
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:GRIN2A-CPPED1 (FusionGDB2 ID:34802) |
Fusion Gene Summary for GRIN2A-CPPED1 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: GRIN2A-CPPED1 | Fusion gene ID: 34802 | Hgene | Tgene | Gene symbol | GRIN2A | CPPED1 | Gene ID | 2903 | 55313 |
| Gene name | glutamate ionotropic receptor NMDA type subunit 2A | calcineurin like phosphoesterase domain containing 1 | |
| Synonyms | EPND|FESD|GluN2A|LKS|NMDAR2A|NR2A | CSTP1 | |
| Cytomap | 16p13.2 | 16p13.12 | |
| Type of gene | protein-coding | protein-coding | |
| Description | glutamate receptor ionotropic, NMDA 2AN-methyl D-aspartate receptor subtype 2AN-methyl-D-aspartate receptor channel, subunit epsilon-1N-methyl-D-aspartate receptor subunit 2Aglutamate receptor, ionotropic, N-methyl D-aspartate 2A | serine/threonine-protein phosphatase CPPED1calcineurin-like phosphoesterase domain-containing protein 1complete S-transactivated protein 1 | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | Q12879 | Q9BRF8 | |
| Ensembl transtripts involved in fusion gene | ENST00000330684, ENST00000396573, ENST00000396575, ENST00000404927, ENST00000535259, ENST00000562109, ENST00000566670, | ENST00000261660, ENST00000433677, ENST00000381774, | |
| Fusion gene scores | * DoF score | 8 X 7 X 5=280 | 7 X 7 X 4=196 |
| # samples | 7 | 7 | |
| ** MAII score | log2(7/280*10)=-2 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(7/196*10)=-1.48542682717024 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: GRIN2A [Title/Abstract] AND CPPED1 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | GRIN2A(9934504)-CPPED1(12798906), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | GRIN2A-CPPED1 seems lost the major protein functional domain in Hgene partner, which is a CGC due to the frame-shifted ORF. GRIN2A-CPPED1 seems lost the major protein functional domain in Hgene partner, which is a essential gene due to the frame-shifted ORF. GRIN2A-CPPED1 seems lost the major protein functional domain in Hgene partner, which is a IUPHAR drug target due to the frame-shifted ORF. GRIN2A-CPPED1 seems lost the major protein functional domain in Hgene partner, which is a tumor suppressor due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | GRIN2A | GO:0045471 | response to ethanol | 18445116 |
| Hgene | GRIN2A | GO:0097553 | calcium ion transmembrane import into cytosol | 26875626 |
 Fusion gene breakpoints across GRIN2A (5'-gene) Fusion gene breakpoints across GRIN2A (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
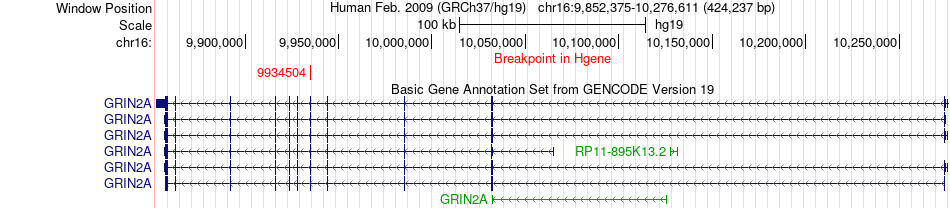 |
 Fusion gene breakpoints across CPPED1 (3'-gene) Fusion gene breakpoints across CPPED1 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
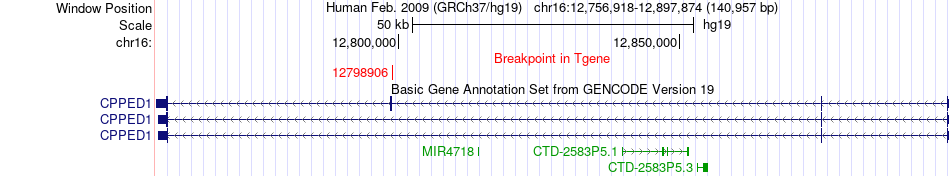 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | CESC | TCGA-DG-A2KH-01A | GRIN2A | chr16 | 9934504 | - | CPPED1 | chr16 | 12798906 | - |
Top |
Fusion Gene ORF analysis for GRIN2A-CPPED1 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-intron | ENST00000330684 | ENST00000261660 | GRIN2A | chr16 | 9934504 | - | CPPED1 | chr16 | 12798906 | - |
| 5CDS-intron | ENST00000330684 | ENST00000433677 | GRIN2A | chr16 | 9934504 | - | CPPED1 | chr16 | 12798906 | - |
| 5CDS-intron | ENST00000396573 | ENST00000261660 | GRIN2A | chr16 | 9934504 | - | CPPED1 | chr16 | 12798906 | - |
| 5CDS-intron | ENST00000396573 | ENST00000433677 | GRIN2A | chr16 | 9934504 | - | CPPED1 | chr16 | 12798906 | - |
| 5CDS-intron | ENST00000396575 | ENST00000261660 | GRIN2A | chr16 | 9934504 | - | CPPED1 | chr16 | 12798906 | - |
| 5CDS-intron | ENST00000396575 | ENST00000433677 | GRIN2A | chr16 | 9934504 | - | CPPED1 | chr16 | 12798906 | - |
| 5CDS-intron | ENST00000404927 | ENST00000261660 | GRIN2A | chr16 | 9934504 | - | CPPED1 | chr16 | 12798906 | - |
| 5CDS-intron | ENST00000404927 | ENST00000433677 | GRIN2A | chr16 | 9934504 | - | CPPED1 | chr16 | 12798906 | - |
| 5CDS-intron | ENST00000535259 | ENST00000261660 | GRIN2A | chr16 | 9934504 | - | CPPED1 | chr16 | 12798906 | - |
| 5CDS-intron | ENST00000535259 | ENST00000433677 | GRIN2A | chr16 | 9934504 | - | CPPED1 | chr16 | 12798906 | - |
| 5CDS-intron | ENST00000562109 | ENST00000261660 | GRIN2A | chr16 | 9934504 | - | CPPED1 | chr16 | 12798906 | - |
| 5CDS-intron | ENST00000562109 | ENST00000433677 | GRIN2A | chr16 | 9934504 | - | CPPED1 | chr16 | 12798906 | - |
| Frame-shift | ENST00000330684 | ENST00000381774 | GRIN2A | chr16 | 9934504 | - | CPPED1 | chr16 | 12798906 | - |
| Frame-shift | ENST00000396573 | ENST00000381774 | GRIN2A | chr16 | 9934504 | - | CPPED1 | chr16 | 12798906 | - |
| Frame-shift | ENST00000404927 | ENST00000381774 | GRIN2A | chr16 | 9934504 | - | CPPED1 | chr16 | 12798906 | - |
| Frame-shift | ENST00000535259 | ENST00000381774 | GRIN2A | chr16 | 9934504 | - | CPPED1 | chr16 | 12798906 | - |
| In-frame | ENST00000396575 | ENST00000381774 | GRIN2A | chr16 | 9934504 | - | CPPED1 | chr16 | 12798906 | - |
| In-frame | ENST00000562109 | ENST00000381774 | GRIN2A | chr16 | 9934504 | - | CPPED1 | chr16 | 12798906 | - |
| intron-3CDS | ENST00000566670 | ENST00000381774 | GRIN2A | chr16 | 9934504 | - | CPPED1 | chr16 | 12798906 | - |
| intron-intron | ENST00000566670 | ENST00000261660 | GRIN2A | chr16 | 9934504 | - | CPPED1 | chr16 | 12798906 | - |
| intron-intron | ENST00000566670 | ENST00000433677 | GRIN2A | chr16 | 9934504 | - | CPPED1 | chr16 | 12798906 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000562109 | GRIN2A | chr16 | 9934504 | - | ENST00000381774 | CPPED1 | chr16 | 12798906 | - | 4152 | 1672 | 21 | 2327 | 768 |
| ENST00000396575 | GRIN2A | chr16 | 9934504 | - | ENST00000381774 | CPPED1 | chr16 | 12798906 | - | 4131 | 1651 | 0 | 2306 | 768 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000562109 | ENST00000381774 | GRIN2A | chr16 | 9934504 | - | CPPED1 | chr16 | 12798906 | - | 0.001262478 | 0.9987375 |
| ENST00000396575 | ENST00000381774 | GRIN2A | chr16 | 9934504 | - | CPPED1 | chr16 | 12798906 | - | 0.001234797 | 0.99876523 |
Top |
Fusion Genomic Features for GRIN2A-CPPED1 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
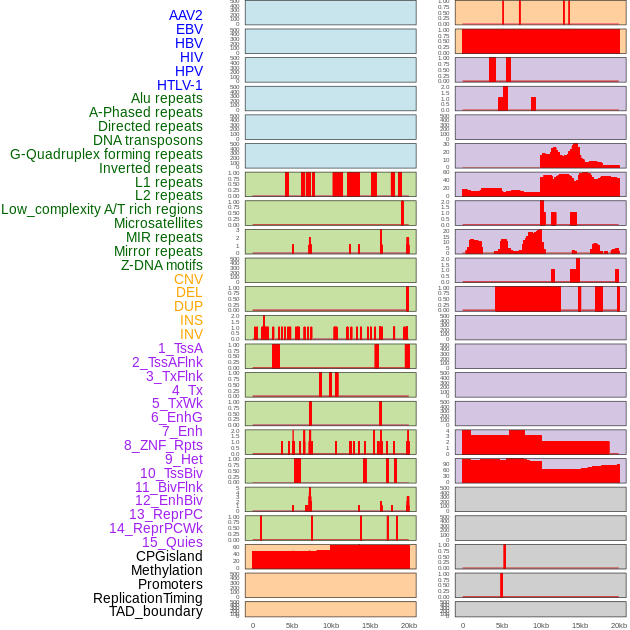 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for GRIN2A-CPPED1 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr16:9934504/chr16:12798906) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| GRIN2A | CPPED1 |
| FUNCTION: Component of NMDA receptor complexes that function as heterotetrameric, ligand-gated ion channels with high calcium permeability and voltage-dependent sensitivity to magnesium. Channel activation requires binding of the neurotransmitter glutamate to the epsilon subunit, glycine binding to the zeta subunit, plus membrane depolarization to eliminate channel inhibition by Mg(2+) (PubMed:8768735, PubMed:26919761, PubMed:26875626, PubMed:28105280). Sensitivity to glutamate and channel kinetics depend on the subunit composition; channels containing GRIN1 and GRIN2A have lower sensitivity to glutamate and faster deactivation kinetics than channels formed by GRIN1 and GRIN2B (PubMed:26919761, PubMed:26875626). Contributes to the slow phase of excitatory postsynaptic current, long-term synaptic potentiation, and learning (By similarity). {ECO:0000250|UniProtKB:P35436, ECO:0000250|UniProtKB:Q00959, ECO:0000269|PubMed:26875626, ECO:0000269|PubMed:26919761, ECO:0000269|PubMed:28105280, ECO:0000269|PubMed:8768735}. | FUNCTION: Protein phosphatase that dephosphorylates AKT family kinase specifically at 'Ser-473', blocking cell cycle progression and promoting cell apoptosis. May play an inhibitory role in glucose uptake by adipocytes. {ECO:0000269|PubMed:23799035, ECO:0000269|PubMed:23939394}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | GRIN2A | chr16:9934504 | chr16:12798906 | ENST00000330684 | - | 7 | 13 | 511_513 | 550 | 1465.0 | Region | Glutamate binding |
| Hgene | GRIN2A | chr16:9934504 | chr16:12798906 | ENST00000396573 | - | 8 | 14 | 511_513 | 550 | 1465.0 | Region | Glutamate binding |
| Hgene | GRIN2A | chr16:9934504 | chr16:12798906 | ENST00000396575 | - | 6 | 12 | 511_513 | 550 | 1465.0 | Region | Glutamate binding |
| Hgene | GRIN2A | chr16:9934504 | chr16:12798906 | ENST00000404927 | - | 7 | 14 | 511_513 | 550 | 1282.0 | Region | Glutamate binding |
| Hgene | GRIN2A | chr16:9934504 | chr16:12798906 | ENST00000562109 | - | 6 | 13 | 511_513 | 550 | 1282.0 | Region | Glutamate binding |
| Tgene | CPPED1 | chr16:9934504 | chr16:12798906 | ENST00000433677 | 0 | 3 | 47_250 | 0 | 173.0 | Region | Note=Catalytic |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | GRIN2A | chr16:9934504 | chr16:12798906 | ENST00000330684 | - | 7 | 13 | 601_620 | 550 | 1465.0 | Intramembrane | Discontinuously helical |
| Hgene | GRIN2A | chr16:9934504 | chr16:12798906 | ENST00000396573 | - | 8 | 14 | 601_620 | 550 | 1465.0 | Intramembrane | Discontinuously helical |
| Hgene | GRIN2A | chr16:9934504 | chr16:12798906 | ENST00000396575 | - | 6 | 12 | 601_620 | 550 | 1465.0 | Intramembrane | Discontinuously helical |
| Hgene | GRIN2A | chr16:9934504 | chr16:12798906 | ENST00000404927 | - | 7 | 14 | 601_620 | 550 | 1282.0 | Intramembrane | Discontinuously helical |
| Hgene | GRIN2A | chr16:9934504 | chr16:12798906 | ENST00000562109 | - | 6 | 13 | 601_620 | 550 | 1282.0 | Intramembrane | Discontinuously helical |
| Hgene | GRIN2A | chr16:9934504 | chr16:12798906 | ENST00000330684 | - | 7 | 13 | 1462_1464 | 550 | 1465.0 | Motif | PDZ-binding |
| Hgene | GRIN2A | chr16:9934504 | chr16:12798906 | ENST00000396573 | - | 8 | 14 | 1462_1464 | 550 | 1465.0 | Motif | PDZ-binding |
| Hgene | GRIN2A | chr16:9934504 | chr16:12798906 | ENST00000396575 | - | 6 | 12 | 1462_1464 | 550 | 1465.0 | Motif | PDZ-binding |
| Hgene | GRIN2A | chr16:9934504 | chr16:12798906 | ENST00000404927 | - | 7 | 14 | 1462_1464 | 550 | 1282.0 | Motif | PDZ-binding |
| Hgene | GRIN2A | chr16:9934504 | chr16:12798906 | ENST00000562109 | - | 6 | 13 | 1462_1464 | 550 | 1282.0 | Motif | PDZ-binding |
| Hgene | GRIN2A | chr16:9934504 | chr16:12798906 | ENST00000330684 | - | 7 | 13 | 599_620 | 550 | 1465.0 | Region | Pore-forming |
| Hgene | GRIN2A | chr16:9934504 | chr16:12798906 | ENST00000330684 | - | 7 | 13 | 689_690 | 550 | 1465.0 | Region | Glutamate binding |
| Hgene | GRIN2A | chr16:9934504 | chr16:12798906 | ENST00000330684 | - | 7 | 13 | 730_731 | 550 | 1465.0 | Region | Glutamate binding |
| Hgene | GRIN2A | chr16:9934504 | chr16:12798906 | ENST00000396573 | - | 8 | 14 | 599_620 | 550 | 1465.0 | Region | Pore-forming |
| Hgene | GRIN2A | chr16:9934504 | chr16:12798906 | ENST00000396573 | - | 8 | 14 | 689_690 | 550 | 1465.0 | Region | Glutamate binding |
| Hgene | GRIN2A | chr16:9934504 | chr16:12798906 | ENST00000396573 | - | 8 | 14 | 730_731 | 550 | 1465.0 | Region | Glutamate binding |
| Hgene | GRIN2A | chr16:9934504 | chr16:12798906 | ENST00000396575 | - | 6 | 12 | 599_620 | 550 | 1465.0 | Region | Pore-forming |
| Hgene | GRIN2A | chr16:9934504 | chr16:12798906 | ENST00000396575 | - | 6 | 12 | 689_690 | 550 | 1465.0 | Region | Glutamate binding |
| Hgene | GRIN2A | chr16:9934504 | chr16:12798906 | ENST00000396575 | - | 6 | 12 | 730_731 | 550 | 1465.0 | Region | Glutamate binding |
| Hgene | GRIN2A | chr16:9934504 | chr16:12798906 | ENST00000404927 | - | 7 | 14 | 599_620 | 550 | 1282.0 | Region | Pore-forming |
| Hgene | GRIN2A | chr16:9934504 | chr16:12798906 | ENST00000404927 | - | 7 | 14 | 689_690 | 550 | 1282.0 | Region | Glutamate binding |
| Hgene | GRIN2A | chr16:9934504 | chr16:12798906 | ENST00000404927 | - | 7 | 14 | 730_731 | 550 | 1282.0 | Region | Glutamate binding |
| Hgene | GRIN2A | chr16:9934504 | chr16:12798906 | ENST00000562109 | - | 6 | 13 | 599_620 | 550 | 1282.0 | Region | Pore-forming |
| Hgene | GRIN2A | chr16:9934504 | chr16:12798906 | ENST00000562109 | - | 6 | 13 | 689_690 | 550 | 1282.0 | Region | Glutamate binding |
| Hgene | GRIN2A | chr16:9934504 | chr16:12798906 | ENST00000562109 | - | 6 | 13 | 730_731 | 550 | 1282.0 | Region | Glutamate binding |
| Hgene | GRIN2A | chr16:9934504 | chr16:12798906 | ENST00000330684 | - | 7 | 13 | 23_555 | 550 | 1465.0 | Topological domain | Extracellular |
| Hgene | GRIN2A | chr16:9934504 | chr16:12798906 | ENST00000330684 | - | 7 | 13 | 577_600 | 550 | 1465.0 | Topological domain | Cytoplasmic |
| Hgene | GRIN2A | chr16:9934504 | chr16:12798906 | ENST00000330684 | - | 7 | 13 | 621_625 | 550 | 1465.0 | Topological domain | Cytoplasmic |
| Hgene | GRIN2A | chr16:9934504 | chr16:12798906 | ENST00000330684 | - | 7 | 13 | 646_816 | 550 | 1465.0 | Topological domain | Extracellular |
| Hgene | GRIN2A | chr16:9934504 | chr16:12798906 | ENST00000330684 | - | 7 | 13 | 838_1464 | 550 | 1465.0 | Topological domain | Cytoplasmic |
| Hgene | GRIN2A | chr16:9934504 | chr16:12798906 | ENST00000396573 | - | 8 | 14 | 23_555 | 550 | 1465.0 | Topological domain | Extracellular |
| Hgene | GRIN2A | chr16:9934504 | chr16:12798906 | ENST00000396573 | - | 8 | 14 | 577_600 | 550 | 1465.0 | Topological domain | Cytoplasmic |
| Hgene | GRIN2A | chr16:9934504 | chr16:12798906 | ENST00000396573 | - | 8 | 14 | 621_625 | 550 | 1465.0 | Topological domain | Cytoplasmic |
| Hgene | GRIN2A | chr16:9934504 | chr16:12798906 | ENST00000396573 | - | 8 | 14 | 646_816 | 550 | 1465.0 | Topological domain | Extracellular |
| Hgene | GRIN2A | chr16:9934504 | chr16:12798906 | ENST00000396573 | - | 8 | 14 | 838_1464 | 550 | 1465.0 | Topological domain | Cytoplasmic |
| Hgene | GRIN2A | chr16:9934504 | chr16:12798906 | ENST00000396575 | - | 6 | 12 | 23_555 | 550 | 1465.0 | Topological domain | Extracellular |
| Hgene | GRIN2A | chr16:9934504 | chr16:12798906 | ENST00000396575 | - | 6 | 12 | 577_600 | 550 | 1465.0 | Topological domain | Cytoplasmic |
| Hgene | GRIN2A | chr16:9934504 | chr16:12798906 | ENST00000396575 | - | 6 | 12 | 621_625 | 550 | 1465.0 | Topological domain | Cytoplasmic |
| Hgene | GRIN2A | chr16:9934504 | chr16:12798906 | ENST00000396575 | - | 6 | 12 | 646_816 | 550 | 1465.0 | Topological domain | Extracellular |
| Hgene | GRIN2A | chr16:9934504 | chr16:12798906 | ENST00000396575 | - | 6 | 12 | 838_1464 | 550 | 1465.0 | Topological domain | Cytoplasmic |
| Hgene | GRIN2A | chr16:9934504 | chr16:12798906 | ENST00000404927 | - | 7 | 14 | 23_555 | 550 | 1282.0 | Topological domain | Extracellular |
| Hgene | GRIN2A | chr16:9934504 | chr16:12798906 | ENST00000404927 | - | 7 | 14 | 577_600 | 550 | 1282.0 | Topological domain | Cytoplasmic |
| Hgene | GRIN2A | chr16:9934504 | chr16:12798906 | ENST00000404927 | - | 7 | 14 | 621_625 | 550 | 1282.0 | Topological domain | Cytoplasmic |
| Hgene | GRIN2A | chr16:9934504 | chr16:12798906 | ENST00000404927 | - | 7 | 14 | 646_816 | 550 | 1282.0 | Topological domain | Extracellular |
| Hgene | GRIN2A | chr16:9934504 | chr16:12798906 | ENST00000404927 | - | 7 | 14 | 838_1464 | 550 | 1282.0 | Topological domain | Cytoplasmic |
| Hgene | GRIN2A | chr16:9934504 | chr16:12798906 | ENST00000562109 | - | 6 | 13 | 23_555 | 550 | 1282.0 | Topological domain | Extracellular |
| Hgene | GRIN2A | chr16:9934504 | chr16:12798906 | ENST00000562109 | - | 6 | 13 | 577_600 | 550 | 1282.0 | Topological domain | Cytoplasmic |
| Hgene | GRIN2A | chr16:9934504 | chr16:12798906 | ENST00000562109 | - | 6 | 13 | 621_625 | 550 | 1282.0 | Topological domain | Cytoplasmic |
| Hgene | GRIN2A | chr16:9934504 | chr16:12798906 | ENST00000562109 | - | 6 | 13 | 646_816 | 550 | 1282.0 | Topological domain | Extracellular |
| Hgene | GRIN2A | chr16:9934504 | chr16:12798906 | ENST00000562109 | - | 6 | 13 | 838_1464 | 550 | 1282.0 | Topological domain | Cytoplasmic |
| Hgene | GRIN2A | chr16:9934504 | chr16:12798906 | ENST00000330684 | - | 7 | 13 | 556_576 | 550 | 1465.0 | Transmembrane | Helical |
| Hgene | GRIN2A | chr16:9934504 | chr16:12798906 | ENST00000330684 | - | 7 | 13 | 626_645 | 550 | 1465.0 | Transmembrane | Helical |
| Hgene | GRIN2A | chr16:9934504 | chr16:12798906 | ENST00000330684 | - | 7 | 13 | 817_837 | 550 | 1465.0 | Transmembrane | Helical |
| Hgene | GRIN2A | chr16:9934504 | chr16:12798906 | ENST00000396573 | - | 8 | 14 | 556_576 | 550 | 1465.0 | Transmembrane | Helical |
| Hgene | GRIN2A | chr16:9934504 | chr16:12798906 | ENST00000396573 | - | 8 | 14 | 626_645 | 550 | 1465.0 | Transmembrane | Helical |
| Hgene | GRIN2A | chr16:9934504 | chr16:12798906 | ENST00000396573 | - | 8 | 14 | 817_837 | 550 | 1465.0 | Transmembrane | Helical |
| Hgene | GRIN2A | chr16:9934504 | chr16:12798906 | ENST00000396575 | - | 6 | 12 | 556_576 | 550 | 1465.0 | Transmembrane | Helical |
| Hgene | GRIN2A | chr16:9934504 | chr16:12798906 | ENST00000396575 | - | 6 | 12 | 626_645 | 550 | 1465.0 | Transmembrane | Helical |
| Hgene | GRIN2A | chr16:9934504 | chr16:12798906 | ENST00000396575 | - | 6 | 12 | 817_837 | 550 | 1465.0 | Transmembrane | Helical |
| Hgene | GRIN2A | chr16:9934504 | chr16:12798906 | ENST00000404927 | - | 7 | 14 | 556_576 | 550 | 1282.0 | Transmembrane | Helical |
| Hgene | GRIN2A | chr16:9934504 | chr16:12798906 | ENST00000404927 | - | 7 | 14 | 626_645 | 550 | 1282.0 | Transmembrane | Helical |
| Hgene | GRIN2A | chr16:9934504 | chr16:12798906 | ENST00000404927 | - | 7 | 14 | 817_837 | 550 | 1282.0 | Transmembrane | Helical |
| Hgene | GRIN2A | chr16:9934504 | chr16:12798906 | ENST00000562109 | - | 6 | 13 | 556_576 | 550 | 1282.0 | Transmembrane | Helical |
| Hgene | GRIN2A | chr16:9934504 | chr16:12798906 | ENST00000562109 | - | 6 | 13 | 626_645 | 550 | 1282.0 | Transmembrane | Helical |
| Hgene | GRIN2A | chr16:9934504 | chr16:12798906 | ENST00000562109 | - | 6 | 13 | 817_837 | 550 | 1282.0 | Transmembrane | Helical |
| Tgene | CPPED1 | chr16:9934504 | chr16:12798906 | ENST00000381774 | 1 | 4 | 47_250 | 96 | 315.0 | Region | Note=Catalytic |
Top |
Fusion Gene Sequence for GRIN2A-CPPED1 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >34802_34802_1_GRIN2A-CPPED1_GRIN2A_chr16_9934504_ENST00000396575_CPPED1_chr16_12798906_ENST00000381774_length(transcript)=4131nt_BP=1651nt ATGGGCAGAGTGGGCTATTGGACCCTGCTGGTGCTGCCGGCCCTTCTGGTCTGGCGCGGTCCGGCGCCGAGCGCGGCGGCGGAGAAGGGT CCCCCCGCGCTAAATATTGCGGTGATGCTGGGTCACAGCCACGACGTGACAGAGCGCGAACTTCGAACACTGTGGGGCCCCGAGCAGGCG GCGGGGCTGCCCCTGGACGTGAACGTGGTAGCTCTGCTGATGAACCGCACCGACCCCAAGAGCCTCATCACGCACGTGTGCGACCTCATG TCCGGGGCACGCATCCACGGCCTCGTGTTTGGGGACGACACGGACCAGGAGGCCGTAGCCCAGATGCTGGATTTTATCTCCTCCCACACC TTCGTCCCCATCTTGGGCATTCATGGGGGCGCATCTATGATCATGGCTGACAAGGATCCGACGTCTACCTTCTTCCAGTTTGGAGCGTCC ATCCAGCAGCAAGCCACGGTCATGCTGAAGATCATGCAGGATTATGACTGGCATGTCTTCTCCCTGGTGACCACTATCTTCCCTGGCTAC AGGGAATTCATCAGCTTCGTCAAGACCACAGTGGACAACAGCTTTGTGGGCTGGGACATGCAGAATGTGATCACACTGGACACTTCCTTT GAGGATGCAAAGACACAAGTCCAGCTGAAGAAGATCCACTCTTCTGTCATCTTGCTCTACTGTTCCAAAGACGAGGCTGTTCTCATTCTG AGTGAGGCCCGCTCCCTTGGCCTCACCGGGTATGATTTCTTCTGGATTGTCCCCAGCTTGGTCTCTGGGAACACGGAGCTCATCCCAAAA GAGTTTCCATCGGGACTCATTTCTGTCTCCTACGATGACTGGGACTACAGCCTGGAGGCGAGAGTGAGGGACGGCATTGGCATCCTAACC ACCGCTGCATCTTCTATGCTGGAGAAGTTCTCCTACATCCCCGAGGCCAAGGCCAGCTGCTACGGGCAGATGGAGAGGCCAGAGGTCCCG ATGCACACCTTGCACCCATTTATGGTCAATGTTACATGGGATGGCAAAGACTTATCCTTCACTGAGGAAGGCTACCAGGTGCACCCCAGG CTGGTGGTGATTGTGCTGAACAAAGACCGGGAATGGGAAAAGGTGGGCAAGTGGGAGAACCATACGCTGAGCCTGAGGCACGCCGTGTGG CCCAGGTACAAGTCCTTCTCCGACTGTGAGCCGGATGACAACCATCTCAGCATCGTCACCCTGGAGGAGGCCCCATTCGTCATCGTGGAA GACATAGACCCCCTGACCGAGACGTGTGTGAGGAACACCGTGCCATGTCGGAAGTTCGTCAAAATCAACAATTCAACCAATGAGGGGATG AATGTGAAGAAATGCTGCAAGGGGTTCTGCATTGATATTCTGAAGAAGCTTTCCAGAACTGTGAAGTTTACTTACGACCTCTATCTGGTG ACCAATGGGAAGCATGGCAAGAAAGTTAACAATGTGTGGAATGGAATGATCGGTGAAGTGGTCTATCAACGGGCAGTCATGGCAGTTGGC TCGCTCACCATCAATGAGGAACGTTCTGAAGTGGTGGACTTCTCTGTGCCCTTTGTGGAAACGGGAATCAGTGTCATGGTTTCAAGAAGT AATGGCACCGTCTCACCTTCTGCTTTTCTAGGGAAGCCGTGGCGGACGGAGCAGACGGAGGACCTGAAGCGAGTGCTTAGGGCAGTGGAC AGGGCCATCCCACTGGTCCTTGTCAGCGGCAACCATGACATTGGCAACACCCCCACGGCCGAGACCGTCGAGGAGTTCTGCCGGACTTGG GGAGATGACTACTTCAGCTTCTGGGTCGGGGGCGTCCTGTTCCTGGTCCTCAACTCCCAGTTCTACGAGAACCCCTCCAAATGCCCCAGC CTGAAGCAGGCTCAGGACCAGTGGCTGGACGAGCAGCTGAGCATCGCGAGGCAGCGGCACTGCCAGCATGCCATCGTCTTCCAGCACATC CCGCTGTTCCTGGAGAGCATCGACGAGGACGACGACTACTACTTCAACCTCAGCAAGTCCACTCGGAAGAAGTTGGCAGACAAGTTCATC CACGCAGGTGTCAAAGTCGTGTTCTCAGGCCACTACCACAGGAATGCCGGGGGTACCTACCAGAACCTCGACATGGTGGTGTCATCTGCC ATTGGATGCCAGCTGGGCAGAGACCCCCACGGGCTCCGAGTCGTGGTGGTCACCGCCGAGAAAATTGTTCACCGATACTACAGTCTAGAT GAGCTGAGTGAGAAAGGAATAGAAGACGATCTCATGGATTTGATCAAGAAAAAATGACGCTCCTTCCCGTTCCCGTTCACTTTTCACTTG CACTATTTTTTTATTTTGCCAGAAACAGCAGCTGCACACAACCTCTTGCTGAAATATAAAAATAGCCCAGGCAGGTTTGTGAATTTATGT CCTTTTGCATAAATCAGAGAGCTGGGGCCCTGTCCTGAATTATATTCAAAATAGAACTGCATTTCCTTTAAAATGGAGCGAATGATCCTG GAAATCAGTTTTTAAAATATTAGACTATCTCTGCATGGATGTTTGAATAAATTCTCCTAATGCTGTGTTTCTCAACTCCACTTTTGAATT GGATTGTATTCTGGTTAGCATATGGTCAAAGTCTGCCTATCGACCTCAATTCCAATCTGACAGCCAAGTATGATCTTTGATAATTCCTTA AACAAACTACACCTGATTGTATTAAATTGTGGTCACGATTCCCAAAACGCTAGGCAAAACAAAGTTCATCTCCTTTCCTTTCCAGCGGAA CGAACAGCTGTCTCCCTTGTTGTGAGAGCCATGCAGACTTATTTTGACAAGGAGCCAAGTAGGCACCATTTACTGTTCAATTCTGAGACT TCTGATGATAGGAACAGAATGCAGTTGATACAGCTCGGAGCTTAAATTTGCCTTCCTGGGTGCCTAGAAAAGTCACCTTTGCACATAAAT TGGCCATTTCTGAAAATCTCCTCCAAGAGACTGAAACCAAAGCTCTAAAACGCCGCTTCTTCAGTCTCTGGGGAGCACGTTGAGTTGGTT CACATCCACATTCTGGCTGTGTAGCCACTCCACAGATGGGTGGTCTCTTGGGATCAGAATGCCCCTCCACTCATGAGACTCTTCATTTTG TCCACTTTGACAGGAAAAGTGGGAATGTATGCAGAGCTCTCAAAAGAAACAAAAAAGGCCAAAACGGTGCCTTCAGCCACATCCTCTGAA TTGGCCCTGACTTGGACTAAATGCACTAATGCAAAATCCCTTGACAAAAGCGCATAGGTTATTTCAAACCAGCATTGTTTTTTATGTAAC CTGTTTTACCGCATCTTCTCAGCAGCTTCTGACCACTGCTCAATTTTCTCCTTTACAGCCATTGTTCTGGTGGACAAATAACCTAGGTAC TCCAAATCCTGGCAGGAAAAATATACAGCATTATGAAACAGCACTCAGTAATCCTAAAATGATTTCCAAAGCTGGTTACACATGACCTGC AAAGTCTATTAAATTAAAAGACTTTCTCATTACAGAGTTTAGTCAACGTAGCAAAACCATGGGAAATTAAATGGAAAGATAATTAAATAT GTAAATTCATAAGGAACAAAAGACGAGAAGTTAATTATACAATAGGCTGGTGACACCAGAGTTCATTGTTATGTGCCACAAGAAAATTAA ATTTTTTAAAAAACTAGAACGCTCTTCTTTTAGTGTTTTGCTGAAATCCTGTTTTGTATTTGTTTCTTTACAACAGGTGTAGGTATAGGA GGTCAAGAAAAGGAGTTCGGTAAAGGGCATAGCTAATAACAACCACACATTGGGCCAGGCACAGTGGCTTACGCCTGTAATCCCAGCACT TTGGGAGGCTGAGACGGGCAGATCACCTGAGATCAGGAGTTTGAGAACAGCCTGGCCAACATGGTGAAACTCCATCTCTACTAAAAATAC AAAAATTAGCCAGGCATGCTGGTGCACACCTGTAATCCCAGCTACTCGGGAGGCTGAGGCAGGAGATCGCTGGAACTCAGGAGGCAGAGG >34802_34802_1_GRIN2A-CPPED1_GRIN2A_chr16_9934504_ENST00000396575_CPPED1_chr16_12798906_ENST00000381774_length(amino acids)=768AA_BP=550 MGRVGYWTLLVLPALLVWRGPAPSAAAEKGPPALNIAVMLGHSHDVTERELRTLWGPEQAAGLPLDVNVVALLMNRTDPKSLITHVCDLM SGARIHGLVFGDDTDQEAVAQMLDFISSHTFVPILGIHGGASMIMADKDPTSTFFQFGASIQQQATVMLKIMQDYDWHVFSLVTTIFPGY REFISFVKTTVDNSFVGWDMQNVITLDTSFEDAKTQVQLKKIHSSVILLYCSKDEAVLILSEARSLGLTGYDFFWIVPSLVSGNTELIPK EFPSGLISVSYDDWDYSLEARVRDGIGILTTAASSMLEKFSYIPEAKASCYGQMERPEVPMHTLHPFMVNVTWDGKDLSFTEEGYQVHPR LVVIVLNKDREWEKVGKWENHTLSLRHAVWPRYKSFSDCEPDDNHLSIVTLEEAPFVIVEDIDPLTETCVRNTVPCRKFVKINNSTNEGM NVKKCCKGFCIDILKKLSRTVKFTYDLYLVTNGKHGKKVNNVWNGMIGEVVYQRAVMAVGSLTINEERSEVVDFSVPFVETGISVMVSRS NGTVSPSAFLGKPWRTEQTEDLKRVLRAVDRAIPLVLVSGNHDIGNTPTAETVEEFCRTWGDDYFSFWVGGVLFLVLNSQFYENPSKCPS LKQAQDQWLDEQLSIARQRHCQHAIVFQHIPLFLESIDEDDDYYFNLSKSTRKKLADKFIHAGVKVVFSGHYHRNAGGTYQNLDMVVSSA -------------------------------------------------------------- >34802_34802_2_GRIN2A-CPPED1_GRIN2A_chr16_9934504_ENST00000562109_CPPED1_chr16_12798906_ENST00000381774_length(transcript)=4152nt_BP=1672nt CAGGGACCGTCAGTGGCGACTATGGGCAGAGTGGGCTATTGGACCCTGCTGGTGCTGCCGGCCCTTCTGGTCTGGCGCGGTCCGGCGCCG AGCGCGGCGGCGGAGAAGGGTCCCCCCGCGCTAAATATTGCGGTGATGCTGGGTCACAGCCACGACGTGACAGAGCGCGAACTTCGAACA CTGTGGGGCCCCGAGCAGGCGGCGGGGCTGCCCCTGGACGTGAACGTGGTAGCTCTGCTGATGAACCGCACCGACCCCAAGAGCCTCATC ACGCACGTGTGCGACCTCATGTCCGGGGCACGCATCCACGGCCTCGTGTTTGGGGACGACACGGACCAGGAGGCCGTAGCCCAGATGCTG GATTTTATCTCCTCCCACACCTTCGTCCCCATCTTGGGCATTCATGGGGGCGCATCTATGATCATGGCTGACAAGGATCCGACGTCTACC TTCTTCCAGTTTGGAGCGTCCATCCAGCAGCAAGCCACGGTCATGCTGAAGATCATGCAGGATTATGACTGGCATGTCTTCTCCCTGGTG ACCACTATCTTCCCTGGCTACAGGGAATTCATCAGCTTCGTCAAGACCACAGTGGACAACAGCTTTGTGGGCTGGGACATGCAGAATGTG ATCACACTGGACACTTCCTTTGAGGATGCAAAGACACAAGTCCAGCTGAAGAAGATCCACTCTTCTGTCATCTTGCTCTACTGTTCCAAA GACGAGGCTGTTCTCATTCTGAGTGAGGCCCGCTCCCTTGGCCTCACCGGGTATGATTTCTTCTGGATTGTCCCCAGCTTGGTCTCTGGG AACACGGAGCTCATCCCAAAAGAGTTTCCATCGGGACTCATTTCTGTCTCCTACGATGACTGGGACTACAGCCTGGAGGCGAGAGTGAGG GACGGCATTGGCATCCTAACCACCGCTGCATCTTCTATGCTGGAGAAGTTCTCCTACATCCCCGAGGCCAAGGCCAGCTGCTACGGGCAG ATGGAGAGGCCAGAGGTCCCGATGCACACCTTGCACCCATTTATGGTCAATGTTACATGGGATGGCAAAGACTTATCCTTCACTGAGGAA GGCTACCAGGTGCACCCCAGGCTGGTGGTGATTGTGCTGAACAAAGACCGGGAATGGGAAAAGGTGGGCAAGTGGGAGAACCATACGCTG AGCCTGAGGCACGCCGTGTGGCCCAGGTACAAGTCCTTCTCCGACTGTGAGCCGGATGACAACCATCTCAGCATCGTCACCCTGGAGGAG GCCCCATTCGTCATCGTGGAAGACATAGACCCCCTGACCGAGACGTGTGTGAGGAACACCGTGCCATGTCGGAAGTTCGTCAAAATCAAC AATTCAACCAATGAGGGGATGAATGTGAAGAAATGCTGCAAGGGGTTCTGCATTGATATTCTGAAGAAGCTTTCCAGAACTGTGAAGTTT ACTTACGACCTCTATCTGGTGACCAATGGGAAGCATGGCAAGAAAGTTAACAATGTGTGGAATGGAATGATCGGTGAAGTGGTCTATCAA CGGGCAGTCATGGCAGTTGGCTCGCTCACCATCAATGAGGAACGTTCTGAAGTGGTGGACTTCTCTGTGCCCTTTGTGGAAACGGGAATC AGTGTCATGGTTTCAAGAAGTAATGGCACCGTCTCACCTTCTGCTTTTCTAGGGAAGCCGTGGCGGACGGAGCAGACGGAGGACCTGAAG CGAGTGCTTAGGGCAGTGGACAGGGCCATCCCACTGGTCCTTGTCAGCGGCAACCATGACATTGGCAACACCCCCACGGCCGAGACCGTC GAGGAGTTCTGCCGGACTTGGGGAGATGACTACTTCAGCTTCTGGGTCGGGGGCGTCCTGTTCCTGGTCCTCAACTCCCAGTTCTACGAG AACCCCTCCAAATGCCCCAGCCTGAAGCAGGCTCAGGACCAGTGGCTGGACGAGCAGCTGAGCATCGCGAGGCAGCGGCACTGCCAGCAT GCCATCGTCTTCCAGCACATCCCGCTGTTCCTGGAGAGCATCGACGAGGACGACGACTACTACTTCAACCTCAGCAAGTCCACTCGGAAG AAGTTGGCAGACAAGTTCATCCACGCAGGTGTCAAAGTCGTGTTCTCAGGCCACTACCACAGGAATGCCGGGGGTACCTACCAGAACCTC GACATGGTGGTGTCATCTGCCATTGGATGCCAGCTGGGCAGAGACCCCCACGGGCTCCGAGTCGTGGTGGTCACCGCCGAGAAAATTGTT CACCGATACTACAGTCTAGATGAGCTGAGTGAGAAAGGAATAGAAGACGATCTCATGGATTTGATCAAGAAAAAATGACGCTCCTTCCCG TTCCCGTTCACTTTTCACTTGCACTATTTTTTTATTTTGCCAGAAACAGCAGCTGCACACAACCTCTTGCTGAAATATAAAAATAGCCCA GGCAGGTTTGTGAATTTATGTCCTTTTGCATAAATCAGAGAGCTGGGGCCCTGTCCTGAATTATATTCAAAATAGAACTGCATTTCCTTT AAAATGGAGCGAATGATCCTGGAAATCAGTTTTTAAAATATTAGACTATCTCTGCATGGATGTTTGAATAAATTCTCCTAATGCTGTGTT TCTCAACTCCACTTTTGAATTGGATTGTATTCTGGTTAGCATATGGTCAAAGTCTGCCTATCGACCTCAATTCCAATCTGACAGCCAAGT ATGATCTTTGATAATTCCTTAAACAAACTACACCTGATTGTATTAAATTGTGGTCACGATTCCCAAAACGCTAGGCAAAACAAAGTTCAT CTCCTTTCCTTTCCAGCGGAACGAACAGCTGTCTCCCTTGTTGTGAGAGCCATGCAGACTTATTTTGACAAGGAGCCAAGTAGGCACCAT TTACTGTTCAATTCTGAGACTTCTGATGATAGGAACAGAATGCAGTTGATACAGCTCGGAGCTTAAATTTGCCTTCCTGGGTGCCTAGAA AAGTCACCTTTGCACATAAATTGGCCATTTCTGAAAATCTCCTCCAAGAGACTGAAACCAAAGCTCTAAAACGCCGCTTCTTCAGTCTCT GGGGAGCACGTTGAGTTGGTTCACATCCACATTCTGGCTGTGTAGCCACTCCACAGATGGGTGGTCTCTTGGGATCAGAATGCCCCTCCA CTCATGAGACTCTTCATTTTGTCCACTTTGACAGGAAAAGTGGGAATGTATGCAGAGCTCTCAAAAGAAACAAAAAAGGCCAAAACGGTG CCTTCAGCCACATCCTCTGAATTGGCCCTGACTTGGACTAAATGCACTAATGCAAAATCCCTTGACAAAAGCGCATAGGTTATTTCAAAC CAGCATTGTTTTTTATGTAACCTGTTTTACCGCATCTTCTCAGCAGCTTCTGACCACTGCTCAATTTTCTCCTTTACAGCCATTGTTCTG GTGGACAAATAACCTAGGTACTCCAAATCCTGGCAGGAAAAATATACAGCATTATGAAACAGCACTCAGTAATCCTAAAATGATTTCCAA AGCTGGTTACACATGACCTGCAAAGTCTATTAAATTAAAAGACTTTCTCATTACAGAGTTTAGTCAACGTAGCAAAACCATGGGAAATTA AATGGAAAGATAATTAAATATGTAAATTCATAAGGAACAAAAGACGAGAAGTTAATTATACAATAGGCTGGTGACACCAGAGTTCATTGT TATGTGCCACAAGAAAATTAAATTTTTTAAAAAACTAGAACGCTCTTCTTTTAGTGTTTTGCTGAAATCCTGTTTTGTATTTGTTTCTTT ACAACAGGTGTAGGTATAGGAGGTCAAGAAAAGGAGTTCGGTAAAGGGCATAGCTAATAACAACCACACATTGGGCCAGGCACAGTGGCT TACGCCTGTAATCCCAGCACTTTGGGAGGCTGAGACGGGCAGATCACCTGAGATCAGGAGTTTGAGAACAGCCTGGCCAACATGGTGAAA CTCCATCTCTACTAAAAATACAAAAATTAGCCAGGCATGCTGGTGCACACCTGTAATCCCAGCTACTCGGGAGGCTGAGGCAGGAGATCG CTGGAACTCAGGAGGCAGAGGTTGCAGTGAGCCAAGATCATGCCACTGCACTCCAGCCTGGGCAACAGAGCAAGACTCTGTCTCAAAAAA >34802_34802_2_GRIN2A-CPPED1_GRIN2A_chr16_9934504_ENST00000562109_CPPED1_chr16_12798906_ENST00000381774_length(amino acids)=768AA_BP=550 MGRVGYWTLLVLPALLVWRGPAPSAAAEKGPPALNIAVMLGHSHDVTERELRTLWGPEQAAGLPLDVNVVALLMNRTDPKSLITHVCDLM SGARIHGLVFGDDTDQEAVAQMLDFISSHTFVPILGIHGGASMIMADKDPTSTFFQFGASIQQQATVMLKIMQDYDWHVFSLVTTIFPGY REFISFVKTTVDNSFVGWDMQNVITLDTSFEDAKTQVQLKKIHSSVILLYCSKDEAVLILSEARSLGLTGYDFFWIVPSLVSGNTELIPK EFPSGLISVSYDDWDYSLEARVRDGIGILTTAASSMLEKFSYIPEAKASCYGQMERPEVPMHTLHPFMVNVTWDGKDLSFTEEGYQVHPR LVVIVLNKDREWEKVGKWENHTLSLRHAVWPRYKSFSDCEPDDNHLSIVTLEEAPFVIVEDIDPLTETCVRNTVPCRKFVKINNSTNEGM NVKKCCKGFCIDILKKLSRTVKFTYDLYLVTNGKHGKKVNNVWNGMIGEVVYQRAVMAVGSLTINEERSEVVDFSVPFVETGISVMVSRS NGTVSPSAFLGKPWRTEQTEDLKRVLRAVDRAIPLVLVSGNHDIGNTPTAETVEEFCRTWGDDYFSFWVGGVLFLVLNSQFYENPSKCPS LKQAQDQWLDEQLSIARQRHCQHAIVFQHIPLFLESIDEDDDYYFNLSKSTRKKLADKFIHAGVKVVFSGHYHRNAGGTYQNLDMVVSSA -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for GRIN2A-CPPED1 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for GRIN2A-CPPED1 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for GRIN2A-CPPED1 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
