
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:GRHL2-NRBP2 (FusionGDB2 ID:34713) |
Fusion Gene Summary for GRHL2-NRBP2 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: GRHL2-NRBP2 | Fusion gene ID: 34713 | Hgene | Tgene | Gene symbol | GRHL2 | NRBP2 | Gene ID | 79977 | 340371 |
| Gene name | grainyhead like transcription factor 2 | nuclear receptor binding protein 2 | |
| Synonyms | BOM|DFNA28|ECTDS|PPCD4|TFCP2L3 | TRG16|pp9320 | |
| Cytomap | 8q22.3 | 8q24.3 | |
| Type of gene | protein-coding | protein-coding | |
| Description | grainyhead-like protein 2 homologbrother of mammalian grainyheadgrainyhead-like 2transcription factor CP2-like 3 | nuclear receptor-binding protein 2transformation-related protein 16 | |
| Modification date | 20200313 | 20200320 | |
| UniProtAcc | Q6ISB3 | Q9NSY0 | |
| Ensembl transtripts involved in fusion gene | ENST00000251808, ENST00000395927, ENST00000517674, | ENST00000327830, ENST00000442628, | |
| Fusion gene scores | * DoF score | 25 X 11 X 10=2750 | 3 X 3 X 3=27 |
| # samples | 25 | 3 | |
| ** MAII score | log2(25/2750*10)=-3.4594316186373 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(3/27*10)=0.15200309344505 effective Gene in Pan-Cancer Fusion Genes (eGinPCFGs). DoF>8 and MAII>0 | |
| Context | PubMed: GRHL2 [Title/Abstract] AND NRBP2 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | GRHL2(102661727)-NRBP2(144922401), # samples:2 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | GRHL2 | GO:0008544 | epidermis development | 23254293 |
| Hgene | GRHL2 | GO:0044030 | regulation of DNA methylation | 23254293 |
| Hgene | GRHL2 | GO:0045617 | negative regulation of keratinocyte differentiation | 23254293 |
| Hgene | GRHL2 | GO:0045944 | positive regulation of transcription by RNA polymerase II | 23814079 |
 Fusion gene breakpoints across GRHL2 (5'-gene) Fusion gene breakpoints across GRHL2 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
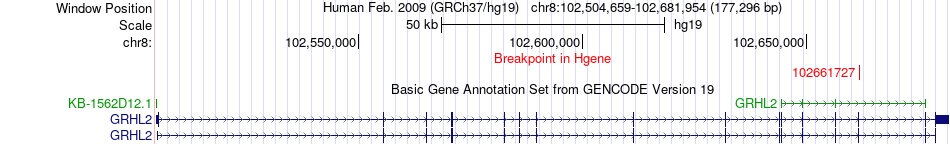 |
 Fusion gene breakpoints across NRBP2 (3'-gene) Fusion gene breakpoints across NRBP2 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
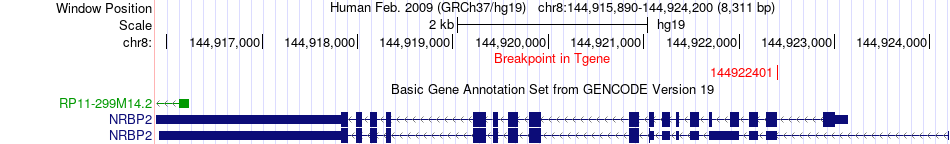 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | BRCA | TCGA-D8-A1JL-01A | GRHL2 | chr8 | 102661727 | + | NRBP2 | chr8 | 144922401 | - |
Top |
Fusion Gene ORF analysis for GRHL2-NRBP2 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-5UTR | ENST00000251808 | ENST00000327830 | GRHL2 | chr8 | 102661727 | + | NRBP2 | chr8 | 144922401 | - |
| 5CDS-5UTR | ENST00000395927 | ENST00000327830 | GRHL2 | chr8 | 102661727 | + | NRBP2 | chr8 | 144922401 | - |
| In-frame | ENST00000251808 | ENST00000442628 | GRHL2 | chr8 | 102661727 | + | NRBP2 | chr8 | 144922401 | - |
| In-frame | ENST00000395927 | ENST00000442628 | GRHL2 | chr8 | 102661727 | + | NRBP2 | chr8 | 144922401 | - |
| intron-3CDS | ENST00000517674 | ENST00000442628 | GRHL2 | chr8 | 102661727 | + | NRBP2 | chr8 | 144922401 | - |
| intron-5UTR | ENST00000517674 | ENST00000327830 | GRHL2 | chr8 | 102661727 | + | NRBP2 | chr8 | 144922401 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000251808 | GRHL2 | chr8 | 102661727 | + | ENST00000442628 | NRBP2 | chr8 | 144922401 | - | 5354 | 2036 | 230 | 3412 | 1060 |
| ENST00000395927 | GRHL2 | chr8 | 102661727 | + | ENST00000442628 | NRBP2 | chr8 | 144922401 | - | 5088 | 1770 | 54 | 3146 | 1030 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000251808 | ENST00000442628 | GRHL2 | chr8 | 102661727 | + | NRBP2 | chr8 | 144922401 | - | 0.003008584 | 0.9969914 |
| ENST00000395927 | ENST00000442628 | GRHL2 | chr8 | 102661727 | + | NRBP2 | chr8 | 144922401 | - | 0.002619032 | 0.9973809 |
Top |
Fusion Genomic Features for GRHL2-NRBP2 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
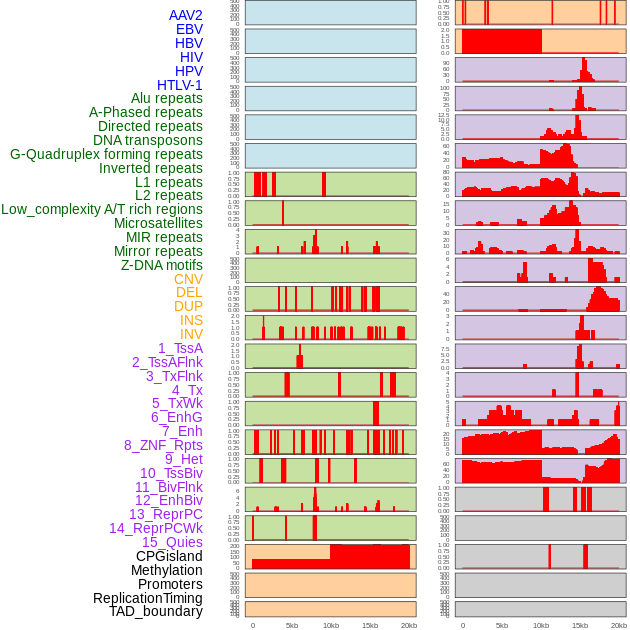 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for GRHL2-NRBP2 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr8:102661727/chr8:144922401) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| GRHL2 | NRBP2 |
| FUNCTION: Transcription factor playing an important role in primary neurulation and in epithelial development (PubMed:29309642, PubMed:25152456). Binds directly to the consensus DNA sequence 5'-AACCGGTT-3' acting as an activator and repressor on distinct target genes (By similarity). During embryogenesis, plays unique and cooperative roles with GRHL3 in establishing distinct zones of primary neurulation. Essential for closure 3 (rostral end of the forebrain), functions cooperatively with GRHL3 in closure 2 (forebrain/midbrain boundary) and posterior neuropore closure (By similarity). Regulates epithelial morphogenesis acting as a target gene-associated transcriptional activator of apical junctional complex components. Up-regulates of CLDN3 and CLDN4, as well as of RAB25, which increases the CLDN4 protein and its localization at tight junctions (By similarity). Comprises an essential component of the transcriptional machinery that establishes appropriate expression levels of CLDN4 and CDH1 in different types of epithelia. Exhibits functional redundancy with GRHL3 in epidermal morphogenetic events and epidermal wound repair (By similarity). In lung, forms a regulatory loop with NKX2-1 that coordinates lung epithelial cell morphogenesis and differentiation (By similarity). In keratinocytes, plays a role in telomerase activation during cellular proliferation, regulates TERT expression by binding to TERT promoter region and inhibiting DNA methylation at the 5'-CpG island, possibly by interfering with DNMT1 enzyme activity (PubMed:19015635, PubMed:20938050). In addition, impairs keratinocyte differentiation and epidermal function by inhibiting the expression of genes clustered at the epidermal differentiation complex (EDC) as well as GRHL1 and GRHL3 through epigenetic mechanisms (PubMed:23254293). {ECO:0000250|UniProtKB:Q8K5C0, ECO:0000269|PubMed:19015635, ECO:0000269|PubMed:20938050, ECO:0000269|PubMed:20978075, ECO:0000269|PubMed:23254293, ECO:0000269|PubMed:25152456, ECO:0000269|PubMed:29309642, ECO:0000305|PubMed:12175488}. | FUNCTION: May regulate apoptosis of neural progenitor cells during their differentiation. {ECO:0000250}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | GRHL2 | chr8:102661727 | chr8:144922401 | ENST00000251808 | + | 14 | 16 | 1_93 | 566 | 626.0 | Region | Transcription activation |
| Hgene | GRHL2 | chr8:102661727 | chr8:144922401 | ENST00000395927 | + | 14 | 16 | 1_93 | 550 | 610.0 | Region | Transcription activation |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | NRBP2 | chr8:102661727 | chr8:144922401 | ENST00000442628 | 0 | 18 | 38_306 | 43 | 502.0 | Domain | Protein kinase |
Top |
Fusion Gene Sequence for GRHL2-NRBP2 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >34713_34713_1_GRHL2-NRBP2_GRHL2_chr8_102661727_ENST00000251808_NRBP2_chr8_144922401_ENST00000442628_length(transcript)=5354nt_BP=2036nt TCCTTGCGAGAAAGTTACCTGTGGCCGCCCAAGTCCGCCACTTTCTGCTCTGTGTCTGCCCATTGCCACGATCCAGGAGGACTCCGCGCC GCCCGGCCGCCTCCGAGCTCGGGCCCCATGTGAGGGGCCCCCCCTTATCCCACCTTTCCGGCTAGGTGAGGGCGCGAGCGGGCGAGCGAG CGAGAGTGGTGAGGGGGGACGGAAAAGCAGAATTACCTGTAGCTCTTGTTCTGCCATCTCGGGCGCTCTCACACACCTTCACCTGCACAG ACTTGAAAGTCCAGTTTCACCAGAGGCTGAGGCTCCAGGAAAAGCGGAGCAAGTTCATTGGATCAAACATGTCACAAGAGTCGGACAATA ATAAAAGACTAGTGGCCTTAGTGCCCATGCCCAGTGACCCTCCATTCAATACCCGAAGAGCCTACACCAGTGAGGATGAAGCCTGGAAGT CATACTTGGAGAATCCCCTGACAGCAGCCACCAAGGCCATGATGAGCATTAATGGTGATGAGGACAGTGCTGCTGCCCTCGGCCTGCTCT ATGACTACTACAAGGTTCCTCGAGACAAGAGGCTGCTGTCTGTAAGCAAAGCAAGTGACAGCCAAGAAGACCAGGAGAAAAGAAACTGCC TTGGCACCAGTGAAGCCCAGAGTAATTTGAGTGGAGGAGAAAACCGAGTGCAAGTCCTAAAGACTGTTCCAGTGAACCTTTCCCTAAATC AAGATCACCTGGAGAATTCCAAGCGGGAACAGTACAGCATCAGCTTCCCCGAGAGCTCTGCCATCATCCCGGTGTCGGGAATCACGGTGG TGAAAGCTGAAGATTTCACACCAGTTTTCATGGCCCCACCTGTGCACTATCCCCGGGGAGATGGGGAAGAGCAACGAGTGGTTATCTTTG AACAGACTCAGTATGACGTGCCCTCGCTGGCCACCCACAGCGCCTATCTCAAAGACGACCAGCGCAGCACTCCGGACAGCACATACAGCG AGAGCTTCAAGGACGCAGCCACAGAGAAATTTCGGAGTGCTTCAGTTGGGGCTGAGGAGTACATGTATGATCAGACATCAAGTGGCACAT TTCAGTACACCCTGGAAGCCACCAAATCTCTCCGTCAGAAGCAGGGGGAGGGCCCCATGACCTACCTCAACAAAGGACAGTTCTATGCCA TAACACTCAGCGAGACCGGAGACAACAAATGCTTCCGACACCCCATCAGCAAAGTCAGGAGTGTGGTGATGGTGGTCTTCAGTGAAGACA AAAACAGAGATGAACAGCTCAAATACTGGAAATACTGGCACTCTCGGCAGCATACGGCGAAGCAGAGGGTCCTTGACATTGCCGATTACA AGGAGAGCTTTAATACGATTGGAAACATTGAAGAGATTGCATATAATGCTGTTTCCTTTACCTGGGACGTGAATGAAGAGGCGAAGATTT TCATCACCGTGAATTGCTTGAGCACAGATTTCTCCTCCCAAAAAGGGGTGAAAGGACTTCCTTTGATGATTCAGATTGACACATACAGTT ATAACAATCGTAGCAATAAACCCATTCATAGAGCTTATTGCCAGATCAAGGTCTTCTGTGACAAAGGAGCAGAAAGAAAAATCCGAGATG AAGAGCGGAAGCAGAACAGGAAGAAAGGGAAAGGCCAGGCCTCCCAAACTCAATGCAACAGCTCCTCTGATGGGAAGTTGGCTGCCATAC CTTTACAGAAGAAGAGTGACATCACCTACTTCAAAACCATGCCTGATCTCCACTCACAGCCAGTTCTCTTCATACCTGATGTTCACTTTG CAAACCTGCAGAGGACCGGACAGGTGTATTACAACACGGATGATGAACGAGAAGGTGGCAGTGTCCTTGTTAAACGGATGTTCCGGCCCA TGGAAGAGGAGTTTGGTCCAGTGCCTTCAAAGCAGATGAAAGAAGAAGGGACAAAGCGAGTGCTCTTGTACGTGAGGAAGGAGACTGACG ATGTGTTCGATGCATTGATGTTGAAGTCTCCCACAGTGAAGGGCCTGATGGAAGCGGTAAACCAAGGGAACATGCCAGGGCTTCAGAGCA CCTTCCTAGCCATGGACACGGAGGAGGGGGTAGAGGTGGTGTGGAACGAGCTCCACTTCGGAGACAGGAAGGCCTTCGCGGCGCACGAGG AGAAGATCCAGACCGTGTTCGAGCAGCTGGTGCTGGTGGACCACCCGAACATCGTGAAGTTGCACAAGTACTGGCTGGATACCTCTGAGG CCTGCGCGAGGGTCATCTTCATCACAGAGTACGTGTCATCAGGCAGCCTCAAGCAATTCCTCAAAAAGACCAAGAAGAACCACAAGGCCA TGAACGCCCGGGCCTGGAAGCGCTGGTGCACGCAGATCCTGTCTGCGCTCAGCTTCCTGCACGCCTGCAGCCCCCCAATCATCCACGGGA ACCTGACCAGCGACACCATCTTCATTCAGCACAACGGCCTCATCAAGATCGGCTCCGTGTGGCACCGAATCTTCTCCAATGCACTTCCAG ATGATCTCCGAAGCCCCATCCGCGCTGAGCGAGAGGAACTTCGGAACCTGCACTTCTTCCCCCCAGAGTATGGAGAGGTGGCCGATGGGA CCGCTGTGGACATCTTCTCCTTTGGGATGTGTGCGCTGGAGATGGCTGTACTGGAAATCCAGACCAATGGGGACACCCGGGTCACAGAGG AGGCCATTGCTCGCGCCAGGCACTCGCTGAGTGACCCCAACATGCGGGAGTTCATCCTTTGCTGCCTGGCCCGGGACCCTGCCCGCCGGC CCTCTGCCCACAGCCTCCTCTTCCACCGCGTGCTCTTCGAGGTGCACTCGCTGAAGCTCCTGGCAGCCCACTGCTTCATCCAGCACCAGT ACCTCATGCCTGAGAATGTGGTGGAGGAGAAGACCAAGGCCATGGACCTGCACGCGGTCTTGGCGGAGCTTCCCCGGCCCCGCAGGCCCC CGCTGCAGTGGCGGTACTCGGAAGTCTCCTTCATGGAGCTGGACAAATTCCTGGAGGATGTCAGGAATGGAATCTACCCACTGATGAACT TTGCAGCCACTCGACCCCTGGGGCTGCCCCGTGTGCTGGCCCCACCCCCGGAGGAGGTCCAAAAGGCCAAGACCCCGACGCCAGAGCCCT TTGACTCTGAGACCAGAAAGGTCATCCAGATGCAGTGCAACCTGGAGAGAAGCGAGGACAAGGCGCGCTGGCATCTCACTCTGCTTCTGG TGCTGGAAGACCGGCTGCACCGGCAGCTGACCTACGACCTGCTCCCAACGGACAGCGCCCAGGACCTCGCCTCGGAGCTCGTGCACTATG GCTTCCTCCACGAGGACGACCGGATGAAGCTGGCCGCCTTCCTGGAGAGCACCTTCCTCAAGTACCGTGGGACCCAGGCCTGACCCGGAG CCCCAGCCCCAGGGGACCATGCCGGGGTGCTGCCTGGGCAGGCCATGTTGGGGAGACTCCAGCACCGTGGGGCTGCCCTCCTCCATGCGC CTGGGAGCACAAAGGCCCCGGTAGTGAAGGAACCCCCCGTCTCCTGAGAGTGGGGCTGACCCTGCCTTGGGCGCCGAGGGGTTGGGGGGT GGGTGTGGGGGAGCCGTTAGGCCTCCCAGGTCCTTAGGATCAGGGTTGCCCCCAGAACCCCTTCCCATATCCTCCATTCTCCGCCCTGAG TTCCTACCCAGGCTGCCTGGCCGGGGCCACTGCCTCCTCAGCATGCAGGAGGCTGCCCTGTAGGGAACCCCAGCTCTGGGGCTTGGGGGT GAGGGTCAGCCCTGGACAGACCTCTGCCCAGGGAACTGCTCCATGGGGTCTGGGAGAGCAGCCATCCCCTGCTGGCACCATAGACCCACA CAAGGAGCCTGCACAGCAAGCCAGCGGTGACACACCTGCAGGTGTCAGGCATGGCACTGGGCACAACAGGGACCTGGCAGGAGAAACAGA CCACAGAGAGGTCTGGAGTTGAGGCTGTTGTCAGCAAAGCCCCTGGTCCCACACAGCTCTGCCCTAGAGCCACCTCTTTGACCCTTTACC CACCCTGAGACCAGAACTTGCAGCCCCTCTGCAGATCTCCTCTGGCCACTGCAGCCCCTCCAATGGGCTTTTTCTCTCATGCATTCCCTG GCCTGGAGGCGTCAGGGACCCCACATCCTCCCTGCTCCTCAGACTCACAGCCCCTCCATGTTACCTCCCGCACCTCCTCCCTGGGGCAGC TGCTCCCTGGGCCTCTGAGGATGTCAGCTCCTGGCTCCCTGCCTCTCTCCCACTCCACTCCTGGCTCAGTCTTAGAGATTTCTATGCCCT CATGGATTCTACCCCTGCCTTCCTGGCCTCTTGATTCTTGGCTTGCCTCTCCTCCAATTCCAAACTTAGTGAAATGGCCTTAAGCATTTT AAACTGTATGTATACATTAGCGCATTCATGCCTTTCTAAACGCATTTCAAATGTCAACCAGGAAGGCACACCACTGTATTAGTTTTATAC TGCCGCTGTAAAATTTACCACAAACTTAGTGACTTAACACAAATTTATTGCAATTCTGTAGGCTGGAAGTCTGACTATGGGTCTCACTGG ACTAGAATCAAGGCTGGCAGGCTGCCTTCCTTCCTGGAGGTTCTAGGGGAGACTCTGTCTCCTGCTCCTTCAGGCTGCTGGCAGAATCCA CATCCTTTCGGTGGCAGGGCCAAGGTCCCCACTTTCTTGCTGACTGTAAACTAAGGCCACTTCCAGCTTGTAGAGGCTGCCTACATTCCT TGGCTCTTGGCCCCCTCCTCCATCTTCAGAGCTAGCAGGTTCAGTCTGTGTCACGAACCATTTCTCTGGTTCCCTGCAGACAGGAAAGGT TGTCCCTAAGGACTCATGAGATTAGGTTGGGCCCAGCCAGATAATACATGATAATCTCCCTCCTCAAGGTTTTTAATATTAAACACATCT GCAGGACACATTTTGCCATGTAAACTAACATTCACTGGTTCCAGGGATTAAGGAATGAACCTCTTTTGTTGGGGAAGGGTGGCATTCTGC TGACCACAGCACTCCAACCAAAAGCCAAAAACCAAAGCAAGACTTACTAACGCATATCAAATAAATTAAAGGTACAAAATCGTGAATCTC AGTTATCTTAAATATTCCAATACTATTTACAAAATTATTCAAATTCTCACGCCTTCCAACTCAAAATTAGCAATCTAAAGTAATTTCCAT ATCCTAGATGGAAACCCTCATGCTAAACTGTCTGATTATGCATGGTTCTAAATGGTTTCAGTGGCAAATACATAACATTGTACTACTGAT >34713_34713_1_GRHL2-NRBP2_GRHL2_chr8_102661727_ENST00000251808_NRBP2_chr8_144922401_ENST00000442628_length(amino acids)=1060AA_BP=602 MPSRALSHTFTCTDLKVQFHQRLRLQEKRSKFIGSNMSQESDNNKRLVALVPMPSDPPFNTRRAYTSEDEAWKSYLENPLTAATKAMMSI NGDEDSAAALGLLYDYYKVPRDKRLLSVSKASDSQEDQEKRNCLGTSEAQSNLSGGENRVQVLKTVPVNLSLNQDHLENSKREQYSISFP ESSAIIPVSGITVVKAEDFTPVFMAPPVHYPRGDGEEQRVVIFEQTQYDVPSLATHSAYLKDDQRSTPDSTYSESFKDAATEKFRSASVG AEEYMYDQTSSGTFQYTLEATKSLRQKQGEGPMTYLNKGQFYAITLSETGDNKCFRHPISKVRSVVMVVFSEDKNRDEQLKYWKYWHSRQ HTAKQRVLDIADYKESFNTIGNIEEIAYNAVSFTWDVNEEAKIFITVNCLSTDFSSQKGVKGLPLMIQIDTYSYNNRSNKPIHRAYCQIK VFCDKGAERKIRDEERKQNRKKGKGQASQTQCNSSSDGKLAAIPLQKKSDITYFKTMPDLHSQPVLFIPDVHFANLQRTGQVYYNTDDER EGGSVLVKRMFRPMEEEFGPVPSKQMKEEGTKRVLLYVRKETDDVFDALMLKSPTVKGLMEAVNQGNMPGLQSTFLAMDTEEGVEVVWNE LHFGDRKAFAAHEEKIQTVFEQLVLVDHPNIVKLHKYWLDTSEACARVIFITEYVSSGSLKQFLKKTKKNHKAMNARAWKRWCTQILSAL SFLHACSPPIIHGNLTSDTIFIQHNGLIKIGSVWHRIFSNALPDDLRSPIRAEREELRNLHFFPPEYGEVADGTAVDIFSFGMCALEMAV LEIQTNGDTRVTEEAIARARHSLSDPNMREFILCCLARDPARRPSAHSLLFHRVLFEVHSLKLLAAHCFIQHQYLMPENVVEEKTKAMDL HAVLAELPRPRRPPLQWRYSEVSFMELDKFLEDVRNGIYPLMNFAATRPLGLPRVLAPPPEEVQKAKTPTPEPFDSETRKVIQMQCNLER -------------------------------------------------------------- >34713_34713_2_GRHL2-NRBP2_GRHL2_chr8_102661727_ENST00000395927_NRBP2_chr8_144922401_ENST00000442628_length(transcript)=5088nt_BP=1770nt ATTGGATCAAACATGTCACAAGAGTCGGACAAGTAAGTGGATCACACGCGCCGGCTGCTGCTACTACTACCACTTTGGGCTGATGGCAAC TGTAATAAAAGACTAGTGGCCTTAGTGCCCATGCCCAGTGACCCTCCATTCAATACCCGAAGAGCCTACACCAGTGAGGATGAAGCCTGG AAGTCATACTTGGAGAATCCCCTGACAGCAGCCACCAAGGCCATGATGAGCATTAATGGTGATGAGGACAGTGCTGCTGCCCTCGGCCTG CTCTATGACTACTACAAGGTTCCTCGAGACAAGAGGCTGCTGTCTGTAAGCAAAGCAAGTGACAGCCAAGAAGACCAGGAGAAAAGAAAC TGCCTTGGCACCAGTGAAGCCCAGAGTAATTTGAGTGGAGGAGAAAACCGAGTGCAAGTCCTAAAGACTGTTCCAGTGAACCTTTCCCTA AATCAAGATCACCTGGAGAATTCCAAGCGGGAACAGTACAGCATCAGCTTCCCCGAGAGCTCTGCCATCATCCCGGTGTCGGGAATCACG GTGGTGAAAGCTGAAGATTTCACACCAGTTTTCATGGCCCCACCTGTGCACTATCCCCGGGGAGATGGGGAAGAGCAACGAGTGGTTATC TTTGAACAGACTCAGTATGACGTGCCCTCGCTGGCCACCCACAGCGCCTATCTCAAAGACGACCAGCGCAGCACTCCGGACAGCACATAC AGCGAGAGCTTCAAGGACGCAGCCACAGAGAAATTTCGGAGTGCTTCAGTTGGGGCTGAGGAGTACATGTATGATCAGACATCAAGTGGC ACATTTCAGTACACCCTGGAAGCCACCAAATCTCTCCGTCAGAAGCAGGGGGAGGGCCCCATGACCTACCTCAACAAAGGACAGTTCTAT GCCATAACACTCAGCGAGACCGGAGACAACAAATGCTTCCGACACCCCATCAGCAAAGTCAGGAGTGTGGTGATGGTGGTCTTCAGTGAA GACAAAAACAGAGATGAACAGCTCAAATACTGGAAATACTGGCACTCTCGGCAGCATACGGCGAAGCAGAGGGTCCTTGACATTGCCGAT TACAAGGAGAGCTTTAATACGATTGGAAACATTGAAGAGATTGCATATAATGCTGTTTCCTTTACCTGGGACGTGAATGAAGAGGCGAAG ATTTTCATCACCGTGAATTGCTTGAGCACAGATTTCTCCTCCCAAAAAGGGGTGAAAGGACTTCCTTTGATGATTCAGATTGACACATAC AGTTATAACAATCGTAGCAATAAACCCATTCATAGAGCTTATTGCCAGATCAAGGTCTTCTGTGACAAAGGAGCAGAAAGAAAAATCCGA GATGAAGAGCGGAAGCAGAACAGGAAGAAAGGGAAAGGCCAGGCCTCCCAAACTCAATGCAACAGCTCCTCTGATGGGAAGTTGGCTGCC ATACCTTTACAGAAGAAGAGTGACATCACCTACTTCAAAACCATGCCTGATCTCCACTCACAGCCAGTTCTCTTCATACCTGATGTTCAC TTTGCAAACCTGCAGAGGACCGGACAGGTGTATTACAACACGGATGATGAACGAGAAGGTGGCAGTGTCCTTGTTAAACGGATGTTCCGG CCCATGGAAGAGGAGTTTGGTCCAGTGCCTTCAAAGCAGATGAAAGAAGAAGGGACAAAGCGAGTGCTCTTGTACGTGAGGAAGGAGACT GACGATGTGTTCGATGCATTGATGTTGAAGTCTCCCACAGTGAAGGGCCTGATGGAAGCGGTAAACCAAGGGAACATGCCAGGGCTTCAG AGCACCTTCCTAGCCATGGACACGGAGGAGGGGGTAGAGGTGGTGTGGAACGAGCTCCACTTCGGAGACAGGAAGGCCTTCGCGGCGCAC GAGGAGAAGATCCAGACCGTGTTCGAGCAGCTGGTGCTGGTGGACCACCCGAACATCGTGAAGTTGCACAAGTACTGGCTGGATACCTCT GAGGCCTGCGCGAGGGTCATCTTCATCACAGAGTACGTGTCATCAGGCAGCCTCAAGCAATTCCTCAAAAAGACCAAGAAGAACCACAAG GCCATGAACGCCCGGGCCTGGAAGCGCTGGTGCACGCAGATCCTGTCTGCGCTCAGCTTCCTGCACGCCTGCAGCCCCCCAATCATCCAC GGGAACCTGACCAGCGACACCATCTTCATTCAGCACAACGGCCTCATCAAGATCGGCTCCGTGTGGCACCGAATCTTCTCCAATGCACTT CCAGATGATCTCCGAAGCCCCATCCGCGCTGAGCGAGAGGAACTTCGGAACCTGCACTTCTTCCCCCCAGAGTATGGAGAGGTGGCCGAT GGGACCGCTGTGGACATCTTCTCCTTTGGGATGTGTGCGCTGGAGATGGCTGTACTGGAAATCCAGACCAATGGGGACACCCGGGTCACA GAGGAGGCCATTGCTCGCGCCAGGCACTCGCTGAGTGACCCCAACATGCGGGAGTTCATCCTTTGCTGCCTGGCCCGGGACCCTGCCCGC CGGCCCTCTGCCCACAGCCTCCTCTTCCACCGCGTGCTCTTCGAGGTGCACTCGCTGAAGCTCCTGGCAGCCCACTGCTTCATCCAGCAC CAGTACCTCATGCCTGAGAATGTGGTGGAGGAGAAGACCAAGGCCATGGACCTGCACGCGGTCTTGGCGGAGCTTCCCCGGCCCCGCAGG CCCCCGCTGCAGTGGCGGTACTCGGAAGTCTCCTTCATGGAGCTGGACAAATTCCTGGAGGATGTCAGGAATGGAATCTACCCACTGATG AACTTTGCAGCCACTCGACCCCTGGGGCTGCCCCGTGTGCTGGCCCCACCCCCGGAGGAGGTCCAAAAGGCCAAGACCCCGACGCCAGAG CCCTTTGACTCTGAGACCAGAAAGGTCATCCAGATGCAGTGCAACCTGGAGAGAAGCGAGGACAAGGCGCGCTGGCATCTCACTCTGCTT CTGGTGCTGGAAGACCGGCTGCACCGGCAGCTGACCTACGACCTGCTCCCAACGGACAGCGCCCAGGACCTCGCCTCGGAGCTCGTGCAC TATGGCTTCCTCCACGAGGACGACCGGATGAAGCTGGCCGCCTTCCTGGAGAGCACCTTCCTCAAGTACCGTGGGACCCAGGCCTGACCC GGAGCCCCAGCCCCAGGGGACCATGCCGGGGTGCTGCCTGGGCAGGCCATGTTGGGGAGACTCCAGCACCGTGGGGCTGCCCTCCTCCAT GCGCCTGGGAGCACAAAGGCCCCGGTAGTGAAGGAACCCCCCGTCTCCTGAGAGTGGGGCTGACCCTGCCTTGGGCGCCGAGGGGTTGGG GGGTGGGTGTGGGGGAGCCGTTAGGCCTCCCAGGTCCTTAGGATCAGGGTTGCCCCCAGAACCCCTTCCCATATCCTCCATTCTCCGCCC TGAGTTCCTACCCAGGCTGCCTGGCCGGGGCCACTGCCTCCTCAGCATGCAGGAGGCTGCCCTGTAGGGAACCCCAGCTCTGGGGCTTGG GGGTGAGGGTCAGCCCTGGACAGACCTCTGCCCAGGGAACTGCTCCATGGGGTCTGGGAGAGCAGCCATCCCCTGCTGGCACCATAGACC CACACAAGGAGCCTGCACAGCAAGCCAGCGGTGACACACCTGCAGGTGTCAGGCATGGCACTGGGCACAACAGGGACCTGGCAGGAGAAA CAGACCACAGAGAGGTCTGGAGTTGAGGCTGTTGTCAGCAAAGCCCCTGGTCCCACACAGCTCTGCCCTAGAGCCACCTCTTTGACCCTT TACCCACCCTGAGACCAGAACTTGCAGCCCCTCTGCAGATCTCCTCTGGCCACTGCAGCCCCTCCAATGGGCTTTTTCTCTCATGCATTC CCTGGCCTGGAGGCGTCAGGGACCCCACATCCTCCCTGCTCCTCAGACTCACAGCCCCTCCATGTTACCTCCCGCACCTCCTCCCTGGGG CAGCTGCTCCCTGGGCCTCTGAGGATGTCAGCTCCTGGCTCCCTGCCTCTCTCCCACTCCACTCCTGGCTCAGTCTTAGAGATTTCTATG CCCTCATGGATTCTACCCCTGCCTTCCTGGCCTCTTGATTCTTGGCTTGCCTCTCCTCCAATTCCAAACTTAGTGAAATGGCCTTAAGCA TTTTAAACTGTATGTATACATTAGCGCATTCATGCCTTTCTAAACGCATTTCAAATGTCAACCAGGAAGGCACACCACTGTATTAGTTTT ATACTGCCGCTGTAAAATTTACCACAAACTTAGTGACTTAACACAAATTTATTGCAATTCTGTAGGCTGGAAGTCTGACTATGGGTCTCA CTGGACTAGAATCAAGGCTGGCAGGCTGCCTTCCTTCCTGGAGGTTCTAGGGGAGACTCTGTCTCCTGCTCCTTCAGGCTGCTGGCAGAA TCCACATCCTTTCGGTGGCAGGGCCAAGGTCCCCACTTTCTTGCTGACTGTAAACTAAGGCCACTTCCAGCTTGTAGAGGCTGCCTACAT TCCTTGGCTCTTGGCCCCCTCCTCCATCTTCAGAGCTAGCAGGTTCAGTCTGTGTCACGAACCATTTCTCTGGTTCCCTGCAGACAGGAA AGGTTGTCCCTAAGGACTCATGAGATTAGGTTGGGCCCAGCCAGATAATACATGATAATCTCCCTCCTCAAGGTTTTTAATATTAAACAC ATCTGCAGGACACATTTTGCCATGTAAACTAACATTCACTGGTTCCAGGGATTAAGGAATGAACCTCTTTTGTTGGGGAAGGGTGGCATT CTGCTGACCACAGCACTCCAACCAAAAGCCAAAAACCAAAGCAAGACTTACTAACGCATATCAAATAAATTAAAGGTACAAAATCGTGAA TCTCAGTTATCTTAAATATTCCAATACTATTTACAAAATTATTCAAATTCTCACGCCTTCCAACTCAAAATTAGCAATCTAAAGTAATTT CCATATCCTAGATGGAAACCCTCATGCTAAACTGTCTGATTATGCATGGTTCTAAATGGTTTCAGTGGCAAATACATAACATTGTACTAC >34713_34713_2_GRHL2-NRBP2_GRHL2_chr8_102661727_ENST00000395927_NRBP2_chr8_144922401_ENST00000442628_length(amino acids)=1030AA_BP=572 MLLLLPLWADGNCNKRLVALVPMPSDPPFNTRRAYTSEDEAWKSYLENPLTAATKAMMSINGDEDSAAALGLLYDYYKVPRDKRLLSVSK ASDSQEDQEKRNCLGTSEAQSNLSGGENRVQVLKTVPVNLSLNQDHLENSKREQYSISFPESSAIIPVSGITVVKAEDFTPVFMAPPVHY PRGDGEEQRVVIFEQTQYDVPSLATHSAYLKDDQRSTPDSTYSESFKDAATEKFRSASVGAEEYMYDQTSSGTFQYTLEATKSLRQKQGE GPMTYLNKGQFYAITLSETGDNKCFRHPISKVRSVVMVVFSEDKNRDEQLKYWKYWHSRQHTAKQRVLDIADYKESFNTIGNIEEIAYNA VSFTWDVNEEAKIFITVNCLSTDFSSQKGVKGLPLMIQIDTYSYNNRSNKPIHRAYCQIKVFCDKGAERKIRDEERKQNRKKGKGQASQT QCNSSSDGKLAAIPLQKKSDITYFKTMPDLHSQPVLFIPDVHFANLQRTGQVYYNTDDEREGGSVLVKRMFRPMEEEFGPVPSKQMKEEG TKRVLLYVRKETDDVFDALMLKSPTVKGLMEAVNQGNMPGLQSTFLAMDTEEGVEVVWNELHFGDRKAFAAHEEKIQTVFEQLVLVDHPN IVKLHKYWLDTSEACARVIFITEYVSSGSLKQFLKKTKKNHKAMNARAWKRWCTQILSALSFLHACSPPIIHGNLTSDTIFIQHNGLIKI GSVWHRIFSNALPDDLRSPIRAEREELRNLHFFPPEYGEVADGTAVDIFSFGMCALEMAVLEIQTNGDTRVTEEAIARARHSLSDPNMRE FILCCLARDPARRPSAHSLLFHRVLFEVHSLKLLAAHCFIQHQYLMPENVVEEKTKAMDLHAVLAELPRPRRPPLQWRYSEVSFMELDKF LEDVRNGIYPLMNFAATRPLGLPRVLAPPPEEVQKAKTPTPEPFDSETRKVIQMQCNLERSEDKARWHLTLLLVLEDRLHRQLTYDLLPT -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for GRHL2-NRBP2 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for GRHL2-NRBP2 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for GRHL2-NRBP2 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
