCAGGCTGGTCTGGAACTCCTGACCTCAGGTGATCCACCTGCCTCGGTCTCCCAAAGTGCTGGGATTACAGACGTGTATACACAAGA CACAAAGATCAGCTCAGGATGGGTCATAGGCCTAATGGTAAAAGGTAAAACTATGAAGCAGGCTGGGCATGATGTCTCATAGCTATAATC CTAACACTCTAGGAGGCCAAAGCTGGAGGACACTGTAGCCCAGAAGTGTGAGACCAGCCCACACAGCATAGTGAAGCCCCGTCTGTACAA >34124_34124_2_GPATCH8-NPLOC4_GPATCH8_chr17_42512433_ENST00000591680_NPLOC4_chr17_79596831_ENST00000374747_length(amino acids)=728AA_BP=116 MADRFSRFNEDRDFQGNHFDQYEEGHLEIEQASLDKPIESDNIGHRLLQKHGWKLGQGLGKSLQGRTDPIPIVVKYDVMGMGRMEMELDY AEDATERRRVLEVEKEDTEELRQKYKIIRVQSPDGVKRITATKRETAATFLKKVAKEFGFQNNGFSVYINRNKTGEITASSNKSLNLLKI KHGDLLFLFPSSLAGPSSEMETSVPPGFKVFGAPNVVEDEIDQYLSKQDGKIYRSRDPQLCRHGPLGKCVHCVPLEPFDEDYLNHLEPPV KHMSFHAYIRKLTGGADKGKFVALENISCKIKSGCEGHLPWPNGICTKCQPSAITLNRQKYRHVDNIMFENHTVADRFLDFWRKTGNQHF GYLYGRYTEHKDIPLGIRAEVAAIYEPPQIGTQNSLELLEDPKAEVVDEIAAKLGLRKVGWIFTDLVSEDTRKGTVRYSRNKDTYFLSSE ECITAGDFQNKHPNMCRLSPDGHFGSKFVTAVATGGPDNQVHFEGYQVSNQCMALVRDECLLPCKDAPELGYAKESSSEQYVPDVFYKDV DKFGNEITQLARPLPVEYLIIDITTTFPKDPVYTFSISQNPFPIENRDVLGETQDFHSLATYLSQNTSSVFLDTISDFHLLLFLVTNEVM PLQDSISLLLEAVRTRNEELAQTWKRSEQWATIEQLCSEYPHPLPRHPVAGAGEQPTLHSSPLPVVPWIPHPAASWQVPSAMQRVETRPP -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for GPATCH8-NPLOC4 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for GPATCH8-NPLOC4 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for GPATCH8-NPLOC4 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
