
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:GPI-COLGALT1 (FusionGDB2 ID:34282) |
Fusion Gene Summary for GPI-COLGALT1 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: GPI-COLGALT1 | Fusion gene ID: 34282 | Hgene | Tgene | Gene symbol | GPI | COLGALT1 | Gene ID | 10007 | 79709 |
| Gene name | glucosamine-6-phosphate deaminase 1 | collagen beta(1-O)galactosyltransferase 1 | |
| Synonyms | GNP1|GNPDA|GNPI|GPI|HLN | BSVD3|ColGalT 1|GLT25D1 | |
| Cytomap | 5q31.3 | 19p13.11 | |
| Type of gene | protein-coding | protein-coding | |
| Description | glucosamine-6-phosphate isomerase 1GNPDA 1glcN6P deaminase 1oscillin | procollagen galactosyltransferase 1glycosyltransferase 25 domain containing 1glycosyltransferase 25 family member 1hydroxylysine galactosyltransferase 1 | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | PIGT | Q8NBJ5 | |
| Ensembl transtripts involved in fusion gene | ENST00000356487, ENST00000415930, ENST00000586425, | ENST00000601354, ENST00000252599, | |
| Fusion gene scores | * DoF score | 27 X 23 X 11=6831 | 4 X 4 X 2=32 |
| # samples | 28 | 4 | |
| ** MAII score | log2(28/6831*10)=-4.60859805991281 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(4/32*10)=0.321928094887362 effective Gene in Pan-Cancer Fusion Genes (eGinPCFGs). DoF>8 and MAII>0 | |
| Context | PubMed: GPI [Title/Abstract] AND COLGALT1 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | GPI(34859607)-COLGALT1(17678215), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
 Fusion gene breakpoints across GPI (5'-gene) Fusion gene breakpoints across GPI (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
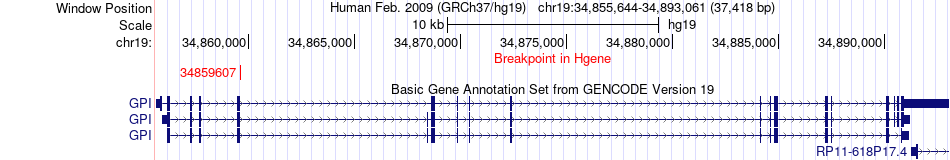 |
 Fusion gene breakpoints across COLGALT1 (3'-gene) Fusion gene breakpoints across COLGALT1 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
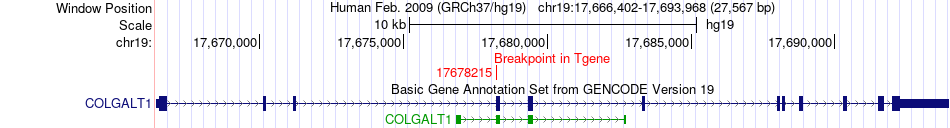 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | STAD | TCGA-BR-8372-01A | GPI | chr19 | 34859607 | + | COLGALT1 | chr19 | 17678215 | + |
Top |
Fusion Gene ORF analysis for GPI-COLGALT1 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-3UTR | ENST00000356487 | ENST00000601354 | GPI | chr19 | 34859607 | + | COLGALT1 | chr19 | 17678215 | + |
| 5CDS-3UTR | ENST00000415930 | ENST00000601354 | GPI | chr19 | 34859607 | + | COLGALT1 | chr19 | 17678215 | + |
| 5CDS-3UTR | ENST00000586425 | ENST00000601354 | GPI | chr19 | 34859607 | + | COLGALT1 | chr19 | 17678215 | + |
| In-frame | ENST00000356487 | ENST00000252599 | GPI | chr19 | 34859607 | + | COLGALT1 | chr19 | 17678215 | + |
| In-frame | ENST00000415930 | ENST00000252599 | GPI | chr19 | 34859607 | + | COLGALT1 | chr19 | 17678215 | + |
| In-frame | ENST00000586425 | ENST00000252599 | GPI | chr19 | 34859607 | + | COLGALT1 | chr19 | 17678215 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000415930 | GPI | chr19 | 34859607 | + | ENST00000252599 | COLGALT1 | chr19 | 17678215 | + | 3784 | 689 | 170 | 2068 | 632 |
| ENST00000356487 | GPI | chr19 | 34859607 | + | ENST00000252599 | COLGALT1 | chr19 | 17678215 | + | 3738 | 643 | 172 | 2022 | 616 |
| ENST00000586425 | GPI | chr19 | 34859607 | + | ENST00000252599 | COLGALT1 | chr19 | 17678215 | + | 3522 | 427 | 25 | 1806 | 593 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000415930 | ENST00000252599 | GPI | chr19 | 34859607 | + | COLGALT1 | chr19 | 17678215 | + | 0.003706087 | 0.99629384 |
| ENST00000356487 | ENST00000252599 | GPI | chr19 | 34859607 | + | COLGALT1 | chr19 | 17678215 | + | 0.003730214 | 0.9962698 |
| ENST00000586425 | ENST00000252599 | GPI | chr19 | 34859607 | + | COLGALT1 | chr19 | 17678215 | + | 0.003716876 | 0.9962831 |
Top |
Fusion Genomic Features for GPI-COLGALT1 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| GPI | chr19 | 34859607 | + | COLGALT1 | chr19 | 17678214 | + | 5.15E-14 | 1 |
| GPI | chr19 | 34859607 | + | COLGALT1 | chr19 | 17678214 | + | 5.15E-14 | 1 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
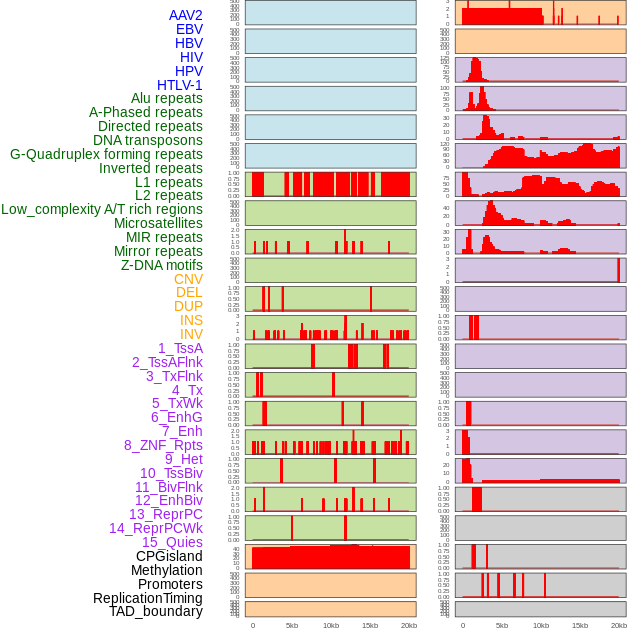 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
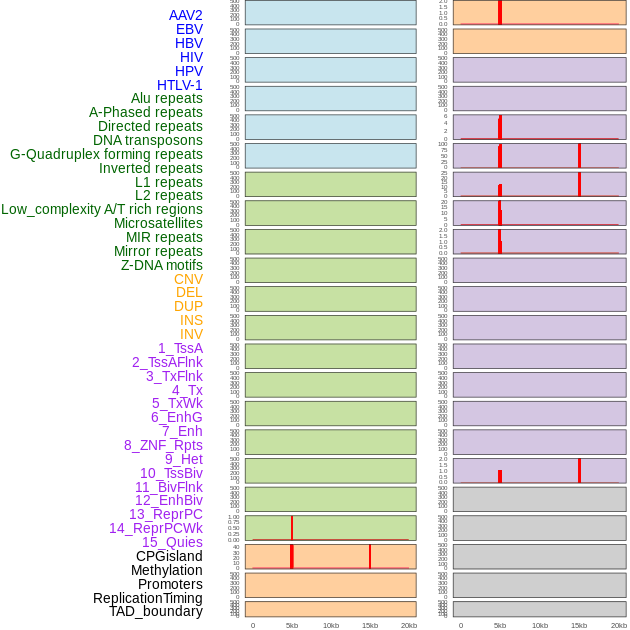 |
Top |
Fusion Protein Features for GPI-COLGALT1 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr19:34859607/chr19:17678215) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| GPI | COLGALT1 |
| 578 | FUNCTION: Beta-galactosyltransferase that transfers beta-galactose to hydroxylysine residues of type I collagen (PubMed:19075007, PubMed:22216269, PubMed:27402836). By acting on collagen glycosylation, facilitates the formation of collagen triple helix (PubMed:27402836). Also involved in the biosynthesis of collagen type IV (PubMed:30412317). {ECO:0000269|PubMed:19075007, ECO:0000269|PubMed:22216269, ECO:0000269|PubMed:27402836, ECO:0000269|PubMed:30412317}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | GPI | chr19:34859607 | chr19:17678215 | ENST00000415930 | + | 5 | 18 | 159_160 | 173 | 570.0 | Region | Glucose-6-phosphate binding |
| Tgene | COLGALT1 | chr19:34859607 | chr19:17678215 | ENST00000252599 | 2 | 12 | 619_622 | 163 | 623.0 | Motif | Endoplasmic reticulum retention motif |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | GPI | chr19:34859607 | chr19:17678215 | ENST00000356487 | + | 4 | 18 | 159_160 | 134 | 559.0 | Region | Glucose-6-phosphate binding |
| Hgene | GPI | chr19:34859607 | chr19:17678215 | ENST00000356487 | + | 4 | 18 | 210_215 | 134 | 559.0 | Region | Glucose-6-phosphate binding |
| Hgene | GPI | chr19:34859607 | chr19:17678215 | ENST00000415930 | + | 5 | 18 | 210_215 | 173 | 570.0 | Region | Glucose-6-phosphate binding |
Top |
Fusion Gene Sequence for GPI-COLGALT1 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >34282_34282_1_GPI-COLGALT1_GPI_chr19_34859607_ENST00000356487_COLGALT1_chr19_17678215_ENST00000252599_length(transcript)=3738nt_BP=643nt AGAGAGGAGCTGAGGCCCCAGATCAGCGGCCGCGGGCAAGGTCGCTCAGCGGGCACCCGGCCTGGGTATCGGGGCGCGGGTCGGGGGCGG GGCCGGGGCTCAGGGGTGGGGCCGGGCCGGGCCGGGCCGGGCGCCTGCGCCATAAAGGCCGCCGCGCGCCCACGCGCCTCGCTTGCTGCG CGCTGCCGGCGCTCCTTCCTCCTCGGCTCGCGTCTCACTCAGTGTACCTTCTAGTCCCGCCATGGCCGCTCTCACCCGGGACCCCCAGTT CCAGAAGCTGCAGCAATGGTACCGCGAGCACCGCTCCGAGCTGAACCTGCGCCGCCTCTTCGATGCCAACAAGGACCGCTTCAACCACTT CAGCTTGACCCTCAACACCAACCATGGGCATATCCTGGTGGATTACTCCAAGAACCTGGTGACGGAGGACGTGATGCGGATGCTGGTGGA CTTGGCCAAGTCCAGGGGCGTGGAGGCCGCCCGGGAGCGGATGTTCAATGGTGAGAAGATCAACTACACCGAGGGTCGAGCCGTGCTGCA CGTGGCTCTGCGGAACCGGTCAAACACACCCATCCTGGTAGACGGCAAGGATGTGATGCCAGAGGTCAACAAGGTTCTGGACAAGATGAA GTCTTTCTGCCAGTTTGTAGATGCGGACAACCTGATCCTCAACCCTGACACACTGAGCCTGCTCATCGCTGAGAACAAGACGGTGGTCGC CCCCATGCTGGATTCCCGGGCTGCGTACTCCAACTTCTGGTGTGGAATGACTTCCCAGGGCTACTACAAGCGCACACCTGCCTACATCCC TATCCGCAAGCGAGACCGCCGGGGCTGCTTTGCAGTTCCCATGGTGCACTCGACCTTCCTGATCGACCTGCGGAAGGCGGCGTCCAGGAA CCTGGCCTTCTACCCACCTCACCCTGACTACACCTGGTCCTTTGACGACATCATCGTCTTTGCCTTCTCCTGCAAGCAGGCAGAGGTTCA GATGTATGTGTGCAACAAGGAGGAGTACGGATTCTTGCCAGTGCCATTGCGCGCCCACAGCACCCTCCAGGATGAGGCCGAGAGCTTCAT GCATGTGCAGCTGGAGGTCATGGTGAAGCACCCGCCCGCAGAGCCCTCCCGCTTCATCTCGGCTCCCACCAAGACACCGGACAAGATGGG CTTCGACGAGGTCTTCATGATCAACCTGAGGCGGCGGCAGGACCGGCGGGAGCGCATGCTGCGGGCGCTGCAGGCACAGGAGATCGAGTG CCGGCTGGTGGAGGCCGTGGACGGCAAAGCCATGAACACCAGCCAGGTGGAGGCGCTGGGGATCCAGATGCTGCCTGGCTACCGGGACCC CTACCACGGCCGGCCCCTCACCAAGGGTGAGCTGGGCTGCTTCCTGAGCCACTACAACATCTGGAAGGAGGTGGTGGACCGGGGGCTGCA GAAATCGCTTGTGTTTGAGGATGACCTGCGTTTTGAGATCTTCTTCAAGAGACGTCTGATGAACCTCATGCGGGATGTGGAGCGGGAGGG CCTGGACTGGGACCTCATCTATGTGGGCCGGAAGCGGATGCAGGTGGAGCACCCCGAGAAGGCTGTGCCTCGCGTGAGGAACCTGGTGGA GGCCGACTATTCCTACTGGACCCTGGCCTACGTGATCTCCCTGCAAGGCGCCCGCAAACTGCTGGCTGCTGAGCCGCTCTCCAAGATGCT GCCTGTGGACGAGTTCCTGCCCGTCATGTTCGACAAACACCCAGTGTCCGAGTACAAGGCCCACTTCTCCCTCCGCAACCTGCATGCCTT CTCTGTGGAGCCGCTGCTCATCTACCCCACACACTACACAGGAGACGATGGCTATGTGAGTGACACCGAGACCTCAGTCGTATGGAACAA TGAGCACGTCAAGACCGACTGGGACCGCGCCAAGTCCCAGAAGATGCGGGAGCAGCAGGCACTGAGCCGTGAGGCCAAGAACTCGGACGT GCTCCAGTCCCCACTGGACAGTGCTGCCCGGGATGAACTCTGAGGGGTAGCAGCCAGAAAGCCAAAGCAGCCATCGGTGGCCCAGGCTCC ACGTGCTTACTGAGGACATCAGGTCCACCTCTGGACCCCTTGGCAGGCCACAGAGGGCTCTCGTGTGGGGTGGTGTCCAGCCAGCTCTTG CTAAGCAATCACGTGCACACAGGCAGCATTAATGGAGTGCCTACTGCATGCCAGCAACAGGGCTTGGCCCTGGGGAATTGGGAGGAACCA AGCCCTCTTCATCTGTTCATGTGCCCAGCATTTATTAAGCACCTGCTGTATGCAAGGTTCCCATGTTACGGCAGTGAATGAGGCATAATT GTTCCCTCCATCAGCGATTGATTCAGTCATCAAGCAGTTACTGATCAGATTAAGAATCAGGCACTAGTGATACACATTCATTTTTAAAAT TCATTCAAGGATTTATTGAGTGCCTACTGTGTGTTGGGTGCCATTCCAGGCTCTGGGATTTTTTTTTTTTTTTTTTTAAGAGTAGAGTCT GTCTCTGTCACCCAGGCTGGAGTGCAGTGGTGTGACGGCTCACTGCAGCCTGCGCCTCCCAGCGTCCAGCAATTCTTGTTTCTCGGCCTC CCAAGTAGCTGGGACTATAGGTGCGTGCCATCACATCTGGCTAGTTTTTGTATTTTTAGTAGAGACGGGGTTTCACCATGTTGGCCAGGC TGGTCTCGAACTCCTGACCTCAAGTGATCTGCCCGCCTTGGCCACCCAAAGTGCTAGGGTTACAGGCATGAGCCACTGTGCCCAGCCCAG GCTCTGGGAATATTGAGATAAATAAGATGCTTCTATCCATTTATTCAAGCACATATTGGTCACCTACTGTGTGCCTGGCACTCATGTCAC AAAGATAAGTTCCTGATTCGGTACACTTACTGAGCACCTGCTGTGTGCAGGGAGCTGAGCTATGGGATGGGAATGGGAGTAAACAAGGTA CTTTTTACTTTTTTCTTTTTTTCCTCACTGCTAGACGGTGTGGGAACTTCTCACTCATTGGCTTCTTTCCCACACACCTGAAGAGCACTG ACTGTGTGCCGGGCACTAGTGATACAAAAGAGTGTGACAGTTGTTCAGTCTGCATTTTCGATCATGGGCTACATGCCGAGTGCTGGGGCA CAGAGATGAACAAGATCGGTTCCTTCACTTCTTCATGCCACAAGTGTTTATTGAGCACCTGTGTGCCAGGCCTCACAGACTCCCAGTTGG GTTGAAGAATGGTTGACTGAGTTTGATTCTTCCTGTACCCTCGGTCGTCTGAGCTGTGTGCAGACAACATCCCCCCACCACCCAAGAGGG AGGGTAGCTCTTCCGCCACCAGGGGCAAGCACAGGTCCTGGTGGCCCCACGCCACATGTTAGCCCCCCTGGAGGGGGCGCCAGTTGGAGA CGGGGGCTGGGTGTCCCTGGCCCACTCCCGGTCCCCTGTGCTTTACCTCCTTGCCCTTGTGTCTCAGGTGTGGTCCCTGCCTGCTTGATG AAGTTGCTCTGTTCAAGCCTTTGGTGGGATCATGTGTTTGGGGGCTTTTAGGGGACCCAGCTGCACTGGGGCACTGCCCGTGGCCTGGGT AGGACATTTCCCAGCAAGGGCTGGAGGAGTTGCCGTGCCTTCAGCCTGAATCGAATGTCAGAACCAGCCAGCGGTGCTTCACCCTCTTGG >34282_34282_1_GPI-COLGALT1_GPI_chr19_34859607_ENST00000356487_COLGALT1_chr19_17678215_ENST00000252599_length(amino acids)=616AA_BP=156 MLRAAGAPSSSARVSLSVPSSPAMAALTRDPQFQKLQQWYREHRSELNLRRLFDANKDRFNHFSLTLNTNHGHILVDYSKNLVTEDVMRM LVDLAKSRGVEAARERMFNGEKINYTEGRAVLHVALRNRSNTPILVDGKDVMPEVNKVLDKMKSFCQFVDADNLILNPDTLSLLIAENKT VVAPMLDSRAAYSNFWCGMTSQGYYKRTPAYIPIRKRDRRGCFAVPMVHSTFLIDLRKAASRNLAFYPPHPDYTWSFDDIIVFAFSCKQA EVQMYVCNKEEYGFLPVPLRAHSTLQDEAESFMHVQLEVMVKHPPAEPSRFISAPTKTPDKMGFDEVFMINLRRRQDRRERMLRALQAQE IECRLVEAVDGKAMNTSQVEALGIQMLPGYRDPYHGRPLTKGELGCFLSHYNIWKEVVDRGLQKSLVFEDDLRFEIFFKRRLMNLMRDVE REGLDWDLIYVGRKRMQVEHPEKAVPRVRNLVEADYSYWTLAYVISLQGARKLLAAEPLSKMLPVDEFLPVMFDKHPVSEYKAHFSLRNL -------------------------------------------------------------- >34282_34282_2_GPI-COLGALT1_GPI_chr19_34859607_ENST00000415930_COLGALT1_chr19_17678215_ENST00000252599_length(transcript)=3784nt_BP=689nt AATAGCCCTTACCACCAGCAGACACACATCATCTGTTGTACTTGCTTATTTGGCACATATGTATCCACAGCGCCTAGAACACTGCCTGTA ACGTGGAAGGTGTTCGATCTATAGAGTTTTGTCGAATGAATGAATGAAGCCGACTAGTGCACAGGGAGTGCAGCGGCGCGATGGTAGCTC TCTGCAGCCTCCAACACCTGGGCTCCAGTGATCCCCGGGCTCTGCCCACCCTCCCCACTGCCACTTCCGGGCAGAGGCCAGCAAAGCGGC GGCGCAAGAGTCCCGCCATGGCCGCTCTCACCCGGGACCCCCAGTTCCAGAAGCTGCAGCAATGGTACCGCGAGCACCGCTCCGAGCTGA ACCTGCGCCGCCTCTTCGATGCCAACAAGGACCGCTTCAACCACTTCAGCTTGACCCTCAACACCAACCATGGGCATATCCTGGTGGATT ACTCCAAGAACCTGGTGACGGAGGACGTGATGCGGATGCTGGTGGACTTGGCCAAGTCCAGGGGCGTGGAGGCCGCCCGGGAGCGGATGT TCAATGGTGAGAAGATCAACTACACCGAGGGTCGAGCCGTGCTGCACGTGGCTCTGCGGAACCGGTCAAACACACCCATCCTGGTAGACG GCAAGGATGTGATGCCAGAGGTCAACAAGGTTCTGGACAAGATGAAGTCTTTCTGCCAGTTTGTAGATGCGGACAACCTGATCCTCAACC CTGACACACTGAGCCTGCTCATCGCTGAGAACAAGACGGTGGTCGCCCCCATGCTGGATTCCCGGGCTGCGTACTCCAACTTCTGGTGTG GAATGACTTCCCAGGGCTACTACAAGCGCACACCTGCCTACATCCCTATCCGCAAGCGAGACCGCCGGGGCTGCTTTGCAGTTCCCATGG TGCACTCGACCTTCCTGATCGACCTGCGGAAGGCGGCGTCCAGGAACCTGGCCTTCTACCCACCTCACCCTGACTACACCTGGTCCTTTG ACGACATCATCGTCTTTGCCTTCTCCTGCAAGCAGGCAGAGGTTCAGATGTATGTGTGCAACAAGGAGGAGTACGGATTCTTGCCAGTGC CATTGCGCGCCCACAGCACCCTCCAGGATGAGGCCGAGAGCTTCATGCATGTGCAGCTGGAGGTCATGGTGAAGCACCCGCCCGCAGAGC CCTCCCGCTTCATCTCGGCTCCCACCAAGACACCGGACAAGATGGGCTTCGACGAGGTCTTCATGATCAACCTGAGGCGGCGGCAGGACC GGCGGGAGCGCATGCTGCGGGCGCTGCAGGCACAGGAGATCGAGTGCCGGCTGGTGGAGGCCGTGGACGGCAAAGCCATGAACACCAGCC AGGTGGAGGCGCTGGGGATCCAGATGCTGCCTGGCTACCGGGACCCCTACCACGGCCGGCCCCTCACCAAGGGTGAGCTGGGCTGCTTCC TGAGCCACTACAACATCTGGAAGGAGGTGGTGGACCGGGGGCTGCAGAAATCGCTTGTGTTTGAGGATGACCTGCGTTTTGAGATCTTCT TCAAGAGACGTCTGATGAACCTCATGCGGGATGTGGAGCGGGAGGGCCTGGACTGGGACCTCATCTATGTGGGCCGGAAGCGGATGCAGG TGGAGCACCCCGAGAAGGCTGTGCCTCGCGTGAGGAACCTGGTGGAGGCCGACTATTCCTACTGGACCCTGGCCTACGTGATCTCCCTGC AAGGCGCCCGCAAACTGCTGGCTGCTGAGCCGCTCTCCAAGATGCTGCCTGTGGACGAGTTCCTGCCCGTCATGTTCGACAAACACCCAG TGTCCGAGTACAAGGCCCACTTCTCCCTCCGCAACCTGCATGCCTTCTCTGTGGAGCCGCTGCTCATCTACCCCACACACTACACAGGAG ACGATGGCTATGTGAGTGACACCGAGACCTCAGTCGTATGGAACAATGAGCACGTCAAGACCGACTGGGACCGCGCCAAGTCCCAGAAGA TGCGGGAGCAGCAGGCACTGAGCCGTGAGGCCAAGAACTCGGACGTGCTCCAGTCCCCACTGGACAGTGCTGCCCGGGATGAACTCTGAG GGGTAGCAGCCAGAAAGCCAAAGCAGCCATCGGTGGCCCAGGCTCCACGTGCTTACTGAGGACATCAGGTCCACCTCTGGACCCCTTGGC AGGCCACAGAGGGCTCTCGTGTGGGGTGGTGTCCAGCCAGCTCTTGCTAAGCAATCACGTGCACACAGGCAGCATTAATGGAGTGCCTAC TGCATGCCAGCAACAGGGCTTGGCCCTGGGGAATTGGGAGGAACCAAGCCCTCTTCATCTGTTCATGTGCCCAGCATTTATTAAGCACCT GCTGTATGCAAGGTTCCCATGTTACGGCAGTGAATGAGGCATAATTGTTCCCTCCATCAGCGATTGATTCAGTCATCAAGCAGTTACTGA TCAGATTAAGAATCAGGCACTAGTGATACACATTCATTTTTAAAATTCATTCAAGGATTTATTGAGTGCCTACTGTGTGTTGGGTGCCAT TCCAGGCTCTGGGATTTTTTTTTTTTTTTTTTTAAGAGTAGAGTCTGTCTCTGTCACCCAGGCTGGAGTGCAGTGGTGTGACGGCTCACT GCAGCCTGCGCCTCCCAGCGTCCAGCAATTCTTGTTTCTCGGCCTCCCAAGTAGCTGGGACTATAGGTGCGTGCCATCACATCTGGCTAG TTTTTGTATTTTTAGTAGAGACGGGGTTTCACCATGTTGGCCAGGCTGGTCTCGAACTCCTGACCTCAAGTGATCTGCCCGCCTTGGCCA CCCAAAGTGCTAGGGTTACAGGCATGAGCCACTGTGCCCAGCCCAGGCTCTGGGAATATTGAGATAAATAAGATGCTTCTATCCATTTAT TCAAGCACATATTGGTCACCTACTGTGTGCCTGGCACTCATGTCACAAAGATAAGTTCCTGATTCGGTACACTTACTGAGCACCTGCTGT GTGCAGGGAGCTGAGCTATGGGATGGGAATGGGAGTAAACAAGGTACTTTTTACTTTTTTCTTTTTTTCCTCACTGCTAGACGGTGTGGG AACTTCTCACTCATTGGCTTCTTTCCCACACACCTGAAGAGCACTGACTGTGTGCCGGGCACTAGTGATACAAAAGAGTGTGACAGTTGT TCAGTCTGCATTTTCGATCATGGGCTACATGCCGAGTGCTGGGGCACAGAGATGAACAAGATCGGTTCCTTCACTTCTTCATGCCACAAG TGTTTATTGAGCACCTGTGTGCCAGGCCTCACAGACTCCCAGTTGGGTTGAAGAATGGTTGACTGAGTTTGATTCTTCCTGTACCCTCGG TCGTCTGAGCTGTGTGCAGACAACATCCCCCCACCACCCAAGAGGGAGGGTAGCTCTTCCGCCACCAGGGGCAAGCACAGGTCCTGGTGG CCCCACGCCACATGTTAGCCCCCCTGGAGGGGGCGCCAGTTGGAGACGGGGGCTGGGTGTCCCTGGCCCACTCCCGGTCCCCTGTGCTTT ACCTCCTTGCCCTTGTGTCTCAGGTGTGGTCCCTGCCTGCTTGATGAAGTTGCTCTGTTCAAGCCTTTGGTGGGATCATGTGTTTGGGGG CTTTTAGGGGACCCAGCTGCACTGGGGCACTGCCCGTGGCCTGGGTAGGACATTTCCCAGCAAGGGCTGGAGGAGTTGCCGTGCCTTCAG CCTGAATCGAATGTCAGAACCAGCCAGCGGTGCTTCACCCTCTTGGGGATAACTTGCTTAGTTTTTTAATAAATGTTCCTGGTTGGTTTT >34282_34282_2_GPI-COLGALT1_GPI_chr19_34859607_ENST00000415930_COLGALT1_chr19_17678215_ENST00000252599_length(amino acids)=632AA_BP=172 MVALCSLQHLGSSDPRALPTLPTATSGQRPAKRRRKSPAMAALTRDPQFQKLQQWYREHRSELNLRRLFDANKDRFNHFSLTLNTNHGHI LVDYSKNLVTEDVMRMLVDLAKSRGVEAARERMFNGEKINYTEGRAVLHVALRNRSNTPILVDGKDVMPEVNKVLDKMKSFCQFVDADNL ILNPDTLSLLIAENKTVVAPMLDSRAAYSNFWCGMTSQGYYKRTPAYIPIRKRDRRGCFAVPMVHSTFLIDLRKAASRNLAFYPPHPDYT WSFDDIIVFAFSCKQAEVQMYVCNKEEYGFLPVPLRAHSTLQDEAESFMHVQLEVMVKHPPAEPSRFISAPTKTPDKMGFDEVFMINLRR RQDRRERMLRALQAQEIECRLVEAVDGKAMNTSQVEALGIQMLPGYRDPYHGRPLTKGELGCFLSHYNIWKEVVDRGLQKSLVFEDDLRF EIFFKRRLMNLMRDVEREGLDWDLIYVGRKRMQVEHPEKAVPRVRNLVEADYSYWTLAYVISLQGARKLLAAEPLSKMLPVDEFLPVMFD KHPVSEYKAHFSLRNLHAFSVEPLLIYPTHYTGDDGYVSDTETSVVWNNEHVKTDWDRAKSQKMREQQALSREAKNSDVLQSPLDSAARD -------------------------------------------------------------- >34282_34282_3_GPI-COLGALT1_GPI_chr19_34859607_ENST00000586425_COLGALT1_chr19_17678215_ENST00000252599_length(transcript)=3522nt_BP=427nt ACTCAGTGTACCTTCTAGTCCCGCCATGGCCGCTCTCACCCGGGACCCCCAGTTCCAGAAGCTGCAGCAATGGTACCGCGAGCACCGCTC CGAGCTGAACCTGCGCCGCCTCTTCGATGCCAACAAGGACCGCTTCAACCACTTCAGCTTGACCCTCAACACCAACCATGGGCATATCCT GGTGGATTACTCCAAGAACCTGGTGACGGAGGACGTGATGCGGATGCTGGTGGACTTGGCCAAGTCCAGGGGCGTGGAGGCCGCCCGGGA GCGGATGTTCAATGGTGAGAAGATCAACTACACCGAGGGTCGAGCCGTGCTGCACGTGGCTCTGCGGAACCGGTCAAACACACCCATCCT GGTAGACGGCAAGGATGTGATGCCAGAGGTCAACAAGGTTCTGGACAAGATGAAGTCTTTCTGCCAGTTTGTAGATGCGGACAACCTGAT CCTCAACCCTGACACACTGAGCCTGCTCATCGCTGAGAACAAGACGGTGGTCGCCCCCATGCTGGATTCCCGGGCTGCGTACTCCAACTT CTGGTGTGGAATGACTTCCCAGGGCTACTACAAGCGCACACCTGCCTACATCCCTATCCGCAAGCGAGACCGCCGGGGCTGCTTTGCAGT TCCCATGGTGCACTCGACCTTCCTGATCGACCTGCGGAAGGCGGCGTCCAGGAACCTGGCCTTCTACCCACCTCACCCTGACTACACCTG GTCCTTTGACGACATCATCGTCTTTGCCTTCTCCTGCAAGCAGGCAGAGGTTCAGATGTATGTGTGCAACAAGGAGGAGTACGGATTCTT GCCAGTGCCATTGCGCGCCCACAGCACCCTCCAGGATGAGGCCGAGAGCTTCATGCATGTGCAGCTGGAGGTCATGGTGAAGCACCCGCC CGCAGAGCCCTCCCGCTTCATCTCGGCTCCCACCAAGACACCGGACAAGATGGGCTTCGACGAGGTCTTCATGATCAACCTGAGGCGGCG GCAGGACCGGCGGGAGCGCATGCTGCGGGCGCTGCAGGCACAGGAGATCGAGTGCCGGCTGGTGGAGGCCGTGGACGGCAAAGCCATGAA CACCAGCCAGGTGGAGGCGCTGGGGATCCAGATGCTGCCTGGCTACCGGGACCCCTACCACGGCCGGCCCCTCACCAAGGGTGAGCTGGG CTGCTTCCTGAGCCACTACAACATCTGGAAGGAGGTGGTGGACCGGGGGCTGCAGAAATCGCTTGTGTTTGAGGATGACCTGCGTTTTGA GATCTTCTTCAAGAGACGTCTGATGAACCTCATGCGGGATGTGGAGCGGGAGGGCCTGGACTGGGACCTCATCTATGTGGGCCGGAAGCG GATGCAGGTGGAGCACCCCGAGAAGGCTGTGCCTCGCGTGAGGAACCTGGTGGAGGCCGACTATTCCTACTGGACCCTGGCCTACGTGAT CTCCCTGCAAGGCGCCCGCAAACTGCTGGCTGCTGAGCCGCTCTCCAAGATGCTGCCTGTGGACGAGTTCCTGCCCGTCATGTTCGACAA ACACCCAGTGTCCGAGTACAAGGCCCACTTCTCCCTCCGCAACCTGCATGCCTTCTCTGTGGAGCCGCTGCTCATCTACCCCACACACTA CACAGGAGACGATGGCTATGTGAGTGACACCGAGACCTCAGTCGTATGGAACAATGAGCACGTCAAGACCGACTGGGACCGCGCCAAGTC CCAGAAGATGCGGGAGCAGCAGGCACTGAGCCGTGAGGCCAAGAACTCGGACGTGCTCCAGTCCCCACTGGACAGTGCTGCCCGGGATGA ACTCTGAGGGGTAGCAGCCAGAAAGCCAAAGCAGCCATCGGTGGCCCAGGCTCCACGTGCTTACTGAGGACATCAGGTCCACCTCTGGAC CCCTTGGCAGGCCACAGAGGGCTCTCGTGTGGGGTGGTGTCCAGCCAGCTCTTGCTAAGCAATCACGTGCACACAGGCAGCATTAATGGA GTGCCTACTGCATGCCAGCAACAGGGCTTGGCCCTGGGGAATTGGGAGGAACCAAGCCCTCTTCATCTGTTCATGTGCCCAGCATTTATT AAGCACCTGCTGTATGCAAGGTTCCCATGTTACGGCAGTGAATGAGGCATAATTGTTCCCTCCATCAGCGATTGATTCAGTCATCAAGCA GTTACTGATCAGATTAAGAATCAGGCACTAGTGATACACATTCATTTTTAAAATTCATTCAAGGATTTATTGAGTGCCTACTGTGTGTTG GGTGCCATTCCAGGCTCTGGGATTTTTTTTTTTTTTTTTTTAAGAGTAGAGTCTGTCTCTGTCACCCAGGCTGGAGTGCAGTGGTGTGAC GGCTCACTGCAGCCTGCGCCTCCCAGCGTCCAGCAATTCTTGTTTCTCGGCCTCCCAAGTAGCTGGGACTATAGGTGCGTGCCATCACAT CTGGCTAGTTTTTGTATTTTTAGTAGAGACGGGGTTTCACCATGTTGGCCAGGCTGGTCTCGAACTCCTGACCTCAAGTGATCTGCCCGC CTTGGCCACCCAAAGTGCTAGGGTTACAGGCATGAGCCACTGTGCCCAGCCCAGGCTCTGGGAATATTGAGATAAATAAGATGCTTCTAT CCATTTATTCAAGCACATATTGGTCACCTACTGTGTGCCTGGCACTCATGTCACAAAGATAAGTTCCTGATTCGGTACACTTACTGAGCA CCTGCTGTGTGCAGGGAGCTGAGCTATGGGATGGGAATGGGAGTAAACAAGGTACTTTTTACTTTTTTCTTTTTTTCCTCACTGCTAGAC GGTGTGGGAACTTCTCACTCATTGGCTTCTTTCCCACACACCTGAAGAGCACTGACTGTGTGCCGGGCACTAGTGATACAAAAGAGTGTG ACAGTTGTTCAGTCTGCATTTTCGATCATGGGCTACATGCCGAGTGCTGGGGCACAGAGATGAACAAGATCGGTTCCTTCACTTCTTCAT GCCACAAGTGTTTATTGAGCACCTGTGTGCCAGGCCTCACAGACTCCCAGTTGGGTTGAAGAATGGTTGACTGAGTTTGATTCTTCCTGT ACCCTCGGTCGTCTGAGCTGTGTGCAGACAACATCCCCCCACCACCCAAGAGGGAGGGTAGCTCTTCCGCCACCAGGGGCAAGCACAGGT CCTGGTGGCCCCACGCCACATGTTAGCCCCCCTGGAGGGGGCGCCAGTTGGAGACGGGGGCTGGGTGTCCCTGGCCCACTCCCGGTCCCC TGTGCTTTACCTCCTTGCCCTTGTGTCTCAGGTGTGGTCCCTGCCTGCTTGATGAAGTTGCTCTGTTCAAGCCTTTGGTGGGATCATGTG TTTGGGGGCTTTTAGGGGACCCAGCTGCACTGGGGCACTGCCCGTGGCCTGGGTAGGACATTTCCCAGCAAGGGCTGGAGGAGTTGCCGT GCCTTCAGCCTGAATCGAATGTCAGAACCAGCCAGCGGTGCTTCACCCTCTTGGGGATAACTTGCTTAGTTTTTTAATAAATGTTCCTGG >34282_34282_3_GPI-COLGALT1_GPI_chr19_34859607_ENST00000586425_COLGALT1_chr19_17678215_ENST00000252599_length(amino acids)=593AA_BP=133 MAALTRDPQFQKLQQWYREHRSELNLRRLFDANKDRFNHFSLTLNTNHGHILVDYSKNLVTEDVMRMLVDLAKSRGVEAARERMFNGEKI NYTEGRAVLHVALRNRSNTPILVDGKDVMPEVNKVLDKMKSFCQFVDADNLILNPDTLSLLIAENKTVVAPMLDSRAAYSNFWCGMTSQG YYKRTPAYIPIRKRDRRGCFAVPMVHSTFLIDLRKAASRNLAFYPPHPDYTWSFDDIIVFAFSCKQAEVQMYVCNKEEYGFLPVPLRAHS TLQDEAESFMHVQLEVMVKHPPAEPSRFISAPTKTPDKMGFDEVFMINLRRRQDRRERMLRALQAQEIECRLVEAVDGKAMNTSQVEALG IQMLPGYRDPYHGRPLTKGELGCFLSHYNIWKEVVDRGLQKSLVFEDDLRFEIFFKRRLMNLMRDVEREGLDWDLIYVGRKRMQVEHPEK AVPRVRNLVEADYSYWTLAYVISLQGARKLLAAEPLSKMLPVDEFLPVMFDKHPVSEYKAHFSLRNLHAFSVEPLLIYPTHYTGDDGYVS -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for GPI-COLGALT1 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for GPI-COLGALT1 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for GPI-COLGALT1 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
