
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:GRID1-AFAP1L2 (FusionGDB2 ID:34741) |
Fusion Gene Summary for GRID1-AFAP1L2 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: GRID1-AFAP1L2 | Fusion gene ID: 34741 | Hgene | Tgene | Gene symbol | GRID1 | AFAP1L2 | Gene ID | 2894 | 84632 |
| Gene name | glutamate ionotropic receptor delta type subunit 1 | actin filament associated protein 1 like 2 | |
| Synonyms | GluD1 | CTB-1144G6.4|KIAA1914|XB130 | |
| Cytomap | 10q23.1-q23.2 | 10q25.3 | |
| Type of gene | protein-coding | protein-coding | |
| Description | glutamate receptor ionotropic, delta-1gluR delta-1 subunitglutamate receptor delta-1 subunit | actin filament-associated protein 1-like 2AFAP1-like protein 2CTB-1144G6.6 | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | Q9ULK0 | Q8N4X5 | |
| Ensembl transtripts involved in fusion gene | ENST00000327946, ENST00000536331, ENST00000552278, | ENST00000491814, ENST00000304129, ENST00000369271, ENST00000545353, | |
| Fusion gene scores | * DoF score | 10 X 8 X 6=480 | 5 X 4 X 4=80 |
| # samples | 11 | 5 | |
| ** MAII score | log2(11/480*10)=-2.12553088208386 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(5/80*10)=-0.678071905112638 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: GRID1 [Title/Abstract] AND AFAP1L2 [Title/Abstract] AND fusion [Title/Abstract] | ||
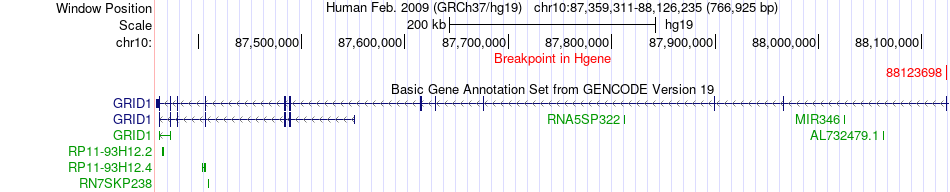
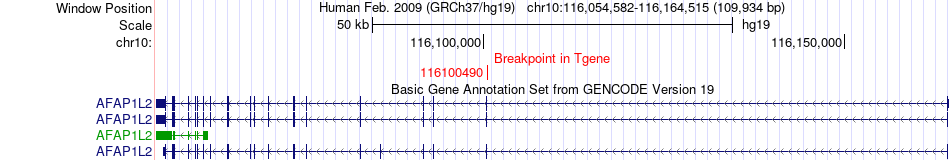
| Most frequent breakpoint | GRID1(88123698)-AFAP1L2(116100490), # samples:2 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Tgene | AFAP1L2 | GO:0006954 | inflammatory response | 17412687 |
| Tgene | AFAP1L2 | GO:0007346 | regulation of mitotic cell cycle | 17412687 |
| Tgene | AFAP1L2 | GO:0032675 | regulation of interleukin-6 production | 17412687 |
| Tgene | AFAP1L2 | GO:0032757 | positive regulation of interleukin-8 production | 17412687 |
| Tgene | AFAP1L2 | GO:0045742 | positive regulation of epidermal growth factor receptor signaling pathway | 17412687 |
| Tgene | AFAP1L2 | GO:0045893 | positive regulation of transcription, DNA-templated | 17412687 |
 Fusion gene breakpoints across GRID1 (5'-gene) Fusion gene breakpoints across GRID1 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
 |
 Fusion gene breakpoints across AFAP1L2 (3'-gene) Fusion gene breakpoints across AFAP1L2 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | SARC | TCGA-DX-A6YR-01A | GRID1 | chr10 | 88123698 | - | AFAP1L2 | chr10 | 116100490 | - |
Top |
Fusion Gene ORF analysis for GRID1-AFAP1L2 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-intron | ENST00000327946 | ENST00000491814 | GRID1 | chr10 | 88123698 | - | AFAP1L2 | chr10 | 116100490 | - |
| In-frame | ENST00000327946 | ENST00000304129 | GRID1 | chr10 | 88123698 | - | AFAP1L2 | chr10 | 116100490 | - |
| In-frame | ENST00000327946 | ENST00000369271 | GRID1 | chr10 | 88123698 | - | AFAP1L2 | chr10 | 116100490 | - |
| In-frame | ENST00000327946 | ENST00000545353 | GRID1 | chr10 | 88123698 | - | AFAP1L2 | chr10 | 116100490 | - |
| intron-3CDS | ENST00000536331 | ENST00000304129 | GRID1 | chr10 | 88123698 | - | AFAP1L2 | chr10 | 116100490 | - |
| intron-3CDS | ENST00000536331 | ENST00000369271 | GRID1 | chr10 | 88123698 | - | AFAP1L2 | chr10 | 116100490 | - |
| intron-3CDS | ENST00000536331 | ENST00000545353 | GRID1 | chr10 | 88123698 | - | AFAP1L2 | chr10 | 116100490 | - |
| intron-3CDS | ENST00000552278 | ENST00000304129 | GRID1 | chr10 | 88123698 | - | AFAP1L2 | chr10 | 116100490 | - |
| intron-3CDS | ENST00000552278 | ENST00000369271 | GRID1 | chr10 | 88123698 | - | AFAP1L2 | chr10 | 116100490 | - |
| intron-3CDS | ENST00000552278 | ENST00000545353 | GRID1 | chr10 | 88123698 | - | AFAP1L2 | chr10 | 116100490 | - |
| intron-intron | ENST00000536331 | ENST00000491814 | GRID1 | chr10 | 88123698 | - | AFAP1L2 | chr10 | 116100490 | - |
| intron-intron | ENST00000552278 | ENST00000491814 | GRID1 | chr10 | 88123698 | - | AFAP1L2 | chr10 | 116100490 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000327946 | GRID1 | chr10 | 88123698 | - | ENST00000304129 | AFAP1L2 | chr10 | 116100490 | - | 3980 | 321 | 86 | 2761 | 891 |
| ENST00000327946 | GRID1 | chr10 | 88123698 | - | ENST00000369271 | AFAP1L2 | chr10 | 116100490 | - | 3968 | 321 | 86 | 2749 | 887 |
| ENST00000327946 | GRID1 | chr10 | 88123698 | - | ENST00000545353 | AFAP1L2 | chr10 | 116100490 | - | 3191 | 321 | 86 | 2920 | 944 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000327946 | ENST00000304129 | GRID1 | chr10 | 88123698 | - | AFAP1L2 | chr10 | 116100490 | - | 0.001486317 | 0.99851364 |
| ENST00000327946 | ENST00000369271 | GRID1 | chr10 | 88123698 | - | AFAP1L2 | chr10 | 116100490 | - | 0.001516158 | 0.99848384 |
| ENST00000327946 | ENST00000545353 | GRID1 | chr10 | 88123698 | - | AFAP1L2 | chr10 | 116100490 | - | 0.004197712 | 0.99580234 |
Top |
Fusion Genomic Features for GRID1-AFAP1L2 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
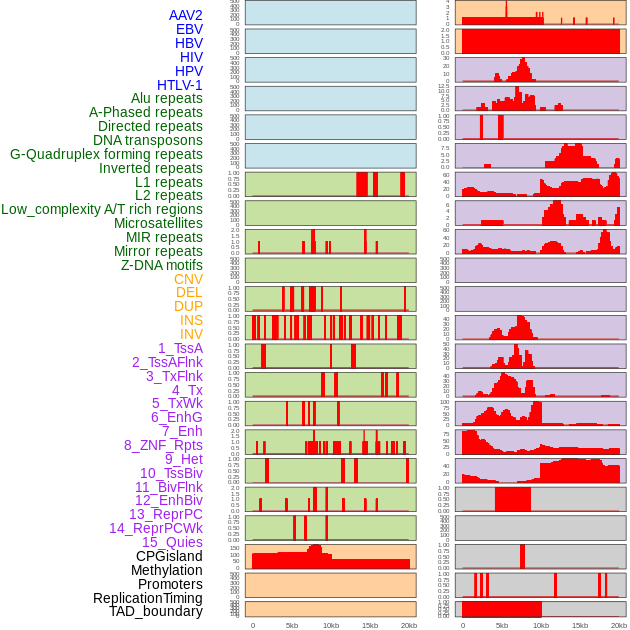 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for GRID1-AFAP1L2 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr10:88123698/chr10:116100490) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| GRID1 | AFAP1L2 |
| FUNCTION: Receptor for glutamate. L-glutamate acts as an excitatory neurotransmitter at many synapses in the central nervous system. The postsynaptic actions of Glu are mediated by a variety of receptors that are named according to their selective agonists. | FUNCTION: May play a role in a signaling cascade by enhancing the kinase activity of SRC. Contributes to SRC-regulated transcription activation. {ECO:0000269|PubMed:17412687}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | AFAP1L2 | chr10:88123698 | chr10:116100490 | ENST00000304129 | 0 | 19 | 652_749 | 5 | 819.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | AFAP1L2 | chr10:88123698 | chr10:116100490 | ENST00000369271 | 0 | 19 | 652_749 | 5 | 815.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | AFAP1L2 | chr10:88123698 | chr10:116100490 | ENST00000304129 | 0 | 19 | 175_271 | 5 | 819.0 | Domain | PH 1 | |
| Tgene | AFAP1L2 | chr10:88123698 | chr10:116100490 | ENST00000304129 | 0 | 19 | 353_447 | 5 | 819.0 | Domain | PH 2 | |
| Tgene | AFAP1L2 | chr10:88123698 | chr10:116100490 | ENST00000369271 | 0 | 19 | 175_271 | 5 | 815.0 | Domain | PH 1 | |
| Tgene | AFAP1L2 | chr10:88123698 | chr10:116100490 | ENST00000369271 | 0 | 19 | 353_447 | 5 | 815.0 | Domain | PH 2 |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | GRID1 | chr10:88123698 | chr10:116100490 | ENST00000327946 | - | 2 | 16 | 21_562 | 78 | 1010.0 | Topological domain | Extracellular |
| Hgene | GRID1 | chr10:88123698 | chr10:116100490 | ENST00000327946 | - | 2 | 16 | 584_637 | 78 | 1010.0 | Topological domain | Cytoplasmic |
| Hgene | GRID1 | chr10:88123698 | chr10:116100490 | ENST00000327946 | - | 2 | 16 | 659_830 | 78 | 1010.0 | Topological domain | Extracellular |
| Hgene | GRID1 | chr10:88123698 | chr10:116100490 | ENST00000327946 | - | 2 | 16 | 852_1009 | 78 | 1010.0 | Topological domain | Cytoplasmic |
| Hgene | GRID1 | chr10:88123698 | chr10:116100490 | ENST00000327946 | - | 2 | 16 | 563_583 | 78 | 1010.0 | Transmembrane | Helical |
| Hgene | GRID1 | chr10:88123698 | chr10:116100490 | ENST00000327946 | - | 2 | 16 | 638_658 | 78 | 1010.0 | Transmembrane | Helical |
| Hgene | GRID1 | chr10:88123698 | chr10:116100490 | ENST00000327946 | - | 2 | 16 | 831_851 | 78 | 1010.0 | Transmembrane | Helical |
Top |
Fusion Gene Sequence for GRID1-AFAP1L2 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >34741_34741_1_GRID1-AFAP1L2_GRID1_chr10_88123698_ENST00000327946_AFAP1L2_chr10_116100490_ENST00000304129_length(transcript)=3980nt_BP=321nt ACTGCACCGGGACCAGCGCCTCCCCGCTTCGCGCTGCCCTCGGCCTCGCCCCGGGCCCGGGTGGATGAGCCGCGCGCCCGGGGGACATGG AAGCGCTGACGCTGTGGCTTCTCCCCTGGATATGCCAGTGCGTGTCGGTGCGGGCCGACTCCATCATCCACATCGGTGCCATCTTCGAGG AGAACGCGGCCAAGGACGACAGGGTGTTCCAGTTGGCGGTATCCGACCTGAGCCTCAACGATGACATCCTGCAGAGCGAGAAGATCACCT ACTCCATCAAGGTCATCGAGGCCAACAACCCATTCCAGGCTGTGCAGGAAGCCCTGGAACAGCTGCTGACAGAGTTGGATGACTTCCTCA AGATTCTTGACCAGGAGAACCTGAGCAGCACAGCACTGGTGAAGAAGAGCTGCCTGGCGGAGCTCCTCCGGCTTTACACCAAAAGCAGCA GCTCTGATGAGGAGTACATTTATATGAACAAAGTGACCATCAACAAGCAACAGAATGCAGAGTCTCAAGGCAAAGCGCCTGAGGAGCAGG GCCTGCTACCCAATGGGGAGCCCAGCCAGCACTCCTCGGCCCCTCAGAAGAGCCTTCCAGACCTCCCGCCACCCAAGATGATTCCAGAAC GGAAACAGCTTGCCATCCCAAAGACGGAGTCTCCAGAGGGCTACTATGAAGAGGCTGAGCCATATGACACATCCCTCAATGAGGACGGAG AGGCTGTGAGCAGCTCCTACGAGTCCTACGATGAAGAGGACGGCAGCAAGGGCAAGTCGGCCCCTTACCAGTGGCCCTCGCCGGAGGCCG GCATCGAGCTGATGCGTGACGCCCGCATCTGCGCCTTCCTGTGGCGCAAGAAGTGGCTGGGACAGTGGGCCAAGCAGCTCTGTGTCATCA AGGACAACAGGCTTCTGTGCTACAAATCCTCCAAGGACCACAGCCCTCAGCTGGACGTGAACCTACTGGGCAGCAGCGTCATTCACAAGG AGAAGCAAGTGCGGAAGAAGGAGCACAAGCTGAAGATCACGCCGATGAATGCCGATGTGATTGTGCTGGGCCTGCAGAGCAAGGACCAGG CTGAGCAGTGGCTCAGGGTCATCCAGGAAGTGAGCGGCCTGCCTTCCGAAGGAGCATCTGAAGGAAACCAGTACACCCCGGATGCCCAGC GCTTTAACTGCCAGAAACCAGATATAGCTGAGAAGTACCTGTCGGCTTCAGAGTATGGGAGCTCCGTGGATGGCCACCCTGAGGTCCCAG AAACCAAAGACGTCAAGAAGAAATGTTCTGCTGGCCTCAAACTGAGCAACCTAATGAATCTGGGCAGGAAGAAATCCACCTCACTGGAGC CTGTGGAGAGGTCCCTCGAGACATCCAGTTACCTGAACGTGCTGGTGAACAGCCAGTGGAAGTCTCGCTGGTGCTCTGTCAGGGACAATC ACCTGCACTTCTACCAGGACCGGAACCGGAGCAAGGTGGCCCAGCAACCCCTCAGCCTGGTGGGCTGCGAGGTGGTCCCAGACCCCAGCC CCGACCACCTCTACTCCTTCCGCATCCTCCACAAGGGCGAGGAGCTGGCCAAGCTTGAGGCCAAGTCTTCCGAGGAAATGGGCCACTGGC TGGGTCTCCTGCTCTCTGAGTCAGGCTCCAAGACAGACCCAGAAGAGTTCACCTACGACTATGTGGATGCCGATAGGGTCTCCTGTATTG TGAGTGCGGCCAAAAACTCTCTCTTACTGATGCAGAGAAAGTTCTCAGAGCCCAACACTTACATCGATGGCCTGCCTAGCCAGGACCGCC AGGAGGAGCTGTATGACGACGTGGACCTGTCAGAGCTCACAGCTGCGGTGGAGCCTACCGAGGAAGCCACCCCTGTTGCAGATGACCCAA ATGAGAGAGAATCTGACCGAGTGTACCTGGACCTCACACCTGTCAAGTCCTTTCTGCATGGCCCCAGCAGTGCACAGGCCCAGGCCTCCT CCCCGACGTTGTCCTGCCTGGACAATGCAACTGAGGCCCTCCCGGCAGACTCAGGCCCAGGTCCCACCCCAGATGAGCCCTGCATAAAGT GTCCAGAGAACCTGGGAGAACAGCAGCTGGAGAGTTTGGAGCCAGAGGATCCTTCCCTGAGAATCACCACCGTCAAAATCCAGACGGAAC AGCAGAGAATCTCCTTCCCACCGAGCTGCCCGGATGCCGTGGTGGCCACCCCACCTGGTGCCAGCCCACCTGTGAAGGACAGGTTGCGCG TGACCAGTGCAGAGATCAAGCTTGGCAAGAATCGGACAGAAGCTGAGGTGAAGCGGTACACAGAGGAGAAGGAGAGGCTTGAAAAGAAGA AGGAAGAAATCCGGGGGCACCTGGCTCAGCTCCGGAAAGAGAAACGGGAGCTAAAGGAAACCCTACTGAAATGCACAGACAAGGAAGTCC TGGCGAGCCTGGAGCAGAAGCTGAAGGAAATTGACGAGGAGTGCCGGGGCGAGGAGAGCAGGCGCGTGGACCTGGAGCTCAGCATCATGG AGGTGAAGGACAACCTGAAGAAGGCTGAGGCAGGGCCTGTGACGTTAGGCACCACCGTGGACACCACCCACCTGGAGAATGTGAGCCCCC GCCCCAAAGCTGTCACACCTGCCTCTGCCCCAGACTGTACCCCAGTCAACTCTGCAACCACACTCAAGAACAGGCCTCTCTCGGTCGTGG TCACAGGCAAAGGCACTGTACTCCAGAAAGCCAAGGAATGGGAGAAGAAAGGAGCAAGTTAGAAAACAAGCTTCATCTAAAGACTCTCAT GTCAATGTGGACCTTGGTGACAATCCTGCTTTGTTAAAGCAAAAACTATGCGAAAGGGTGAGTCTGTTTAGAAGAAAAAGCAAAGACTGA GGTACTGTGAATGGAGAGCTTCAGCTAAGAGGAGGCTCTGTCCCTTTTCAGAGCCAAAGGAAATAATACAACAAAAAGGAGGCTTCTTTG GAGACCTAAGTCTATTGGATGTAAACAAGACGTTGTATTTAGGGATGTTCTGTGTTTCTTTCTTTTTTGAAGTTGTCATCAATTGCTTTA CTAAGATTTTTAAATAGTGAAAACCTCCTGTTTAGACTTTGGTGGAAGATGAATCAAGGAAGCAGGGCCCTGTCTTATGGGTCACGTGTC TTTGGTGAGTGAGAAGACCTAAACTCCTGGCCATCATCTCTTATCCAATACTTAGCAGTTGGGGATTAAACCATCCTTGCCTTCAGTTCT CTCCAATATTACCAGGCCCAACTCAGTCTTCAGTGATTTTAAACAGCATTGACATCATCTGTAAAACCATCATCTGTAAAACCATCTATG ACATGAGTTTTGAGAAACAATAATGGGGAAAATATTTGGGACCAAGCTGAAGCACTAATCCCACTAAGTTAAAGACTTCTTTCCAGTCCA AGGCAGGCCTGAATCAACTGTCTTTAAATAAAATTTTAAGTGATGCTGTATTATATATAGGAAAAAATGCTTAAAATCCTGTCATTTAGA ACAGTGAAAAGTATCTTTTGAGATTAAAGTGACTCTTTACTGTAGGAAAAATATTACTCTGTGTTTACAGATTCATTGCTGTGGTCAGGC CATTTTTAAGGGAAGAGTTATTTAATATAAATAGTCTCTGATTTTAAGTTCTGTTTAATGTTCATTCTCCTTCCAAGAACAAAGTGGTGA TTTTTGGTTAGGGTGATCGCCCTCTTAAAATTGGCAGTGCTGTTCCTTGTGCTGCCCCTGTCTTTTCCTCTGATGGCATTTTTTTTTTTT TTTTTTTAACACAGGTTGAAACATTTCATCTATTATCTCTGCCTCATTTCTGGAGGGTTGTGTATCAGTTCTCTAACACTTGTTCCTGAG AACTAAATGTCTTTTTTATTCTTATTTCCTCTCTCATAAACATTTGGTGACCTTTTACCAAGTGGTGAGTTAGTTAGGTTTTTTAAAATA >34741_34741_1_GRID1-AFAP1L2_GRID1_chr10_88123698_ENST00000327946_AFAP1L2_chr10_116100490_ENST00000304129_length(amino acids)=891AA_BP=78 MEALTLWLLPWICQCVSVRADSIIHIGAIFEENAAKDDRVFQLAVSDLSLNDDILQSEKITYSIKVIEANNPFQAVQEALEQLLTELDDF LKILDQENLSSTALVKKSCLAELLRLYTKSSSSDEEYIYMNKVTINKQQNAESQGKAPEEQGLLPNGEPSQHSSAPQKSLPDLPPPKMIP ERKQLAIPKTESPEGYYEEAEPYDTSLNEDGEAVSSSYESYDEEDGSKGKSAPYQWPSPEAGIELMRDARICAFLWRKKWLGQWAKQLCV IKDNRLLCYKSSKDHSPQLDVNLLGSSVIHKEKQVRKKEHKLKITPMNADVIVLGLQSKDQAEQWLRVIQEVSGLPSEGASEGNQYTPDA QRFNCQKPDIAEKYLSASEYGSSVDGHPEVPETKDVKKKCSAGLKLSNLMNLGRKKSTSLEPVERSLETSSYLNVLVNSQWKSRWCSVRD NHLHFYQDRNRSKVAQQPLSLVGCEVVPDPSPDHLYSFRILHKGEELAKLEAKSSEEMGHWLGLLLSESGSKTDPEEFTYDYVDADRVSC IVSAAKNSLLLMQRKFSEPNTYIDGLPSQDRQEELYDDVDLSELTAAVEPTEEATPVADDPNERESDRVYLDLTPVKSFLHGPSSAQAQA SSPTLSCLDNATEALPADSGPGPTPDEPCIKCPENLGEQQLESLEPEDPSLRITTVKIQTEQQRISFPPSCPDAVVATPPGASPPVKDRL RVTSAEIKLGKNRTEAEVKRYTEEKERLEKKKEEIRGHLAQLRKEKRELKETLLKCTDKEVLASLEQKLKEIDEECRGEESRRVDLELSI -------------------------------------------------------------- >34741_34741_2_GRID1-AFAP1L2_GRID1_chr10_88123698_ENST00000327946_AFAP1L2_chr10_116100490_ENST00000369271_length(transcript)=3968nt_BP=321nt ACTGCACCGGGACCAGCGCCTCCCCGCTTCGCGCTGCCCTCGGCCTCGCCCCGGGCCCGGGTGGATGAGCCGCGCGCCCGGGGGACATGG AAGCGCTGACGCTGTGGCTTCTCCCCTGGATATGCCAGTGCGTGTCGGTGCGGGCCGACTCCATCATCCACATCGGTGCCATCTTCGAGG AGAACGCGGCCAAGGACGACAGGGTGTTCCAGTTGGCGGTATCCGACCTGAGCCTCAACGATGACATCCTGCAGAGCGAGAAGATCACCT ACTCCATCAAGGTCATCGAGGCCAACAACCCATTCCAGGCTGTGCAGGAAGCCCTGGAACAGCTGCTGACAGAGTTGGATGACTTCCTCA AGATTCTTGACCAGGAGAACCTGAGCAGCACAGCACTGGTGAAGAAGAGCTGCCTGGCGGAGCTCCTCCGGCTTTACACCAAAAGCAGCA GCTCTGATGAGGAGTACATTTATATGAACAAAGTGACCATCAACAAGCAACAGAATGCAGAGTCTCAAGGCAAAGCGCCTGAGGAGCAGG GCCTGCTACCCAATGGGGAGCCCAGCCAGCACTCCTCGGCCCCTCAGAAGAGCCTTCCAGACCTCCCGCCACCCAAGATGATTCCAGAAC GGAAACAGCTTGCCATCCCAAAGACGGAGTCTCCAGAGGGCTACTATGAAGAGGCTGAGCCATATGACACATCCCTCAATGAGGACGGAG AGGCTGTGAGCAGCTCCTACGAGTCCTACGATGAAGAGGACGGCAGCAAGGGCAAGTCGGCCCCTTACCAGTGGCCCTCGCCGGAGGCCG GCATCGAGCTGATGCGTGACGCCCGCATCTGCGCCTTCCTGTGGCGCAAGAAGTGGCTGGGACAGTGGGCCAAGCAGCTCTGTGTCATCA AGGACAACAGGCTTCTGTGCTACAAATCCTCCAAGGACCACAGCCCTCAGCTGGACGTGAACCTACTGGGCAGCAGCGTCATTCACAAGG AGAAGCAAGTGCGGAAGAAGGAGCACAAGCTGAAGATCACGCCGATGAATGCCGATGTGATTGTGCTGGGCCTGCAGAGCAAGGACCAGG CTGAGCAGTGGCTCAGGGTCATCCAGGAAGTGAGCGGCCTGCCTTCCGAAGGAGCATCTGAAGGAAACCAGTACACCCCGGATGCCCAGC GCTTTAACTGCCAGAAACCAGATATAGCTGAGAAGTACCTGTCGGCTTCAGAGTATGGGAGCTCCGTGGATGGCCACCCTGAGGTCCCAG AAACCAAAGACGTCAAGAAGAAATGTTCTGCTGGCCTCAAACTGAGCAACCTAATGAATCTGGGCAGGAAGAAATCCACCTCACTGGAGC CTGTGGAGAGGTCCCTCGAGACATCCAGTTACCTGAACGTGCTGGTGAACAGCCAGTGGAAGTCTCGCTGGTGCTCTGTCAGGGACAATC ACCTGCACTTCTACCAGGACCGGAACCGGAGCAAGGTGGCCCAGCAACCCCTCAGCCTGGTGGGCTGCGAGGTGGTCCCAGACCCCAGCC CCGACCACCTCTACTCCTTCCGCATCCTCCACAAGGGCGAGGAGCTGGCCAAGCTTGAGGCCAAGTCTTCCGAGGAAATGGGCCACTGGC TGGGTCTCCTGCTCTCTGAGTCAGGCTCCAAGACAGACCCAGAAGAGTTCACCTACGACTATGTGGATGCCGATAGGGTCTCCTGTATTG TGAGTGCGGCCAAAAACTCTCTCTTACTGATGCAGAGAAAGTTCTCAGAGCCCAACACTTACATCGATGGCCTGCCTAGCCAGGACCGCC AGGAGGAGCTGTATGACGACGTGGACCTGTCAGAGCTCACAGCTGCGGTGGAGCCTACCGAGGAAGCCACCCCTGTTGCAGATGACCCAA ATGAGAGAGAATCTGACCGAGTGTACCTGGACCTCACACCTGTCAAGTCCTTTCTGCATGGCCCCAGCAGTGCACAGGCCCAGGCCTCCT CCCCGACGTTGTCCTGCCTGGACAATGCAACTGAGGCCCTCCCGGCAGACTCAGGCCCAGGTCCCACCCCAGATGAGCCCTGCATAAAGT GTCCAGAGAACCTGGGAGAACAGCAGCTGGAGAGTTTGGAGCCAGAGGATCCTTCCCTGAGAATCACCACCGTCAAAATCCAGACGGAAC AGCAGAGAATCTCCTTCCCACCGAGCTGCCCGGATGCCGTGGTGGCCACCCCACCTGGTGCCAGCCCACCTGTGAAGGACAGGTTGCGCG TGACCAGTGCAGAGATCAAGCTTGGCAAGAATCGGACAGAAGCTGAGGTGAAGCGGTACACAGAGGAGAAGGAGAGGCTTGAAAAGAAGA AGGAAGAAATCCGGGGGCACCTGGCTCAGCTCCGGAAAGAGAAACGGGAGCTAAAGGAAACCCTACTGAAATGCACAGACAAGGAAGTCC TGGCGAGCCTGGAGCAGAAGCTGAAGGAAATTGACGAGGAGTGCCGGGGCGAGGAGAGCAGGCGCGTGGACCTGGAGCTCAGCATCATGG AGGTGAAGGACAACCTGAAGAAGGCTGAGGCAGGGCCTGTGACGTTAGGCACCACCGTGGACACCACCCACCTGGAGAATCCCAAAGCTG TCACACCTGCCTCTGCCCCAGACTGTACCCCAGTCAACTCTGCAACCACACTCAAGAACAGGCCTCTCTCGGTCGTGGTCACAGGCAAAG GCACTGTACTCCAGAAAGCCAAGGAATGGGAGAAGAAAGGAGCAAGTTAGAAAACAAGCTTCATCTAAAGACTCTCATGTCAATGTGGAC CTTGGTGACAATCCTGCTTTGTTAAAGCAAAAACTATGCGAAAGGGTGAGTCTGTTTAGAAGAAAAAGCAAAGACTGAGGTACTGTGAAT GGAGAGCTTCAGCTAAGAGGAGGCTCTGTCCCTTTTCAGAGCCAAAGGAAATAATACAACAAAAAGGAGGCTTCTTTGGAGACCTAAGTC TATTGGATGTAAACAAGACGTTGTATTTAGGGATGTTCTGTGTTTCTTTCTTTTTTGAAGTTGTCATCAATTGCTTTACTAAGATTTTTA AATAGTGAAAACCTCCTGTTTAGACTTTGGTGGAAGATGAATCAAGGAAGCAGGGCCCTGTCTTATGGGTCACGTGTCTTTGGTGAGTGA GAAGACCTAAACTCCTGGCCATCATCTCTTATCCAATACTTAGCAGTTGGGGATTAAACCATCCTTGCCTTCAGTTCTCTCCAATATTAC CAGGCCCAACTCAGTCTTCAGTGATTTTAAACAGCATTGACATCATCTGTAAAACCATCATCTGTAAAACCATCTATGACATGAGTTTTG AGAAACAATAATGGGGAAAATATTTGGGACCAAGCTGAAGCACTAATCCCACTAAGTTAAAGACTTCTTTCCAGTCCAAGGCAGGCCTGA ATCAACTGTCTTTAAATAAAATTTTAAGTGATGCTGTATTATATATAGGAAAAAATGCTTAAAATCCTGTCATTTAGAACAGTGAAAAGT ATCTTTTGAGATTAAAGTGACTCTTTACTGTAGGAAAAATATTACTCTGTGTTTACAGATTCATTGCTGTGGTCAGGCCATTTTTAAGGG AAGAGTTATTTAATATAAATAGTCTCTGATTTTAAGTTCTGTTTAATGTTCATTCTCCTTCCAAGAACAAAGTGGTGATTTTTGGTTAGG GTGATCGCCCTCTTAAAATTGGCAGTGCTGTTCCTTGTGCTGCCCCTGTCTTTTCCTCTGATGGCATTTTTTTTTTTTTTTTTTTAACAC AGGTTGAAACATTTCATCTATTATCTCTGCCTCATTTCTGGAGGGTTGTGTATCAGTTCTCTAACACTTGTTCCTGAGAACTAAATGTCT TTTTTATTCTTATTTCCTCTCTCATAAACATTTGGTGACCTTTTACCAAGTGGTGAGTTAGTTAGGTTTTTTAAAATAAAATGTTCATTG >34741_34741_2_GRID1-AFAP1L2_GRID1_chr10_88123698_ENST00000327946_AFAP1L2_chr10_116100490_ENST00000369271_length(amino acids)=887AA_BP=78 MEALTLWLLPWICQCVSVRADSIIHIGAIFEENAAKDDRVFQLAVSDLSLNDDILQSEKITYSIKVIEANNPFQAVQEALEQLLTELDDF LKILDQENLSSTALVKKSCLAELLRLYTKSSSSDEEYIYMNKVTINKQQNAESQGKAPEEQGLLPNGEPSQHSSAPQKSLPDLPPPKMIP ERKQLAIPKTESPEGYYEEAEPYDTSLNEDGEAVSSSYESYDEEDGSKGKSAPYQWPSPEAGIELMRDARICAFLWRKKWLGQWAKQLCV IKDNRLLCYKSSKDHSPQLDVNLLGSSVIHKEKQVRKKEHKLKITPMNADVIVLGLQSKDQAEQWLRVIQEVSGLPSEGASEGNQYTPDA QRFNCQKPDIAEKYLSASEYGSSVDGHPEVPETKDVKKKCSAGLKLSNLMNLGRKKSTSLEPVERSLETSSYLNVLVNSQWKSRWCSVRD NHLHFYQDRNRSKVAQQPLSLVGCEVVPDPSPDHLYSFRILHKGEELAKLEAKSSEEMGHWLGLLLSESGSKTDPEEFTYDYVDADRVSC IVSAAKNSLLLMQRKFSEPNTYIDGLPSQDRQEELYDDVDLSELTAAVEPTEEATPVADDPNERESDRVYLDLTPVKSFLHGPSSAQAQA SSPTLSCLDNATEALPADSGPGPTPDEPCIKCPENLGEQQLESLEPEDPSLRITTVKIQTEQQRISFPPSCPDAVVATPPGASPPVKDRL RVTSAEIKLGKNRTEAEVKRYTEEKERLEKKKEEIRGHLAQLRKEKRELKETLLKCTDKEVLASLEQKLKEIDEECRGEESRRVDLELSI -------------------------------------------------------------- >34741_34741_3_GRID1-AFAP1L2_GRID1_chr10_88123698_ENST00000327946_AFAP1L2_chr10_116100490_ENST00000545353_length(transcript)=3191nt_BP=321nt ACTGCACCGGGACCAGCGCCTCCCCGCTTCGCGCTGCCCTCGGCCTCGCCCCGGGCCCGGGTGGATGAGCCGCGCGCCCGGGGGACATGG AAGCGCTGACGCTGTGGCTTCTCCCCTGGATATGCCAGTGCGTGTCGGTGCGGGCCGACTCCATCATCCACATCGGTGCCATCTTCGAGG AGAACGCGGCCAAGGACGACAGGGTGTTCCAGTTGGCGGTATCCGACCTGAGCCTCAACGATGACATCCTGCAGAGCGAGAAGATCACCT ACTCCATCAAGGTCATCGAGGCCAACAACCCATTCCAGGCTGTGCAGGAAGCCCTGGAACAGCTGCTGACAGAGTTGGATGACTTCCTCA AGATTCTTGACCAGGAGAACCTGAGCAGCACAGCACTGGTGAAGAAGAGCTGCCTGGCGGAGCTCCTCCGGCTTTACACCAAAAGCAGCA GCTCTGATGAGGAGTACATTTATATGAACAAAGTGACCATCAACAAGCAACAGAATGCAGAGTCTCAAGGCAAAGCGCCTGAGGAGCAGG GCCTGCTACCCAATGGGGAGCCCAGCCAGCACTCCTCGGCCCCTCAGAAGAGCCTTCCAGACCTCCCGCCACCCAAGATGAGAATCCTGA CCTTGGGCCAGATCTCGGGTGCCCAGAGCTTTTCTCCAGATGGGACCCACCTGACGGGAACGGAGTTTCTTGGAGGCTGGAGCTGGAGGC CCAGCCAGATCAGGCTGATGAGCCCTGGGACCCACTGGGGAGACCTTACTGCCACAGAGATTCCAGAACGGAAACAGCTTGCCATCCCAA AGACGGAGTCTCCAGAGGGCTACTATGAAGAGGCTGAGCCATATGACACATCCCTCAATGAGGACGGAGAGGCTGTGAGCAGCTCCTACG AGTCCTACGATGAAGAGGACGGCAGCAAGGGCAAGTCGGCCCCTTACCAGTGGCCCTCGCCGGAGGCCGGCATCGAGCTGATGCGTGACG CCCGCATCTGCGCCTTCCTGTGGCGCAAGAAGTGGCTGGGACAGTGGGCCAAGCAGCTCTGTGTCATCAAGGACAACAGGCTTCTGTGCT ACAAATCCTCCAAGGACCACAGCCCTCAGCTGGACGTGAACCTACTGGGCAGCAGCGTCATTCACAAGGAGAAGCAAGTGCGGAAGAAGG AGCACAAGCTGAAGATCACGCCGATGAATGCCGATGTGATTGTGCTGGGCCTGCAGAGCAAGGACCAGGCTGAGCAGTGGCTCAGGGTCA TCCAGGAAGTGAGCGGCCTGCCTTCCGAAGGAGCATCTGAAGGAAACCAGTACACCCCGGATGCCCAGCGCTTTAACTGCCAGAAACCAG ATATAGCTGAGAAGTACCTGTCGGCTTCAGAGTATGGGAGCTCCGTGGATGGCCACCCTGAGGTCCCAGAAACCAAAGACGTCAAGAAGA AATGTTCTGCTGGCCTCAAACTGAGCAACCTAATGAATCTGGGCAGGAAGAAATCCACCTCACTGGAGCCTGTGGAGAGGTCCCTCGAGA CATCCAGTTACCTGAACGTGCTGGTGAACAGCCAGTGGAAGTCTCGCTGGTGCTCTGTCAGGGACAATCACCTGCACTTCTACCAGGACC GGAACCGGAGCAAGGTGGCCCAGCAACCCCTCAGCCTGGTGGGCTGCGAGGTGGTCCCAGACCCCAGCCCCGACCACCTCTACTCCTTCC GCATCCTCCACAAGGGCGAGGAGCTGGCCAAGCTTGAGGCCAAGTCTTCCGAGGAAATGGGCCACTGGCTGGGTCTCCTGCTCTCTGAGT CAGGCTCCAAGACAGACCCAGAAGAGTTCACCTACGACTATGTGGATGCCGATAGGGTCTCCTGTATTGTGAGTGCGGCCAAAAACTCTC TCTTACTGATGCAGAGAAAGTTCTCAGAGCCCAACACTTACATCGATGGCCTGCCTAGCCAGGACCGCCAGGAGGAGCTGTATGACGACG TGGACCTGTCAGAGCTCACAGCTGCGGTGGAGCCTACCGAGGAAGCCACCCCTGTTGCAGATGACCCAAATGAGAGAGAATCTGACCGAG TGTACCTGGACCTCACACCTGTCAAGTCCTTTCTGCATGGCCCCAGCAGTGCACAGGCCCAGGCCTCCTCCCCGACGTTGTCCTGCCTGG ACAATGCAACTGAGGCCCTCCCGGCAGACTCAGGCCCAGGTCCCACCCCAGATGAGCCCTGCATAAAGTGTCCAGAGAACCTGGGAGAAC AGCAGCTGGAGAGTTTGGAGCCAGAGGATCCTTCCCTGAGAATCACCACCGTCAAAATCCAGACGGAACAGCAGAGAATCTCCTTCCCAC CGAGCTGCCCGGATGCCGTGGTGGCCACCCCACCTGGTGCCAGCCCACCTGTGAAGGACAGGTTGCGCGTGACCAGTGCAGAGATCAAGC TTGGCAAGAATCGGACAGAAGCTGAGGTGAAGCGGTACACAGAGGAGAAGGAGAGGCTTGAAAAGAAGAAGGAAGAAATCCGGGGGCACC TGGCTCAGCTCCGGAAAGAGAAACGGGAGCTAAAGGAAACCCTACTGAAATGCACAGACAAGGAAGTCCTGGCGAGCCTGGAGCAGAAGC TGAAGGAAATTGACGAGGAGTGCCGGGGCGAGGAGAGCAGGCGCGTGGACCTGGAGCTCAGCATCATGGAGGTGAAGGACAACCTGAAGA AGGCTGAGGCAGGGCCTGTGACGTTAGGCACCACCGTGGACACCACCCACCTGGAGAATGTGAGCCCCCGCCCCAAAGCTGTCACACCTG CCTCTGCCCCAGACTGTACCCCAGTCAACTCTGCAACCACACTCAAGAACAGGCCTCTCTCGGTCGTGGTCACAGGCAAAGGCACTGTAC TCCAGAAAGCCAAGGAATGGGAGAAGAAAGGAGCAAGTTAGAAAACAAGCTTCATCTAAAGACTCTCATGTCAATGTGGACCTTGGTGAC AATCCTGCTTTGTTAAAGCAAAAACTATGCGAAAGGGTGAGTCTGTTTAGAAGAAAAAGCAAAGACTGAGGTACTGTGAATGGAGAGCTT CAGCTAAGAGGAGGCTCTGTCCCTTTTCAGAGCCAAAGGAAATAATACAACAAAAAGGAGGCTTCTTTGGAGACCTAAGTCTATTGGATG >34741_34741_3_GRID1-AFAP1L2_GRID1_chr10_88123698_ENST00000327946_AFAP1L2_chr10_116100490_ENST00000545353_length(amino acids)=944AA_BP=78 MEALTLWLLPWICQCVSVRADSIIHIGAIFEENAAKDDRVFQLAVSDLSLNDDILQSEKITYSIKVIEANNPFQAVQEALEQLLTELDDF LKILDQENLSSTALVKKSCLAELLRLYTKSSSSDEEYIYMNKVTINKQQNAESQGKAPEEQGLLPNGEPSQHSSAPQKSLPDLPPPKMRI LTLGQISGAQSFSPDGTHLTGTEFLGGWSWRPSQIRLMSPGTHWGDLTATEIPERKQLAIPKTESPEGYYEEAEPYDTSLNEDGEAVSSS YESYDEEDGSKGKSAPYQWPSPEAGIELMRDARICAFLWRKKWLGQWAKQLCVIKDNRLLCYKSSKDHSPQLDVNLLGSSVIHKEKQVRK KEHKLKITPMNADVIVLGLQSKDQAEQWLRVIQEVSGLPSEGASEGNQYTPDAQRFNCQKPDIAEKYLSASEYGSSVDGHPEVPETKDVK KKCSAGLKLSNLMNLGRKKSTSLEPVERSLETSSYLNVLVNSQWKSRWCSVRDNHLHFYQDRNRSKVAQQPLSLVGCEVVPDPSPDHLYS FRILHKGEELAKLEAKSSEEMGHWLGLLLSESGSKTDPEEFTYDYVDADRVSCIVSAAKNSLLLMQRKFSEPNTYIDGLPSQDRQEELYD DVDLSELTAAVEPTEEATPVADDPNERESDRVYLDLTPVKSFLHGPSSAQAQASSPTLSCLDNATEALPADSGPGPTPDEPCIKCPENLG EQQLESLEPEDPSLRITTVKIQTEQQRISFPPSCPDAVVATPPGASPPVKDRLRVTSAEIKLGKNRTEAEVKRYTEEKERLEKKKEEIRG HLAQLRKEKRELKETLLKCTDKEVLASLEQKLKEIDEECRGEESRRVDLELSIMEVKDNLKKAEAGPVTLGTTVDTTHLENVSPRPKAVT -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for GRID1-AFAP1L2 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
| Hgene | GRID1 | chr10:88123698 | chr10:116100490 | ENST00000327946 | - | 2 | 16 | 21_436 | 78.33333333333333 | 1010.0 | CBLN1 |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for GRID1-AFAP1L2 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for GRID1-AFAP1L2 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
