
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:GPR124-WRN (FusionGDB2 ID:34366) |
Fusion Gene Summary for GPR124-WRN |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: GPR124-WRN | Fusion gene ID: 34366 | Hgene | Tgene | Gene symbol | GPR124 | WRN | Gene ID | 25960 | 7486 |
| Gene name | adhesion G protein-coupled receptor A2 | WRN RecQ like helicase | |
| Synonyms | GPR124|TEM5 | RECQ3|RECQL2|RECQL3 | |
| Cytomap | 8p11.23 | 8p12 | |
| Type of gene | protein-coding | protein-coding | |
| Description | adhesion G protein-coupled receptor A2G-protein coupled receptor 124tumor endothelial marker 5 | Werner syndrome ATP-dependent helicaseDNA helicase, RecQ-like type 3Werner syndrome RecQ like helicaseWerner syndrome, RecQ helicase-likeexonuclease WRNrecQ protein-like 2 | |
| Modification date | 20200313 | 20200320 | |
| UniProtAcc | . | . | |
| Ensembl transtripts involved in fusion gene | ENST00000315215, ENST00000412232, | ENST00000298139, | |
| Fusion gene scores | * DoF score | 3 X 3 X 2=18 | 9 X 10 X 7=630 |
| # samples | 3 | 11 | |
| ** MAII score | log2(3/18*10)=0.736965594166206 effective Gene in Pan-Cancer Fusion Genes (eGinPCFGs). DoF>8 and MAII>0 | log2(11/630*10)=-2.51784830486262 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: GPR124 [Title/Abstract] AND WRN [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | GPR124(37655052)-WRN(30938383), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | GPR124 | GO:0090263 | positive regulation of canonical Wnt signaling pathway | 30026314 |
| Tgene | WRN | GO:0000731 | DNA synthesis involved in DNA repair | 17563354 |
| Tgene | WRN | GO:0006259 | DNA metabolic process | 16622405 |
| Tgene | WRN | GO:0006284 | base-excision repair | 17611195 |
| Tgene | WRN | GO:0006974 | cellular response to DNA damage stimulus | 18203716 |
| Tgene | WRN | GO:0006979 | response to oxidative stress | 17611195 |
| Tgene | WRN | GO:0009267 | cellular response to starvation | 11420665 |
| Tgene | WRN | GO:0010225 | response to UV-C | 17563354 |
| Tgene | WRN | GO:0031297 | replication fork processing | 17115688 |
| Tgene | WRN | GO:0032508 | DNA duplex unwinding | 11735402|26420422 |
| Tgene | WRN | GO:0044806 | G-quadruplex DNA unwinding | 11735402 |
| Tgene | WRN | GO:0051345 | positive regulation of hydrolase activity | 17611195 |
| Tgene | WRN | GO:0061820 | telomeric D-loop disassembly | 15200954|19734539|26420422 |
| Tgene | WRN | GO:0071480 | cellular response to gamma radiation | 21639834 |
| Tgene | WRN | GO:0098530 | positive regulation of strand invasion | 26420422 |
| Tgene | WRN | GO:1902570 | protein localization to nucleolus | 11420665 |
 Fusion gene breakpoints across GPR124 (5'-gene) Fusion gene breakpoints across GPR124 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
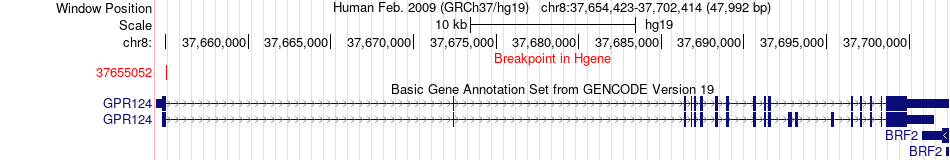 |
 Fusion gene breakpoints across WRN (3'-gene) Fusion gene breakpoints across WRN (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
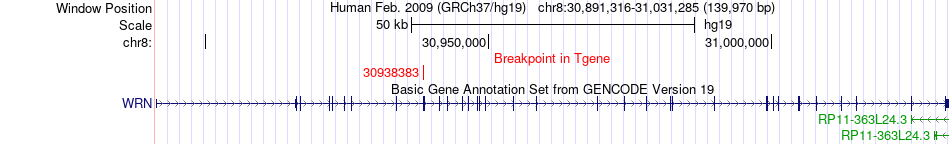 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | STAD | TCGA-HU-A4GJ-01A | GPR124 | chr8 | 37655052 | + | WRN | chr8 | 30938383 | + |
Top |
Fusion Gene ORF analysis for GPR124-WRN |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| In-frame | ENST00000315215 | ENST00000298139 | GPR124 | chr8 | 37655052 | + | WRN | chr8 | 30938383 | + |
| In-frame | ENST00000412232 | ENST00000298139 | GPR124 | chr8 | 37655052 | + | WRN | chr8 | 30938383 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000315215 | GPR124 | chr8 | 37655052 | + | ENST00000298139 | WRN | chr8 | 30938383 | + | 4756 | 629 | 276 | 4088 | 1270 |
| ENST00000412232 | GPR124 | chr8 | 37655052 | + | ENST00000298139 | WRN | chr8 | 30938383 | + | 4406 | 279 | 10 | 3738 | 1242 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000315215 | ENST00000298139 | GPR124 | chr8 | 37655052 | + | WRN | chr8 | 30938383 | + | 0.000169383 | 0.9998306 |
| ENST00000412232 | ENST00000298139 | GPR124 | chr8 | 37655052 | + | WRN | chr8 | 30938383 | + | 0.000107617 | 0.99989235 |
Top |
Fusion Genomic Features for GPR124-WRN |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| GPR124 | chr8 | 37655052 | + | WRN | chr8 | 30938382 | + | 0.3488811 | 0.6511189 |
| GPR124 | chr8 | 37655052 | + | WRN | chr8 | 30938382 | + | 0.3488811 | 0.6511189 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
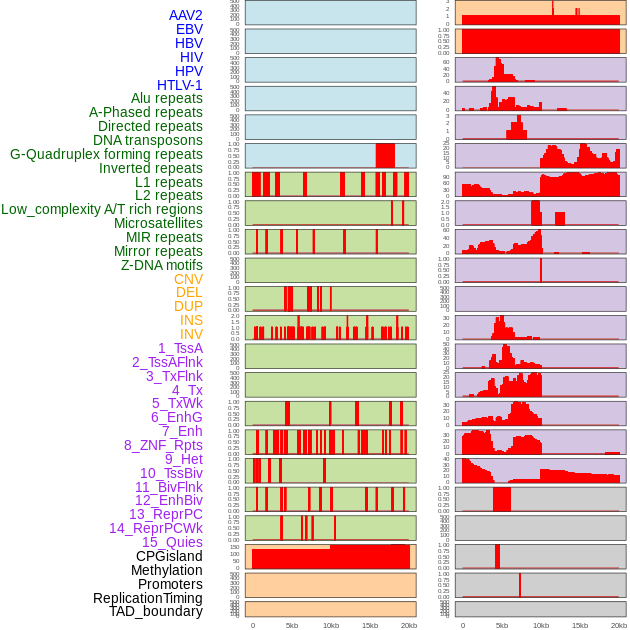 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
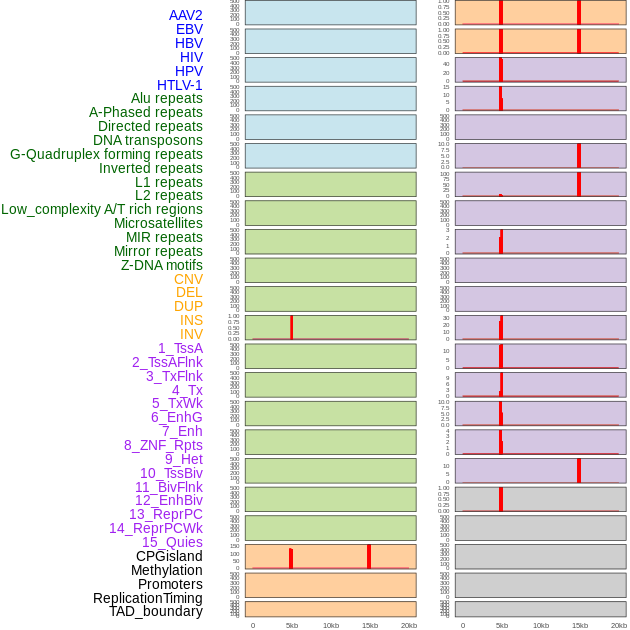 |
Top |
Fusion Protein Features for GPR124-WRN |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr8:37655052/chr8:30938383) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| . | . |
| FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. | FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | WRN | chr8:37655052 | chr8:30938383 | ENST00000298139 | 7 | 35 | 507_510 | 279 | 1433.0 | Compositional bias | Note=Poly-Glu | |
| Tgene | WRN | chr8:37655052 | chr8:30938383 | ENST00000298139 | 7 | 35 | 1150_1229 | 279 | 1433.0 | Domain | HRDC | |
| Tgene | WRN | chr8:37655052 | chr8:30938383 | ENST00000298139 | 7 | 35 | 558_724 | 279 | 1433.0 | Domain | Helicase ATP-binding | |
| Tgene | WRN | chr8:37655052 | chr8:30938383 | ENST00000298139 | 7 | 35 | 749_899 | 279 | 1433.0 | Domain | Helicase C-terminal | |
| Tgene | WRN | chr8:37655052 | chr8:30938383 | ENST00000298139 | 7 | 35 | 668_671 | 279 | 1433.0 | Motif | Note=DEAH box | |
| Tgene | WRN | chr8:37655052 | chr8:30938383 | ENST00000298139 | 7 | 35 | 571_578 | 279 | 1433.0 | Nucleotide binding | ATP | |
| Tgene | WRN | chr8:37655052 | chr8:30938383 | ENST00000298139 | 7 | 35 | 424_477 | 279 | 1433.0 | Region | Note=2 X 27 AA tandem repeats of H-L-S-P-N-D-N-E-N-D-T-S-Y-V-I-E-S-D-E-D-L-E-M-E-M-L-K | |
| Tgene | WRN | chr8:37655052 | chr8:30938383 | ENST00000298139 | 7 | 35 | 424_450 | 279 | 1433.0 | Repeat | 1 | |
| Tgene | WRN | chr8:37655052 | chr8:30938383 | ENST00000298139 | 7 | 35 | 451_477 | 279 | 1433.0 | Repeat | 2 |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | GPR124 | chr8:37655052 | chr8:30938383 | ENST00000315215 | + | 1 | 16 | 190_240 | 88 | 1122.0 | Domain | LRRCT |
| Hgene | GPR124 | chr8:37655052 | chr8:30938383 | ENST00000315215 | + | 1 | 16 | 247_344 | 88 | 1122.0 | Domain | Ig-like |
| Hgene | GPR124 | chr8:37655052 | chr8:30938383 | ENST00000315215 | + | 1 | 16 | 707_758 | 88 | 1122.0 | Domain | GPS |
| Hgene | GPR124 | chr8:37655052 | chr8:30938383 | ENST00000412232 | + | 1 | 19 | 190_240 | 88 | 1339.0 | Domain | LRRCT |
| Hgene | GPR124 | chr8:37655052 | chr8:30938383 | ENST00000412232 | + | 1 | 19 | 247_344 | 88 | 1339.0 | Domain | Ig-like |
| Hgene | GPR124 | chr8:37655052 | chr8:30938383 | ENST00000412232 | + | 1 | 19 | 707_758 | 88 | 1339.0 | Domain | GPS |
| Hgene | GPR124 | chr8:37655052 | chr8:30938383 | ENST00000315215 | + | 1 | 16 | 362_364 | 88 | 1122.0 | Motif | RGD |
| Hgene | GPR124 | chr8:37655052 | chr8:30938383 | ENST00000412232 | + | 1 | 19 | 362_364 | 88 | 1339.0 | Motif | RGD |
| Hgene | GPR124 | chr8:37655052 | chr8:30938383 | ENST00000315215 | + | 1 | 16 | 107_130 | 88 | 1122.0 | Repeat | LRR 2 |
| Hgene | GPR124 | chr8:37655052 | chr8:30938383 | ENST00000315215 | + | 1 | 16 | 131_154 | 88 | 1122.0 | Repeat | LRR 3 |
| Hgene | GPR124 | chr8:37655052 | chr8:30938383 | ENST00000315215 | + | 1 | 16 | 156_178 | 88 | 1122.0 | Repeat | LRR 4 |
| Hgene | GPR124 | chr8:37655052 | chr8:30938383 | ENST00000315215 | + | 1 | 16 | 82_106 | 88 | 1122.0 | Repeat | LRR 1 |
| Hgene | GPR124 | chr8:37655052 | chr8:30938383 | ENST00000412232 | + | 1 | 19 | 107_130 | 88 | 1339.0 | Repeat | LRR 2 |
| Hgene | GPR124 | chr8:37655052 | chr8:30938383 | ENST00000412232 | + | 1 | 19 | 131_154 | 88 | 1339.0 | Repeat | LRR 3 |
| Hgene | GPR124 | chr8:37655052 | chr8:30938383 | ENST00000412232 | + | 1 | 19 | 156_178 | 88 | 1339.0 | Repeat | LRR 4 |
| Hgene | GPR124 | chr8:37655052 | chr8:30938383 | ENST00000412232 | + | 1 | 19 | 82_106 | 88 | 1339.0 | Repeat | LRR 1 |
| Hgene | GPR124 | chr8:37655052 | chr8:30938383 | ENST00000315215 | + | 1 | 16 | 1041_1047 | 88 | 1122.0 | Topological domain | Extracellular |
| Hgene | GPR124 | chr8:37655052 | chr8:30938383 | ENST00000315215 | + | 1 | 16 | 1069_1338 | 88 | 1122.0 | Topological domain | Cytoplasmic |
| Hgene | GPR124 | chr8:37655052 | chr8:30938383 | ENST00000315215 | + | 1 | 16 | 34_771 | 88 | 1122.0 | Topological domain | Extracellular |
| Hgene | GPR124 | chr8:37655052 | chr8:30938383 | ENST00000315215 | + | 1 | 16 | 793_807 | 88 | 1122.0 | Topological domain | Cytoplasmic |
| Hgene | GPR124 | chr8:37655052 | chr8:30938383 | ENST00000315215 | + | 1 | 16 | 829_832 | 88 | 1122.0 | Topological domain | Extracellular |
| Hgene | GPR124 | chr8:37655052 | chr8:30938383 | ENST00000315215 | + | 1 | 16 | 854_886 | 88 | 1122.0 | Topological domain | Cytoplasmic |
| Hgene | GPR124 | chr8:37655052 | chr8:30938383 | ENST00000315215 | + | 1 | 16 | 908_924 | 88 | 1122.0 | Topological domain | Extracellular |
| Hgene | GPR124 | chr8:37655052 | chr8:30938383 | ENST00000315215 | + | 1 | 16 | 946_1019 | 88 | 1122.0 | Topological domain | Cytoplasmic |
| Hgene | GPR124 | chr8:37655052 | chr8:30938383 | ENST00000412232 | + | 1 | 19 | 1041_1047 | 88 | 1339.0 | Topological domain | Extracellular |
| Hgene | GPR124 | chr8:37655052 | chr8:30938383 | ENST00000412232 | + | 1 | 19 | 1069_1338 | 88 | 1339.0 | Topological domain | Cytoplasmic |
| Hgene | GPR124 | chr8:37655052 | chr8:30938383 | ENST00000412232 | + | 1 | 19 | 34_771 | 88 | 1339.0 | Topological domain | Extracellular |
| Hgene | GPR124 | chr8:37655052 | chr8:30938383 | ENST00000412232 | + | 1 | 19 | 793_807 | 88 | 1339.0 | Topological domain | Cytoplasmic |
| Hgene | GPR124 | chr8:37655052 | chr8:30938383 | ENST00000412232 | + | 1 | 19 | 829_832 | 88 | 1339.0 | Topological domain | Extracellular |
| Hgene | GPR124 | chr8:37655052 | chr8:30938383 | ENST00000412232 | + | 1 | 19 | 854_886 | 88 | 1339.0 | Topological domain | Cytoplasmic |
| Hgene | GPR124 | chr8:37655052 | chr8:30938383 | ENST00000412232 | + | 1 | 19 | 908_924 | 88 | 1339.0 | Topological domain | Extracellular |
| Hgene | GPR124 | chr8:37655052 | chr8:30938383 | ENST00000412232 | + | 1 | 19 | 946_1019 | 88 | 1339.0 | Topological domain | Cytoplasmic |
| Hgene | GPR124 | chr8:37655052 | chr8:30938383 | ENST00000315215 | + | 1 | 16 | 1020_1040 | 88 | 1122.0 | Transmembrane | Helical%3B Name%3D6 |
| Hgene | GPR124 | chr8:37655052 | chr8:30938383 | ENST00000315215 | + | 1 | 16 | 1048_1068 | 88 | 1122.0 | Transmembrane | Helical%3B Name%3D7 |
| Hgene | GPR124 | chr8:37655052 | chr8:30938383 | ENST00000315215 | + | 1 | 16 | 772_792 | 88 | 1122.0 | Transmembrane | Helical%3B Name%3D1 |
| Hgene | GPR124 | chr8:37655052 | chr8:30938383 | ENST00000315215 | + | 1 | 16 | 808_828 | 88 | 1122.0 | Transmembrane | Helical%3B Name%3D2 |
| Hgene | GPR124 | chr8:37655052 | chr8:30938383 | ENST00000315215 | + | 1 | 16 | 833_853 | 88 | 1122.0 | Transmembrane | Helical%3B Name%3D3 |
| Hgene | GPR124 | chr8:37655052 | chr8:30938383 | ENST00000315215 | + | 1 | 16 | 887_907 | 88 | 1122.0 | Transmembrane | Helical%3B Name%3D4 |
| Hgene | GPR124 | chr8:37655052 | chr8:30938383 | ENST00000315215 | + | 1 | 16 | 925_945 | 88 | 1122.0 | Transmembrane | Helical%3B Name%3D5 |
| Hgene | GPR124 | chr8:37655052 | chr8:30938383 | ENST00000412232 | + | 1 | 19 | 1020_1040 | 88 | 1339.0 | Transmembrane | Helical%3B Name%3D6 |
| Hgene | GPR124 | chr8:37655052 | chr8:30938383 | ENST00000412232 | + | 1 | 19 | 1048_1068 | 88 | 1339.0 | Transmembrane | Helical%3B Name%3D7 |
| Hgene | GPR124 | chr8:37655052 | chr8:30938383 | ENST00000412232 | + | 1 | 19 | 772_792 | 88 | 1339.0 | Transmembrane | Helical%3B Name%3D1 |
| Hgene | GPR124 | chr8:37655052 | chr8:30938383 | ENST00000412232 | + | 1 | 19 | 808_828 | 88 | 1339.0 | Transmembrane | Helical%3B Name%3D2 |
| Hgene | GPR124 | chr8:37655052 | chr8:30938383 | ENST00000412232 | + | 1 | 19 | 833_853 | 88 | 1339.0 | Transmembrane | Helical%3B Name%3D3 |
| Hgene | GPR124 | chr8:37655052 | chr8:30938383 | ENST00000412232 | + | 1 | 19 | 887_907 | 88 | 1339.0 | Transmembrane | Helical%3B Name%3D4 |
| Hgene | GPR124 | chr8:37655052 | chr8:30938383 | ENST00000412232 | + | 1 | 19 | 925_945 | 88 | 1339.0 | Transmembrane | Helical%3B Name%3D5 |
| Tgene | WRN | chr8:37655052 | chr8:30938383 | ENST00000298139 | 7 | 35 | 60_228 | 279 | 1433.0 | Domain | Note=3'-5' exonuclease |
Top |
Fusion Gene Sequence for GPR124-WRN |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >34366_34366_1_GPR124-WRN_GPR124_chr8_37655052_ENST00000315215_WRN_chr8_30938383_ENST00000298139_length(transcript)=4756nt_BP=629nt GCGGCGGCAGCAGGAGCCCGGCGCGATCCGCTAGGTCCCAGCCCAGCGCCCAGCGAGCAGGCGACGCGGAGGGGCCGGGCCTCCAGTGTC CCGAGGGCCGGGCGCTGAGACTCCGGCCGCGCAGCTGGGAGCTGCCCGCGCTGCGCTGACAGCCGCGCCGACGTCCTCCCCGCCGGGGCG CTCGCAGGACATGCCCCCGGGGCGCGGCGGCGGGGACCCCGGGGCTCGCCTCCGCCCAGGGCCCCCCTCCACGCCCTCGGGAGCCCCGGG CCCCCGCTGAGCACTCCTCCCGCACGCCTGGGTCCCTCCGGCCGGCGCGCAGCCCGGCCCCAGCGCTGTGGGTCCCCGCGGGGCGATGGG TTGATGGGCGCCGGGGGACGCAGGATGCGGGGGGCGCCCGCGCGCCTGCTGCTGCCGCTGCTGCCGTGGCTCCTGCTGCTCCTGGCGCCC GAGGCTCGGGGCGCGCCCGGCTGCCCGCTATCCATCCGCAGCTGCAAGTGCTCGGGGGAGCGGCCCAAGGGGCTGAGCGGCGGCGTCCCT GGCCCGGCTCGGCGGAGGGTGGTGTGCAGCGGCGGGGACCTCCCGGAGCCTCCCGAGCCCGGCCTTCTGCCTAACGGCACCGTTACCCTG GTTTCTATCTTACTAAAGGATATTTCAGAAAATCTATATTCACTGAGGAGGATGATAATTGGGTCTACTAACATTGAGACTGAACTGAGG CCCAGCAATAATTTAAACTTATTATCCTTTGAAGATTCAACTACTGGGGGAGTACAACAGAAACAAATTAGAGAACATGAAGTTTTAATT CACGTTGAAGATGAAACATGGGACCCAACACTTGATCATTTAGCTAAACATGATGGAGAAGATGTACTTGGAAATAAAGTGGAACGAAAA GAAGATGGATTTGAAGATGGAGTAGAAGACAACAAATTGAAAGAGAATATGGAAAGAGCTTGTTTGATGTCGTTAGATATTACAGAACAT GAACTCCAAATTTTGGAACAGCAGTCTCAGGAAGAATATCTTAGTGATATTGCTTATAAATCTACTGAGCATTTATCTCCCAATGATAAT GAAAACGATACGTCCTATGTAATTGAGAGTGATGAAGATTTAGAAATGGAGATGCTTAAGCATTTATCTCCCAATGATAATGAAAACGAT ACGTCCTATGTAATTGAGAGTGATGAAGATTTAGAAATGGAGATGCTTAAGTCTTTAGAAAACCTCAATAGTGGCACGGTAGAACCAACT CATTCTAAATGCTTAAAAATGGAAAGAAATCTGGGTCTTCCTACTAAAGAAGAAGAAGAAGATGATGAAAATGAAGCTAATGAAGGGGAA GAAGATGATGATAAGGACTTTTTGTGGCCAGCACCCAATGAAGAGCAAGTTACTTGCCTCAAGATGTACTTTGGCCATTCCAGTTTTAAA CCAGTTCAGTGGAAAGTGATTCATTCAGTATTAGAAGAAAGAAGAGATAATGTTGCTGTCATGGCAACTGGATATGGAAAGAGTTTGTGC TTCCAGTATCCACCTGTTTATGTAGGCAAGATTGGCCTTGTTATCTCTCCCCTTATTTCTCTGATGGAAGACCAAGTGCTACAGCTTAAA ATGTCCAACATCCCAGCTTGCTTCCTTGGATCAGCACAGTCAGAAAATGTTCTAACAGATATTAAATTAGGTAAATACCGGATTGTATAC GTAACTCCAGAATACTGTTCAGGTAACATGGGCCTGCTCCAGCAACTTGAGGCTGATATTGGTATCACGCTCATTGCTGTGGATGAGGCT CACTGTATTTCTGAGTGGGGGCATGATTTTAGGGATTCATTCAGGAAGTTGGGCTCCCTAAAGACAGCACTGCCAATGGTTCCAATCGTT GCACTTACTGCTACTGCAAGTTCTTCAATCCGGGAAGACATTGTACGTTGCTTAAATCTGAGAAATCCTCAGATCACCTGTACTGGTTTT GATCGACCAAACCTGTATTTAGAAGTTAGGCGAAAAACAGGGAATATCCTTCAGGATCTGCAGCCATTTCTTGTCAAAACAAGTTCCCAC TGGGAATTTGAAGGTCCAACAATCATCTACTGTCCTTCTAGAAAAATGACACAACAAGTTACAGGTGAACTTAGGAAACTGAATCTATCC TGTGGAACATACCATGCGGGCATGAGTTTTAGCACAAGGAAAGACATTCATCATAGGTTTGTAAGAGATGAAATTCAGTGTGTCATAGCT ACCATAGCTTTTGGAATGGGCATTAATAAAGCTGACATTCGCCAAGTCATTCATTACGGTGCTCCTAAGGACATGGAATCATATTATCAG GAGATTGGTAGAGCTGGTCGTGATGGACTTCAAAGTTCTTGTCACGTCCTCTGGGCTCCTGCAGACATTAACTTAAATAGGCACCTTCTT ACTGAGATACGTAATGAGAAGTTTCGATTATACAAATTAAAGATGATGGCAAAGATGGAAAAATATCTTCATTCTAGCAGATGTAGGAGA CAAATCATCTTGTCTCATTTTGAGGACAAACAAGTACAAAAAGCCTCCTTGGGAATTATGGGAACTGAAAAATGCTGTGATAATTGCAGG TCCAGATTGGATCATTGCTATTCCATGGATGACTCAGAGGATACATCCTGGGACTTTGGTCCACAAGCATTTAAGCTTTTGTCTGCTGTG GACATCTTAGGCGAAAAATTTGGAATTGGGCTTCCAATTTTATTTCTCCGAGGATCTAATTCTCAGCGTCTTGCCGATCAATATCGCAGG CACAGTTTATTTGGCACTGGCAAGGATCAAACAGAGAGTTGGTGGAAGGCTTTTTCCCGTCAGCTGATCACTGAGGGATTCTTGGTAGAA GTTTCTCGGTATAACAAATTTATGAAGATTTGCGCCCTTACGAAAAAGGGTAGAAATTGGCTTCATAAAGCTAATACAGAATCTCAGAGC CTCATCCTTCAAGCTAATGAAGAATTGTGTCCAAAGAAGTTGCTTCTGCCTAGTTCGAAAACTGTATCTTCGGGCACCAAAGAGCATTGT TATAATCAAGTACCAGTTGAATTAAGTACAGAGAAGAAGTCTAACTTGGAGAAGTTATATTCTTATAAACCATGTGATAAGATTTCTTCT GGGAGTAACATTTCTAAAAAAAGTATCATGGTACAGTCACCAGAAAAAGCTTACAGTTCCTCACAGCCTGTTATTTCGGCACAAGAGCAG GAGACTCAGATTGTGTTATATGGCAAATTGGTAGAAGCTAGGCAGAAACATGCCAATAAAATGGATGTTCCCCCAGCTATTCTGGCAACA AACAAGATACTGGTGGATATGGCCAAAATGAGACCAACTACGGTTGAAAACGTAAAAAGGATTGATGGTGTTTCTGAAGGCAAAGCTGCC ATGTTGGCCCCTCTGTTGGAAGTCATCAAACATTTCTGCCAAACAAATAGTGTTCAGACAGACCTCTTTTCAAGTACAAAACCTCAAGAA GAACAGAAGACGAGTCTGGTAGCAAAAAATAAAATATGCACACTTTCACAGTCTATGGCCATCACATACTCTTTATTCCAAGAAAAGAAG ATGCCTTTGAAGAGCATAGCTGAGAGCAGGATTCTGCCTCTCATGACAATTGGCATGCACTTATCCCAAGCGGTGAAAGCTGGCTGCCCC CTTGATTTGGAGCGAGCAGGCCTGACTCCAGAGGTTCAGAAGATTATTGCTGATGTTATCCGAAACCCTCCCGTCAACTCAGATATGAGT AAAATTAGCCTAATCAGAATGTTAGTTCCTGAAAACATTGACACGTACCTTATCCACATGGCAATTGAGATCCTTAAACATGGTCCTGAC AGCGGACTTCAACCTTCATGTGATGTCAACAAAAGGAGATGTTTTCCCGGTTCTGAAGAGATCTGTTCAAGTTCTAAGAGAAGCAAGGAA GAAGTAGGCATCAATACTGAGACTTCATCTGCAGAGAGAAAGAGACGATTACCTGTGTGGTTTGCCAAAGGAAGTGATACCAGCAAGAAA TTAATGGACAAAACGAAAAGGGGAGGTCTTTTTAGTTAAGCTGGCAATTACCAGAACAATTATGTTTCTTGCTGTATTATAAGAGGATAG CTATATTTTATTTCTGAAGAGTAAGGAGTAGTATTTTGGCTTAAAAATCATTCTAATTACAAAGTTCACTGTTTATTGAAGAACTGGCAT CTTAAATCAGCCTTCCGCAATTCATGTAGTTTCTGGGTCTTCTGGGAGCCTACGTGAGTACATCACCTAACAGAATATTAAATTAGACTT CCTGTAAGATTGCTTTAAGAAACTGTTACTGTCCTGTTTTCTAATCTCTTTATTAAAACAGTGTATTTGGAAAATGTTATGTGCTCTGAT TTGATATAGATAACAGATTAGTAGTTACATGGTAATTATGTGATATAAAATATTCATATATTATCAAAATTCTGTTTTGTAAATGTAAGA AAGCATAGTTATTTTACAAATTGTTTTTACTGTCTTTTGAAGAAGTTCTTAAATACGTTGTTAAATGGTATTAGTTGACCAGGGCAGTGA AAATGAAACCGCATTTTGGGTGCCATTAAATAGGGAAAAAACATGTAAAAAATGTAAAATGGAGACCAATTGCACTAGGCAAGTGTATAT >34366_34366_1_GPR124-WRN_GPR124_chr8_37655052_ENST00000315215_WRN_chr8_30938383_ENST00000298139_length(amino acids)=1270AA_BP=118 MSTPPARLGPSGRRAARPQRCGSPRGDGLMGAGGRRMRGAPARLLLPLLPWLLLLLAPEARGAPGCPLSIRSCKCSGERPKGLSGGVPGP ARRRVVCSGGDLPEPPEPGLLPNGTVTLVSILLKDISENLYSLRRMIIGSTNIETELRPSNNLNLLSFEDSTTGGVQQKQIREHEVLIHV EDETWDPTLDHLAKHDGEDVLGNKVERKEDGFEDGVEDNKLKENMERACLMSLDITEHELQILEQQSQEEYLSDIAYKSTEHLSPNDNEN DTSYVIESDEDLEMEMLKHLSPNDNENDTSYVIESDEDLEMEMLKSLENLNSGTVEPTHSKCLKMERNLGLPTKEEEEDDENEANEGEED DDKDFLWPAPNEEQVTCLKMYFGHSSFKPVQWKVIHSVLEERRDNVAVMATGYGKSLCFQYPPVYVGKIGLVISPLISLMEDQVLQLKMS NIPACFLGSAQSENVLTDIKLGKYRIVYVTPEYCSGNMGLLQQLEADIGITLIAVDEAHCISEWGHDFRDSFRKLGSLKTALPMVPIVAL TATASSSIREDIVRCLNLRNPQITCTGFDRPNLYLEVRRKTGNILQDLQPFLVKTSSHWEFEGPTIIYCPSRKMTQQVTGELRKLNLSCG TYHAGMSFSTRKDIHHRFVRDEIQCVIATIAFGMGINKADIRQVIHYGAPKDMESYYQEIGRAGRDGLQSSCHVLWAPADINLNRHLLTE IRNEKFRLYKLKMMAKMEKYLHSSRCRRQIILSHFEDKQVQKASLGIMGTEKCCDNCRSRLDHCYSMDDSEDTSWDFGPQAFKLLSAVDI LGEKFGIGLPILFLRGSNSQRLADQYRRHSLFGTGKDQTESWWKAFSRQLITEGFLVEVSRYNKFMKICALTKKGRNWLHKANTESQSLI LQANEELCPKKLLLPSSKTVSSGTKEHCYNQVPVELSTEKKSNLEKLYSYKPCDKISSGSNISKKSIMVQSPEKAYSSSQPVISAQEQET QIVLYGKLVEARQKHANKMDVPPAILATNKILVDMAKMRPTTVENVKRIDGVSEGKAAMLAPLLEVIKHFCQTNSVQTDLFSSTKPQEEQ KTSLVAKNKICTLSQSMAITYSLFQEKKMPLKSIAESRILPLMTIGMHLSQAVKAGCPLDLERAGLTPEVQKIIADVIRNPPVNSDMSKI SLIRMLVPENIDTYLIHMAIEILKHGPDSGLQPSCDVNKRRCFPGSEEICSSSKRSKEEVGINTETSSAERKRRLPVWFAKGSDTSKKLM -------------------------------------------------------------- >34366_34366_2_GPR124-WRN_GPR124_chr8_37655052_ENST00000412232_WRN_chr8_30938383_ENST00000298139_length(transcript)=4406nt_BP=279nt GGGCGATGGGTTGATGGGCGCCGGGGGACGCAGGATGCGGGGGGCGCCCGCGCGCCTGCTGCTGCCGCTGCTGCCGTGGCTCCTGCTGCT CCTGGCGCCCGAGGCTCGGGGCGCGCCCGGCTGCCCGCTATCCATCCGCAGCTGCAAGTGCTCGGGGGAGCGGCCCAAGGGGCTGAGCGG CGGCGTCCCTGGCCCGGCTCGGCGGAGGGTGGTGTGCAGCGGCGGGGACCTCCCGGAGCCTCCCGAGCCCGGCCTTCTGCCTAACGGCAC CGTTACCCTGGTTTCTATCTTACTAAAGGATATTTCAGAAAATCTATATTCACTGAGGAGGATGATAATTGGGTCTACTAACATTGAGAC TGAACTGAGGCCCAGCAATAATTTAAACTTATTATCCTTTGAAGATTCAACTACTGGGGGAGTACAACAGAAACAAATTAGAGAACATGA AGTTTTAATTCACGTTGAAGATGAAACATGGGACCCAACACTTGATCATTTAGCTAAACATGATGGAGAAGATGTACTTGGAAATAAAGT GGAACGAAAAGAAGATGGATTTGAAGATGGAGTAGAAGACAACAAATTGAAAGAGAATATGGAAAGAGCTTGTTTGATGTCGTTAGATAT TACAGAACATGAACTCCAAATTTTGGAACAGCAGTCTCAGGAAGAATATCTTAGTGATATTGCTTATAAATCTACTGAGCATTTATCTCC CAATGATAATGAAAACGATACGTCCTATGTAATTGAGAGTGATGAAGATTTAGAAATGGAGATGCTTAAGCATTTATCTCCCAATGATAA TGAAAACGATACGTCCTATGTAATTGAGAGTGATGAAGATTTAGAAATGGAGATGCTTAAGTCTTTAGAAAACCTCAATAGTGGCACGGT AGAACCAACTCATTCTAAATGCTTAAAAATGGAAAGAAATCTGGGTCTTCCTACTAAAGAAGAAGAAGAAGATGATGAAAATGAAGCTAA TGAAGGGGAAGAAGATGATGATAAGGACTTTTTGTGGCCAGCACCCAATGAAGAGCAAGTTACTTGCCTCAAGATGTACTTTGGCCATTC CAGTTTTAAACCAGTTCAGTGGAAAGTGATTCATTCAGTATTAGAAGAAAGAAGAGATAATGTTGCTGTCATGGCAACTGGATATGGAAA GAGTTTGTGCTTCCAGTATCCACCTGTTTATGTAGGCAAGATTGGCCTTGTTATCTCTCCCCTTATTTCTCTGATGGAAGACCAAGTGCT ACAGCTTAAAATGTCCAACATCCCAGCTTGCTTCCTTGGATCAGCACAGTCAGAAAATGTTCTAACAGATATTAAATTAGGTAAATACCG GATTGTATACGTAACTCCAGAATACTGTTCAGGTAACATGGGCCTGCTCCAGCAACTTGAGGCTGATATTGGTATCACGCTCATTGCTGT GGATGAGGCTCACTGTATTTCTGAGTGGGGGCATGATTTTAGGGATTCATTCAGGAAGTTGGGCTCCCTAAAGACAGCACTGCCAATGGT TCCAATCGTTGCACTTACTGCTACTGCAAGTTCTTCAATCCGGGAAGACATTGTACGTTGCTTAAATCTGAGAAATCCTCAGATCACCTG TACTGGTTTTGATCGACCAAACCTGTATTTAGAAGTTAGGCGAAAAACAGGGAATATCCTTCAGGATCTGCAGCCATTTCTTGTCAAAAC AAGTTCCCACTGGGAATTTGAAGGTCCAACAATCATCTACTGTCCTTCTAGAAAAATGACACAACAAGTTACAGGTGAACTTAGGAAACT GAATCTATCCTGTGGAACATACCATGCGGGCATGAGTTTTAGCACAAGGAAAGACATTCATCATAGGTTTGTAAGAGATGAAATTCAGTG TGTCATAGCTACCATAGCTTTTGGAATGGGCATTAATAAAGCTGACATTCGCCAAGTCATTCATTACGGTGCTCCTAAGGACATGGAATC ATATTATCAGGAGATTGGTAGAGCTGGTCGTGATGGACTTCAAAGTTCTTGTCACGTCCTCTGGGCTCCTGCAGACATTAACTTAAATAG GCACCTTCTTACTGAGATACGTAATGAGAAGTTTCGATTATACAAATTAAAGATGATGGCAAAGATGGAAAAATATCTTCATTCTAGCAG ATGTAGGAGACAAATCATCTTGTCTCATTTTGAGGACAAACAAGTACAAAAAGCCTCCTTGGGAATTATGGGAACTGAAAAATGCTGTGA TAATTGCAGGTCCAGATTGGATCATTGCTATTCCATGGATGACTCAGAGGATACATCCTGGGACTTTGGTCCACAAGCATTTAAGCTTTT GTCTGCTGTGGACATCTTAGGCGAAAAATTTGGAATTGGGCTTCCAATTTTATTTCTCCGAGGATCTAATTCTCAGCGTCTTGCCGATCA ATATCGCAGGCACAGTTTATTTGGCACTGGCAAGGATCAAACAGAGAGTTGGTGGAAGGCTTTTTCCCGTCAGCTGATCACTGAGGGATT CTTGGTAGAAGTTTCTCGGTATAACAAATTTATGAAGATTTGCGCCCTTACGAAAAAGGGTAGAAATTGGCTTCATAAAGCTAATACAGA ATCTCAGAGCCTCATCCTTCAAGCTAATGAAGAATTGTGTCCAAAGAAGTTGCTTCTGCCTAGTTCGAAAACTGTATCTTCGGGCACCAA AGAGCATTGTTATAATCAAGTACCAGTTGAATTAAGTACAGAGAAGAAGTCTAACTTGGAGAAGTTATATTCTTATAAACCATGTGATAA GATTTCTTCTGGGAGTAACATTTCTAAAAAAAGTATCATGGTACAGTCACCAGAAAAAGCTTACAGTTCCTCACAGCCTGTTATTTCGGC ACAAGAGCAGGAGACTCAGATTGTGTTATATGGCAAATTGGTAGAAGCTAGGCAGAAACATGCCAATAAAATGGATGTTCCCCCAGCTAT TCTGGCAACAAACAAGATACTGGTGGATATGGCCAAAATGAGACCAACTACGGTTGAAAACGTAAAAAGGATTGATGGTGTTTCTGAAGG CAAAGCTGCCATGTTGGCCCCTCTGTTGGAAGTCATCAAACATTTCTGCCAAACAAATAGTGTTCAGACAGACCTCTTTTCAAGTACAAA ACCTCAAGAAGAACAGAAGACGAGTCTGGTAGCAAAAAATAAAATATGCACACTTTCACAGTCTATGGCCATCACATACTCTTTATTCCA AGAAAAGAAGATGCCTTTGAAGAGCATAGCTGAGAGCAGGATTCTGCCTCTCATGACAATTGGCATGCACTTATCCCAAGCGGTGAAAGC TGGCTGCCCCCTTGATTTGGAGCGAGCAGGCCTGACTCCAGAGGTTCAGAAGATTATTGCTGATGTTATCCGAAACCCTCCCGTCAACTC AGATATGAGTAAAATTAGCCTAATCAGAATGTTAGTTCCTGAAAACATTGACACGTACCTTATCCACATGGCAATTGAGATCCTTAAACA TGGTCCTGACAGCGGACTTCAACCTTCATGTGATGTCAACAAAAGGAGATGTTTTCCCGGTTCTGAAGAGATCTGTTCAAGTTCTAAGAG AAGCAAGGAAGAAGTAGGCATCAATACTGAGACTTCATCTGCAGAGAGAAAGAGACGATTACCTGTGTGGTTTGCCAAAGGAAGTGATAC CAGCAAGAAATTAATGGACAAAACGAAAAGGGGAGGTCTTTTTAGTTAAGCTGGCAATTACCAGAACAATTATGTTTCTTGCTGTATTAT AAGAGGATAGCTATATTTTATTTCTGAAGAGTAAGGAGTAGTATTTTGGCTTAAAAATCATTCTAATTACAAAGTTCACTGTTTATTGAA GAACTGGCATCTTAAATCAGCCTTCCGCAATTCATGTAGTTTCTGGGTCTTCTGGGAGCCTACGTGAGTACATCACCTAACAGAATATTA AATTAGACTTCCTGTAAGATTGCTTTAAGAAACTGTTACTGTCCTGTTTTCTAATCTCTTTATTAAAACAGTGTATTTGGAAAATGTTAT GTGCTCTGATTTGATATAGATAACAGATTAGTAGTTACATGGTAATTATGTGATATAAAATATTCATATATTATCAAAATTCTGTTTTGT AAATGTAAGAAAGCATAGTTATTTTACAAATTGTTTTTACTGTCTTTTGAAGAAGTTCTTAAATACGTTGTTAAATGGTATTAGTTGACC AGGGCAGTGAAAATGAAACCGCATTTTGGGTGCCATTAAATAGGGAAAAAACATGTAAAAAATGTAAAATGGAGACCAATTGCACTAGGC >34366_34366_2_GPR124-WRN_GPR124_chr8_37655052_ENST00000412232_WRN_chr8_30938383_ENST00000298139_length(amino acids)=1242AA_BP=90 MMGAGGRRMRGAPARLLLPLLPWLLLLLAPEARGAPGCPLSIRSCKCSGERPKGLSGGVPGPARRRVVCSGGDLPEPPEPGLLPNGTVTL VSILLKDISENLYSLRRMIIGSTNIETELRPSNNLNLLSFEDSTTGGVQQKQIREHEVLIHVEDETWDPTLDHLAKHDGEDVLGNKVERK EDGFEDGVEDNKLKENMERACLMSLDITEHELQILEQQSQEEYLSDIAYKSTEHLSPNDNENDTSYVIESDEDLEMEMLKHLSPNDNEND TSYVIESDEDLEMEMLKSLENLNSGTVEPTHSKCLKMERNLGLPTKEEEEDDENEANEGEEDDDKDFLWPAPNEEQVTCLKMYFGHSSFK PVQWKVIHSVLEERRDNVAVMATGYGKSLCFQYPPVYVGKIGLVISPLISLMEDQVLQLKMSNIPACFLGSAQSENVLTDIKLGKYRIVY VTPEYCSGNMGLLQQLEADIGITLIAVDEAHCISEWGHDFRDSFRKLGSLKTALPMVPIVALTATASSSIREDIVRCLNLRNPQITCTGF DRPNLYLEVRRKTGNILQDLQPFLVKTSSHWEFEGPTIIYCPSRKMTQQVTGELRKLNLSCGTYHAGMSFSTRKDIHHRFVRDEIQCVIA TIAFGMGINKADIRQVIHYGAPKDMESYYQEIGRAGRDGLQSSCHVLWAPADINLNRHLLTEIRNEKFRLYKLKMMAKMEKYLHSSRCRR QIILSHFEDKQVQKASLGIMGTEKCCDNCRSRLDHCYSMDDSEDTSWDFGPQAFKLLSAVDILGEKFGIGLPILFLRGSNSQRLADQYRR HSLFGTGKDQTESWWKAFSRQLITEGFLVEVSRYNKFMKICALTKKGRNWLHKANTESQSLILQANEELCPKKLLLPSSKTVSSGTKEHC YNQVPVELSTEKKSNLEKLYSYKPCDKISSGSNISKKSIMVQSPEKAYSSSQPVISAQEQETQIVLYGKLVEARQKHANKMDVPPAILAT NKILVDMAKMRPTTVENVKRIDGVSEGKAAMLAPLLEVIKHFCQTNSVQTDLFSSTKPQEEQKTSLVAKNKICTLSQSMAITYSLFQEKK MPLKSIAESRILPLMTIGMHLSQAVKAGCPLDLERAGLTPEVQKIIADVIRNPPVNSDMSKISLIRMLVPENIDTYLIHMAIEILKHGPD -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for GPR124-WRN |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
| Tgene | WRN | chr8:37655052 | chr8:30938383 | ENST00000298139 | 7 | 35 | 987_993 | 279.6666666666667 | 1433.0 | DNA |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
| Tgene | WRN | chr8:37655052 | chr8:30938383 | ENST00000298139 | 7 | 35 | 2_277 | 279.6666666666667 | 1433.0 | WRNIP1 |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for GPR124-WRN |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for GPR124-WRN |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
