
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:GRM3-CASR (FusionGDB2 ID:34900) |
Fusion Gene Summary for GRM3-CASR |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: GRM3-CASR | Fusion gene ID: 34900 | Hgene | Tgene | Gene symbol | GRM3 | CASR | Gene ID | 2913 | 846 |
| Gene name | glutamate metabotropic receptor 3 | calcium sensing receptor | |
| Synonyms | GLUR3|GPRC1C|MGLUR3|mGlu3 | CAR|EIG8|FHH|FIH|GPRC2A|HHC|HHC1|HYPOC1|NSHPT|PCAR1|hCasR | |
| Cytomap | 7q21.11-q21.12 | 3q13.33-q21.1 | |
| Type of gene | protein-coding | protein-coding | |
| Description | metabotropic glutamate receptor 3glutamate receptor, metabotropic 3 | extracellular calcium-sensing receptorparathyroid Ca(2+)-sensing receptor 1parathyroid cell calcium-sensing receptor 1 | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | Q14832 | P41180 | |
| Ensembl transtripts involved in fusion gene | ENST00000361669, ENST00000394720, ENST00000439827, ENST00000536043, ENST00000546348, | ENST00000296154, ENST00000490131, ENST00000498619, | |
| Fusion gene scores | * DoF score | 4 X 5 X 1=20 | 8 X 9 X 7=504 |
| # samples | 5 | 15 | |
| ** MAII score | log2(5/20*10)=1.32192809488736 effective Gene in Pan-Cancer Fusion Genes (eGinPCFGs). DoF>8 and MAII>0 | log2(15/504*10)=-1.74846123300404 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: GRM3 [Title/Abstract] AND CASR [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | GRM3(86479793)-CASR(122003403), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | GRM3-CASR seems lost the major protein functional domain in Hgene partner, which is a CGC due to the frame-shifted ORF. GRM3-CASR seems lost the major protein functional domain in Hgene partner, which is a essential gene due to the frame-shifted ORF. GRM3-CASR seems lost the major protein functional domain in Hgene partner, which is a IUPHAR drug target due to the frame-shifted ORF. GRM3-CASR seems lost the major protein functional domain in Tgene partner, which is a IUPHAR drug target due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Tgene | CASR | GO:0005513 | detection of calcium ion | 27434672 |
| Tgene | CASR | GO:0006874 | cellular calcium ion homeostasis | 27434672 |
| Tgene | CASR | GO:0007186 | G protein-coupled receptor signaling pathway | 27434672 |
| Tgene | CASR | GO:0070509 | calcium ion import | 20846291 |
 Fusion gene breakpoints across GRM3 (5'-gene) Fusion gene breakpoints across GRM3 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
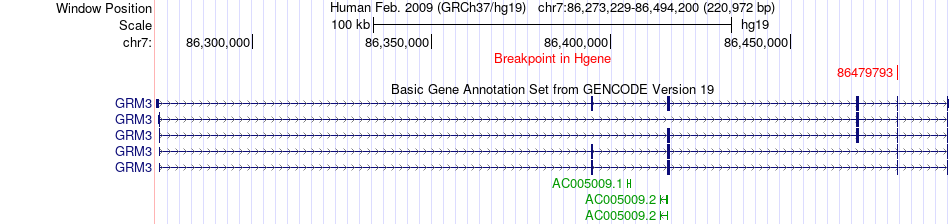 |
 Fusion gene breakpoints across CASR (3'-gene) Fusion gene breakpoints across CASR (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
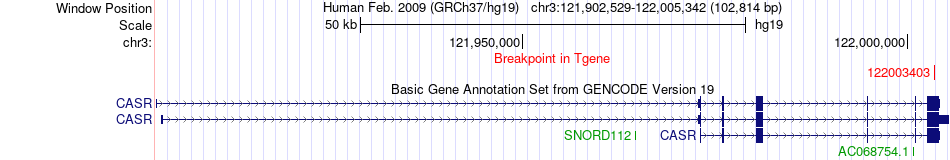 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChiTaRS5.0 | N/A | AX542233 | GRM3 | chr7 | 86479793 | + | CASR | chr3 | 122003403 | + |
Top |
Fusion Gene ORF analysis for GRM3-CASR |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| Frame-shift | ENST00000361669 | ENST00000296154 | GRM3 | chr7 | 86479793 | + | CASR | chr3 | 122003403 | + |
| Frame-shift | ENST00000361669 | ENST00000490131 | GRM3 | chr7 | 86479793 | + | CASR | chr3 | 122003403 | + |
| Frame-shift | ENST00000394720 | ENST00000296154 | GRM3 | chr7 | 86479793 | + | CASR | chr3 | 122003403 | + |
| Frame-shift | ENST00000394720 | ENST00000490131 | GRM3 | chr7 | 86479793 | + | CASR | chr3 | 122003403 | + |
| Frame-shift | ENST00000394720 | ENST00000498619 | GRM3 | chr7 | 86479793 | + | CASR | chr3 | 122003403 | + |
| Frame-shift | ENST00000439827 | ENST00000296154 | GRM3 | chr7 | 86479793 | + | CASR | chr3 | 122003403 | + |
| Frame-shift | ENST00000439827 | ENST00000490131 | GRM3 | chr7 | 86479793 | + | CASR | chr3 | 122003403 | + |
| Frame-shift | ENST00000439827 | ENST00000498619 | GRM3 | chr7 | 86479793 | + | CASR | chr3 | 122003403 | + |
| Frame-shift | ENST00000536043 | ENST00000296154 | GRM3 | chr7 | 86479793 | + | CASR | chr3 | 122003403 | + |
| Frame-shift | ENST00000536043 | ENST00000490131 | GRM3 | chr7 | 86479793 | + | CASR | chr3 | 122003403 | + |
| Frame-shift | ENST00000546348 | ENST00000296154 | GRM3 | chr7 | 86479793 | + | CASR | chr3 | 122003403 | + |
| Frame-shift | ENST00000546348 | ENST00000490131 | GRM3 | chr7 | 86479793 | + | CASR | chr3 | 122003403 | + |
| In-frame | ENST00000361669 | ENST00000498619 | GRM3 | chr7 | 86479793 | + | CASR | chr3 | 122003403 | + |
| In-frame | ENST00000536043 | ENST00000498619 | GRM3 | chr7 | 86479793 | + | CASR | chr3 | 122003403 | + |
| In-frame | ENST00000546348 | ENST00000498619 | GRM3 | chr7 | 86479793 | + | CASR | chr3 | 122003403 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000361669 | GRM3 | chr7 | 86479793 | + | ENST00000498619 | CASR | chr3 | 122003403 | + | 5496 | 3557 | 1054 | 4191 | 1045 |
| ENST00000546348 | GRM3 | chr7 | 86479793 | + | ENST00000498619 | CASR | chr3 | 122003403 | + | 3556 | 1617 | 374 | 2251 | 625 |
| ENST00000536043 | GRM3 | chr7 | 86479793 | + | ENST00000498619 | CASR | chr3 | 122003403 | + | 4166 | 2227 | 153 | 2861 | 902 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000361669 | ENST00000498619 | GRM3 | chr7 | 86479793 | + | CASR | chr3 | 122003403 | + | 0.000652637 | 0.9993474 |
| ENST00000546348 | ENST00000498619 | GRM3 | chr7 | 86479793 | + | CASR | chr3 | 122003403 | + | 0.001623309 | 0.9983767 |
| ENST00000536043 | ENST00000498619 | GRM3 | chr7 | 86479793 | + | CASR | chr3 | 122003403 | + | 0.001073524 | 0.99892646 |
Top |
Fusion Genomic Features for GRM3-CASR |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
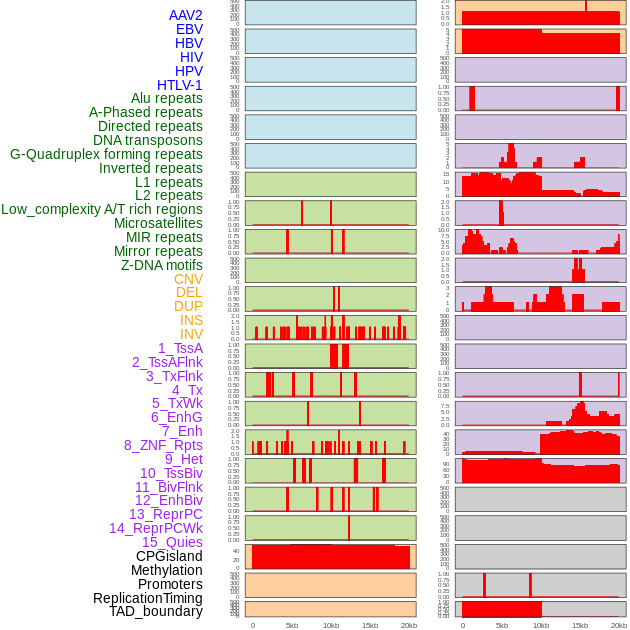 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for GRM3-CASR |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr7:86479793/chr3:122003403) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| GRM3 | CASR |
| FUNCTION: G-protein coupled receptor for glutamate. Ligand binding causes a conformation change that triggers signaling via guanine nucleotide-binding proteins (G proteins) and modulates the activity of down-stream effectors. Signaling inhibits adenylate cyclase activity. {ECO:0000269|PubMed:8840013}. | FUNCTION: G-protein-coupled receptor that senses changes in the extracellular concentration of calcium ions and plays a key role in maintaining calcium homeostasis (PubMed:7759551, PubMed:8702647, PubMed:8636323, PubMed:8878438, PubMed:17555508, PubMed:19789209, PubMed:21566075, PubMed:22114145, PubMed:23966241, PubMed:25292184, PubMed:25104082, PubMed:26386835, PubMed:25766501, PubMed:22789683). Senses fluctuations in the circulating calcium concentration and modulates the production of parathyroid hormone (PTH) in parathyroid glands (By similarity). The activity of this receptor is mediated by a G-protein that activates a phosphatidylinositol-calcium second messenger system (PubMed:7759551). The G-protein-coupled receptor activity is activated by a co-agonist mechanism: aromatic amino acids, such as Trp or Phe, act concertedly with divalent cations, such as calcium or magnesium, to achieve full receptor activation (PubMed:27434672, PubMed:27386547). {ECO:0000250|UniProtKB:Q9QY96, ECO:0000269|PubMed:17555508, ECO:0000269|PubMed:19789209, ECO:0000269|PubMed:21566075, ECO:0000269|PubMed:22114145, ECO:0000269|PubMed:22789683, ECO:0000269|PubMed:23966241, ECO:0000269|PubMed:25104082, ECO:0000269|PubMed:25292184, ECO:0000269|PubMed:25766501, ECO:0000269|PubMed:26386835, ECO:0000269|PubMed:27386547, ECO:0000269|PubMed:27434672, ECO:0000269|PubMed:7759551, ECO:0000269|PubMed:8636323, ECO:0000269|PubMed:8702647, ECO:0000269|PubMed:8878438}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | CASR | chr7:86479793 | chr3:122003403 | ENST00000296154 | 0 | 6 | 189_324 | 0 | 1079.0 | Region | Ligand-binding 2 (LB2) | |
| Tgene | CASR | chr7:86479793 | chr3:122003403 | ENST00000296154 | 0 | 6 | 22_188 | 0 | 1079.0 | Region | Ligand-binding 1 (LB1) | |
| Tgene | CASR | chr7:86479793 | chr3:122003403 | ENST00000296154 | 0 | 6 | 415_417 | 0 | 1079.0 | Region | Anion binding | |
| Tgene | CASR | chr7:86479793 | chr3:122003403 | ENST00000296154 | 0 | 6 | 542_612 | 0 | 1079.0 | Region | Cysteine-rich (CR) | |
| Tgene | CASR | chr7:86479793 | chr3:122003403 | ENST00000296154 | 0 | 6 | 66_70 | 0 | 1079.0 | Region | Anion binding | |
| Tgene | CASR | chr7:86479793 | chr3:122003403 | ENST00000490131 | 0 | 7 | 189_324 | 0 | 1079.0 | Region | Ligand-binding 2 (LB2) | |
| Tgene | CASR | chr7:86479793 | chr3:122003403 | ENST00000490131 | 0 | 7 | 22_188 | 0 | 1079.0 | Region | Ligand-binding 1 (LB1) | |
| Tgene | CASR | chr7:86479793 | chr3:122003403 | ENST00000490131 | 0 | 7 | 415_417 | 0 | 1079.0 | Region | Anion binding | |
| Tgene | CASR | chr7:86479793 | chr3:122003403 | ENST00000490131 | 0 | 7 | 542_612 | 0 | 1079.0 | Region | Cysteine-rich (CR) | |
| Tgene | CASR | chr7:86479793 | chr3:122003403 | ENST00000490131 | 0 | 7 | 66_70 | 0 | 1079.0 | Region | Anion binding | |
| Tgene | CASR | chr7:86479793 | chr3:122003403 | ENST00000296154 | 0 | 6 | 20_612 | 0 | 1079.0 | Topological domain | Extracellular | |
| Tgene | CASR | chr7:86479793 | chr3:122003403 | ENST00000296154 | 0 | 6 | 636_649 | 0 | 1079.0 | Topological domain | Cytoplasmic | |
| Tgene | CASR | chr7:86479793 | chr3:122003403 | ENST00000296154 | 0 | 6 | 671_681 | 0 | 1079.0 | Topological domain | Extracellular | |
| Tgene | CASR | chr7:86479793 | chr3:122003403 | ENST00000296154 | 0 | 6 | 701_724 | 0 | 1079.0 | Topological domain | Cytoplasmic | |
| Tgene | CASR | chr7:86479793 | chr3:122003403 | ENST00000296154 | 0 | 6 | 746_769 | 0 | 1079.0 | Topological domain | Extracellular | |
| Tgene | CASR | chr7:86479793 | chr3:122003403 | ENST00000296154 | 0 | 6 | 793_805 | 0 | 1079.0 | Topological domain | Cytoplasmic | |
| Tgene | CASR | chr7:86479793 | chr3:122003403 | ENST00000296154 | 0 | 6 | 829_836 | 0 | 1079.0 | Topological domain | Extracellular | |
| Tgene | CASR | chr7:86479793 | chr3:122003403 | ENST00000296154 | 0 | 6 | 863_1078 | 0 | 1079.0 | Topological domain | Cytoplasmic | |
| Tgene | CASR | chr7:86479793 | chr3:122003403 | ENST00000490131 | 0 | 7 | 20_612 | 0 | 1079.0 | Topological domain | Extracellular | |
| Tgene | CASR | chr7:86479793 | chr3:122003403 | ENST00000490131 | 0 | 7 | 636_649 | 0 | 1079.0 | Topological domain | Cytoplasmic | |
| Tgene | CASR | chr7:86479793 | chr3:122003403 | ENST00000490131 | 0 | 7 | 671_681 | 0 | 1079.0 | Topological domain | Extracellular | |
| Tgene | CASR | chr7:86479793 | chr3:122003403 | ENST00000490131 | 0 | 7 | 701_724 | 0 | 1079.0 | Topological domain | Cytoplasmic | |
| Tgene | CASR | chr7:86479793 | chr3:122003403 | ENST00000490131 | 0 | 7 | 746_769 | 0 | 1079.0 | Topological domain | Extracellular | |
| Tgene | CASR | chr7:86479793 | chr3:122003403 | ENST00000490131 | 0 | 7 | 793_805 | 0 | 1079.0 | Topological domain | Cytoplasmic | |
| Tgene | CASR | chr7:86479793 | chr3:122003403 | ENST00000490131 | 0 | 7 | 829_836 | 0 | 1079.0 | Topological domain | Extracellular | |
| Tgene | CASR | chr7:86479793 | chr3:122003403 | ENST00000490131 | 0 | 7 | 863_1078 | 0 | 1079.0 | Topological domain | Cytoplasmic | |
| Tgene | CASR | chr7:86479793 | chr3:122003403 | ENST00000296154 | 0 | 6 | 613_635 | 0 | 1079.0 | Transmembrane | Helical%3B Name%3D1 | |
| Tgene | CASR | chr7:86479793 | chr3:122003403 | ENST00000296154 | 0 | 6 | 650_670 | 0 | 1079.0 | Transmembrane | Helical%3B Name%3D2 | |
| Tgene | CASR | chr7:86479793 | chr3:122003403 | ENST00000296154 | 0 | 6 | 682_700 | 0 | 1079.0 | Transmembrane | Helical%3B Name%3D3 | |
| Tgene | CASR | chr7:86479793 | chr3:122003403 | ENST00000296154 | 0 | 6 | 725_745 | 0 | 1079.0 | Transmembrane | Helical%3B Name%3D4 | |
| Tgene | CASR | chr7:86479793 | chr3:122003403 | ENST00000296154 | 0 | 6 | 770_792 | 0 | 1079.0 | Transmembrane | Helical%3B Name%3D5 | |
| Tgene | CASR | chr7:86479793 | chr3:122003403 | ENST00000296154 | 0 | 6 | 806_828 | 0 | 1079.0 | Transmembrane | Helical%3B Name%3D6 | |
| Tgene | CASR | chr7:86479793 | chr3:122003403 | ENST00000296154 | 0 | 6 | 837_862 | 0 | 1079.0 | Transmembrane | Helical%3B Name%3D7 | |
| Tgene | CASR | chr7:86479793 | chr3:122003403 | ENST00000490131 | 0 | 7 | 613_635 | 0 | 1079.0 | Transmembrane | Helical%3B Name%3D1 | |
| Tgene | CASR | chr7:86479793 | chr3:122003403 | ENST00000490131 | 0 | 7 | 650_670 | 0 | 1079.0 | Transmembrane | Helical%3B Name%3D2 | |
| Tgene | CASR | chr7:86479793 | chr3:122003403 | ENST00000490131 | 0 | 7 | 682_700 | 0 | 1079.0 | Transmembrane | Helical%3B Name%3D3 | |
| Tgene | CASR | chr7:86479793 | chr3:122003403 | ENST00000490131 | 0 | 7 | 725_745 | 0 | 1079.0 | Transmembrane | Helical%3B Name%3D4 | |
| Tgene | CASR | chr7:86479793 | chr3:122003403 | ENST00000490131 | 0 | 7 | 770_792 | 0 | 1079.0 | Transmembrane | Helical%3B Name%3D5 | |
| Tgene | CASR | chr7:86479793 | chr3:122003403 | ENST00000490131 | 0 | 7 | 806_828 | 0 | 1079.0 | Transmembrane | Helical%3B Name%3D6 | |
| Tgene | CASR | chr7:86479793 | chr3:122003403 | ENST00000490131 | 0 | 7 | 837_862 | 0 | 1079.0 | Transmembrane | Helical%3B Name%3D7 |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | GRM3 | chr7:86479793 | chr3:122003403 | ENST00000361669 | + | 1 | 6 | 172_174 | 0 | 880.0 | Region | Glutamate binding |
| Hgene | GRM3 | chr7:86479793 | chr3:122003403 | ENST00000394720 | + | 1 | 5 | 172_174 | 0 | 536.0 | Region | Glutamate binding |
| Hgene | GRM3 | chr7:86479793 | chr3:122003403 | ENST00000439827 | + | 1 | 5 | 172_174 | 0 | 538.0 | Region | Glutamate binding |
| Hgene | GRM3 | chr7:86479793 | chr3:122003403 | ENST00000361669 | + | 1 | 6 | 23_576 | 0 | 880.0 | Topological domain | Extracellular |
| Hgene | GRM3 | chr7:86479793 | chr3:122003403 | ENST00000361669 | + | 1 | 6 | 600_613 | 0 | 880.0 | Topological domain | Cytoplasmic |
| Hgene | GRM3 | chr7:86479793 | chr3:122003403 | ENST00000361669 | + | 1 | 6 | 635_645 | 0 | 880.0 | Topological domain | Extracellular |
| Hgene | GRM3 | chr7:86479793 | chr3:122003403 | ENST00000361669 | + | 1 | 6 | 665_688 | 0 | 880.0 | Topological domain | Cytoplasmic |
| Hgene | GRM3 | chr7:86479793 | chr3:122003403 | ENST00000361669 | + | 1 | 6 | 710_734 | 0 | 880.0 | Topological domain | Extracellular |
| Hgene | GRM3 | chr7:86479793 | chr3:122003403 | ENST00000361669 | + | 1 | 6 | 757_769 | 0 | 880.0 | Topological domain | Cytoplasmic |
| Hgene | GRM3 | chr7:86479793 | chr3:122003403 | ENST00000361669 | + | 1 | 6 | 793_802 | 0 | 880.0 | Topological domain | Extracellular |
| Hgene | GRM3 | chr7:86479793 | chr3:122003403 | ENST00000361669 | + | 1 | 6 | 829_879 | 0 | 880.0 | Topological domain | Cytoplasmic |
| Hgene | GRM3 | chr7:86479793 | chr3:122003403 | ENST00000394720 | + | 1 | 5 | 23_576 | 0 | 536.0 | Topological domain | Extracellular |
| Hgene | GRM3 | chr7:86479793 | chr3:122003403 | ENST00000394720 | + | 1 | 5 | 600_613 | 0 | 536.0 | Topological domain | Cytoplasmic |
| Hgene | GRM3 | chr7:86479793 | chr3:122003403 | ENST00000394720 | + | 1 | 5 | 635_645 | 0 | 536.0 | Topological domain | Extracellular |
| Hgene | GRM3 | chr7:86479793 | chr3:122003403 | ENST00000394720 | + | 1 | 5 | 665_688 | 0 | 536.0 | Topological domain | Cytoplasmic |
| Hgene | GRM3 | chr7:86479793 | chr3:122003403 | ENST00000394720 | + | 1 | 5 | 710_734 | 0 | 536.0 | Topological domain | Extracellular |
| Hgene | GRM3 | chr7:86479793 | chr3:122003403 | ENST00000394720 | + | 1 | 5 | 757_769 | 0 | 536.0 | Topological domain | Cytoplasmic |
| Hgene | GRM3 | chr7:86479793 | chr3:122003403 | ENST00000394720 | + | 1 | 5 | 793_802 | 0 | 536.0 | Topological domain | Extracellular |
| Hgene | GRM3 | chr7:86479793 | chr3:122003403 | ENST00000394720 | + | 1 | 5 | 829_879 | 0 | 536.0 | Topological domain | Cytoplasmic |
| Hgene | GRM3 | chr7:86479793 | chr3:122003403 | ENST00000439827 | + | 1 | 5 | 23_576 | 0 | 538.0 | Topological domain | Extracellular |
| Hgene | GRM3 | chr7:86479793 | chr3:122003403 | ENST00000439827 | + | 1 | 5 | 600_613 | 0 | 538.0 | Topological domain | Cytoplasmic |
| Hgene | GRM3 | chr7:86479793 | chr3:122003403 | ENST00000439827 | + | 1 | 5 | 635_645 | 0 | 538.0 | Topological domain | Extracellular |
| Hgene | GRM3 | chr7:86479793 | chr3:122003403 | ENST00000439827 | + | 1 | 5 | 665_688 | 0 | 538.0 | Topological domain | Cytoplasmic |
| Hgene | GRM3 | chr7:86479793 | chr3:122003403 | ENST00000439827 | + | 1 | 5 | 710_734 | 0 | 538.0 | Topological domain | Extracellular |
| Hgene | GRM3 | chr7:86479793 | chr3:122003403 | ENST00000439827 | + | 1 | 5 | 757_769 | 0 | 538.0 | Topological domain | Cytoplasmic |
| Hgene | GRM3 | chr7:86479793 | chr3:122003403 | ENST00000439827 | + | 1 | 5 | 793_802 | 0 | 538.0 | Topological domain | Extracellular |
| Hgene | GRM3 | chr7:86479793 | chr3:122003403 | ENST00000439827 | + | 1 | 5 | 829_879 | 0 | 538.0 | Topological domain | Cytoplasmic |
| Hgene | GRM3 | chr7:86479793 | chr3:122003403 | ENST00000361669 | + | 1 | 6 | 577_599 | 0 | 880.0 | Transmembrane | Helical%3B Name%3D1 |
| Hgene | GRM3 | chr7:86479793 | chr3:122003403 | ENST00000361669 | + | 1 | 6 | 614_634 | 0 | 880.0 | Transmembrane | Helical%3B Name%3D2 |
| Hgene | GRM3 | chr7:86479793 | chr3:122003403 | ENST00000361669 | + | 1 | 6 | 646_664 | 0 | 880.0 | Transmembrane | Helical%3B Name%3D3 |
| Hgene | GRM3 | chr7:86479793 | chr3:122003403 | ENST00000361669 | + | 1 | 6 | 689_709 | 0 | 880.0 | Transmembrane | Helical%3B Name%3D4 |
| Hgene | GRM3 | chr7:86479793 | chr3:122003403 | ENST00000361669 | + | 1 | 6 | 735_756 | 0 | 880.0 | Transmembrane | Helical%3B Name%3D5 |
| Hgene | GRM3 | chr7:86479793 | chr3:122003403 | ENST00000361669 | + | 1 | 6 | 770_792 | 0 | 880.0 | Transmembrane | Helical%3B Name%3D6 |
| Hgene | GRM3 | chr7:86479793 | chr3:122003403 | ENST00000361669 | + | 1 | 6 | 803_828 | 0 | 880.0 | Transmembrane | Helical%3B Name%3D7 |
| Hgene | GRM3 | chr7:86479793 | chr3:122003403 | ENST00000394720 | + | 1 | 5 | 577_599 | 0 | 536.0 | Transmembrane | Helical%3B Name%3D1 |
| Hgene | GRM3 | chr7:86479793 | chr3:122003403 | ENST00000394720 | + | 1 | 5 | 614_634 | 0 | 536.0 | Transmembrane | Helical%3B Name%3D2 |
| Hgene | GRM3 | chr7:86479793 | chr3:122003403 | ENST00000394720 | + | 1 | 5 | 646_664 | 0 | 536.0 | Transmembrane | Helical%3B Name%3D3 |
| Hgene | GRM3 | chr7:86479793 | chr3:122003403 | ENST00000394720 | + | 1 | 5 | 689_709 | 0 | 536.0 | Transmembrane | Helical%3B Name%3D4 |
| Hgene | GRM3 | chr7:86479793 | chr3:122003403 | ENST00000394720 | + | 1 | 5 | 735_756 | 0 | 536.0 | Transmembrane | Helical%3B Name%3D5 |
| Hgene | GRM3 | chr7:86479793 | chr3:122003403 | ENST00000394720 | + | 1 | 5 | 770_792 | 0 | 536.0 | Transmembrane | Helical%3B Name%3D6 |
| Hgene | GRM3 | chr7:86479793 | chr3:122003403 | ENST00000394720 | + | 1 | 5 | 803_828 | 0 | 536.0 | Transmembrane | Helical%3B Name%3D7 |
| Hgene | GRM3 | chr7:86479793 | chr3:122003403 | ENST00000439827 | + | 1 | 5 | 577_599 | 0 | 538.0 | Transmembrane | Helical%3B Name%3D1 |
| Hgene | GRM3 | chr7:86479793 | chr3:122003403 | ENST00000439827 | + | 1 | 5 | 614_634 | 0 | 538.0 | Transmembrane | Helical%3B Name%3D2 |
| Hgene | GRM3 | chr7:86479793 | chr3:122003403 | ENST00000439827 | + | 1 | 5 | 646_664 | 0 | 538.0 | Transmembrane | Helical%3B Name%3D3 |
| Hgene | GRM3 | chr7:86479793 | chr3:122003403 | ENST00000439827 | + | 1 | 5 | 689_709 | 0 | 538.0 | Transmembrane | Helical%3B Name%3D4 |
| Hgene | GRM3 | chr7:86479793 | chr3:122003403 | ENST00000439827 | + | 1 | 5 | 735_756 | 0 | 538.0 | Transmembrane | Helical%3B Name%3D5 |
| Hgene | GRM3 | chr7:86479793 | chr3:122003403 | ENST00000439827 | + | 1 | 5 | 770_792 | 0 | 538.0 | Transmembrane | Helical%3B Name%3D6 |
| Hgene | GRM3 | chr7:86479793 | chr3:122003403 | ENST00000439827 | + | 1 | 5 | 803_828 | 0 | 538.0 | Transmembrane | Helical%3B Name%3D7 |
Top |
Fusion Gene Sequence for GRM3-CASR |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >34900_34900_1_GRM3-CASR_GRM3_chr7_86479793_ENST00000361669_CASR_chr3_122003403_ENST00000498619_length(transcript)=5496nt_BP=3557nt GTGCAGTTGAGTCGCGAGTACGGCTGAGCTGCGTACCGGCCTCCCTGGCTCTCACACTCCCTCTCTGCTCCCGCTCTCCTAATCTCCTCT GGCATGCGGTCAGCCCCCTGCCCAGGGACCACAGGAGAGTTCTTGTAAGGACTGTTAGTCCCTGCTTACCTGAAAGCCAAGCGCTCTAGC AGAGCTTTAAAGTTGGAGCCGCCACCCTCCCTACCGCCCCATGCCCCTTCACCCCACTCCGAAATTCACCGACCTTTGCATGCACTGCCT AAGGATTTCAGAGTGAGGCAAAGCAGTCGGCAAATCTACCCTGGCTTTTCGTATAAAAATCCTCTCGTCTAGGTACCCTGGCTCACTGAA GACTCTGCAGATATACCCTTATAAGAGGGAGGGTGGGGGAGGGAAAAGAACGAGAGAGGGAGGAAAGAATGAAAAGGAGAGGATGCCAGG AGGTCCGTGCTTCTGCCAAGAGTCCCAATTAGATGCGACGGCTTCAGCCTGGTCAAGGTGAAGGAAAGTTGCTTCCGCGCCTAGGAAGTG GGTTTGCCTGATAAGAGAAGGAGGAGGGGACTCGGCTGGGAAGAGCTCCCCTCCCCTCCGCGGAAGACCACTGGGTCCCCTCTTTCCCCA ACCTCCTCCCTCTCTTCTACTCCACCCCTCCGTTTTCCCACTCCCCACTGACTCGGATGCCTGGATGTTCTGCCACCGGGCAGTGGTCCA GCGTGCAGCCGGGAGGGGGCAGGGGCAGGGGGCACTGTGACAGGAAGCTGCGCGCACAAGTTGGCCATTTCGAGGGCAAAATAAGTTCTC CCTTGGATTTGGAAAGGACAAAGCCAGTAAGCTACCTCTTTTGTGTCGGATGAGGAGGACCAACCATGAGCCAGAGCCCGGGTGCAGGCT CACCGCCGCCGCTGCCACCGCGGTCAGCTCCAGTTCCTGCCAGGAGTTGTCGGTGCGAGGAATTTTGTGACAGGCTCTGTTAGTCTGTTC CTCCCTTATTTGAAGGACAGGCCAAAGATCCAGTTTGGAAATGAGAGAGGACTAGCATGACACATTGGCTCCACCATTGATATCTCCCAG AGGTACAGAAACAGGATTCATGAAGATGTTGACAAGACTGCAAGTTCTTACCTTAGCTTTGTTTTCAAAGGGATTTTTACTCTCTTTAGG GGACCATAACTTTCTAAGGAGAGAGATTAAAATAGAAGGTGACCTTGTTTTAGGGGGCCTGTTTCCTATTAACGAAAAAGGCACTGGAAC TGAAGAATGTGGGCGAATCAATGAAGACCGAGGGATTCAACGCCTGGAAGCCATGTTGTTTGCTATTGATGAAATCAACAAAGATGATTA CTTGCTACCAGGAGTGAAGTTGGGTGTTCACATTTTGGATACATGTTCAAGGGATACCTATGCATTGGAGCAATCACTGGAGTTTGTCAG GGCATCTTTGACAAAAGTGGATGAAGCTGAGTATATGTGTCCTGATGGATCCTATGCCATTCAAGAAAACATCCCACTTCTCATTGCAGG GGTCATTGGTGGCTCTTATAGCAGTGTTTCCATACAGGTGGCAAACCTGCTGCGGCTCTTCCAGATCCCTCAGATCAGCTACGCATCCAC CAGCGCCAAACTCAGTGATAAGTCGCGCTATGATTACTTTGCCAGGACCGTGCCCCCCGACTTCTACCAGGCCAAAGCCATGGCTGAGAT CTTGCGCTTCTTCAACTGGACCTACGTGTCCACAGTAGCCTCCGAGGGTGATTACGGGGAGACAGGGATCGAGGCCTTCGAGCAGGAAGC CCGCCTGCGCAACATCTGCATCGCTACGGCGGAGAAGGTGGGCCGCTCCAACATCCGCAAGTCCTACGACAGCGTGATCCGAGAACTGTT GCAGAAGCCCAACGCGCGCGTCGTGGTCCTCTTCATGCGCAGCGACGACTCGCGGGAGCTCATTGCAGCCGCCAGCCGCGCCAATGCCTC CTTCACCTGGGTGGCCAGCGACGGCTGGGGCGCGCAGGAGAGCATCATCAAGGGCAGCGAGCATGTGGCCTACGGCGCCATCACCCTGGA GCTGGCCTCCCAGCCTGTCCGCCAGTTCGACCGCTACTTCCAGAGCCTCAACCCCTACAACAACCACCGCAACCCCTGGTTCCGGGACTT CTGGGAGCAAAAGTTTCAGTGCAGCCTCCAGAACAAACGCAACCACAGGCGCGTCTGCGACAAGCACCTGGCCATCGACAGCAGCAACTA CGAGCAAGAGTCCAAGATCATGTTTGTGGTGAACGCGGTGTATGCCATGGCCCACGCTTTGCACAAAATGCAGCGCACCCTCTGTCCCAA CACTACCAAGCTTTGTGATGCTATGAAGATCCTGGATGGGAAGAAGTTGTACAAGGATTACTTGCTGAAAATCAACTTCACGGCTCCATT CAACCCAAATAAAGATGCAGATAGCATAGTCAAGTTTGACACTTTTGGAGATGGAATGGGGCGATACAACGTGTTCAATTTCCAAAATGT AGGTGGAAAGTATTCCTACTTGAAAGTTGGTCACTGGGCAGAAACCTTATCGCTAGATGTCAACTCTATCCACTGGTCCCGGAACTCAGT CCCCACTTCCCAGTGCAGCGACCCCTGTGCCCCCAATGAAATGAAGAATATGCAACCAGGGGATGTCTGCTGCTGGATTTGCATCCCCTG TGAACCCTACGAATACCTGGCTGATGAGTTTACCTGTATGGATTGTGGGTCTGGACAGTGGCCCACTGCAGACCTAACTGGATGCTATGA CCTTCCTGAGGACTACATCAGGTGGGAAGACGCCTGGGCCATTGGCCCAGTCACCATTGCCTGTCTGGGTTTTATGTGTACATGCATGGT TGTAACTGTTTTTATCAAGCACAACAACACACCCTTGGTCAAAGCATCGGGCCGAGAACTCTGCTACATCTTATTGTTTGGGGTTGGCCT GTCATACTGCATGACATTCTTCTTCATTGCCAAGCCATCACCAGTCATCTGTGCATTGCGCCGACTCGGGCTGGGGAGTTCCTTCGCTAT CTGTTACTCAGCCCTGCTGACCAAGACAAACTGCATTGCCCGCATCTTCGATGGGGTCAAGAATGGCGCTCAGAGGCCAAAATTCATCAG CCCCAGTTCTCAGGTTTTCATCTGCCTGGGTCTGATCCTGGTGCAAATTGTGATGGTGTCTGTGTGGCTCATCCTGGAGGCCCCAGGCAC CAGGAGGTATACCCTTGCAGAGAAGCGGGAAACAGTCATCCTAAAATGCAATGTCAAAGATTCCAGCATGTTGATCTCTCTTACCTACGA TGTGATCCTGGTGATCTTATGCACTGTGTACGCCTTCAAAACGCGGAAGTGCCCAGAAAATTTCAACGAAGCTAAGTTCATAGGTTTTAC CATGTACACCACGTGCATCATCTGGTTGGCCTTCCTCCCTATATTTTATGTGACATCAAGTGACTACAGAGTTGTCACACACAGACTGCA CCTCAACAGGTTCAGTGTCAGTGGAACTGGGACCACATACTCTCAGTCCATCGAGGAGGTGCGTTGCAGCACCGCAGCTCACGCTTTCAA GGTGGCTGCCCGGGCCACGCTGCGCCGCAGCAACGTCTCCCGCAAGCGGTCCAGCAGCCTTGGAGGCTCCACGGGATCCACCCCCTCCTC CTCCATCAGCAGCAAGAGCAACAGCGAAGACCCATTCCCACAGCCCGAGAGGCAGAAGCAGCAGCAGCCGCTGGCCCTAACCCAGCAAGA GCAGCAGCAGCAGCCCCTGACCCTCCCACAGCAGCAACGATCTCAGCAGCAGCCCAGATGCAAGCAGAAGGTCATCTTTGGCAGCGGCAC GGTCACCTTCTCACTGAGCTTTGATGAGCCTCAGAAGAACGCCATGGCCCACAGGAATTCTACGCACCAGAACTCCCTGGAGGCCCAGAA AAGCAGCGATACGCTGACCCGACACGAGCCATTACTCCCGCTGCAGTGCGGGGAAACGGACTTAGATCTGACCGTCCAGGAAACAGGTCT GCAAGGACCTGTGGGTGGAGACCAGCGGCCAGAGGTGGAGGACCCTGAAGAGTTGTCCCCAGCACTTGTAGTGTCCAGTTCACAGAGCTT TGTCATCAGTGGTGGAGGCAGCACTGTTACAGAAAACGTAGTGAATTCATAAAATGGAAGGAGAAGACTGGGCTAGGGAGAATGCAGAGA GGTTTCTTGGGGTCCCAGGGAAGAGGAATCGCCCCAGACTCCTTTCCTCTGAGGAAGAAGGGATAATAGACACATCAAATGCCCCGAATT TAGTCACACCATCTTAAATGACAGTGAATTGACCCATGTTCCCTTTAAAATTAAAAAAAAGAAGAGCCTTGTGTTTCTGTGGTTGCATTT GTCAAAGCATTGAGATCTCCACGGTCAGATTTGCTGTTCACCCACATCTAATGTCTCTTCCTCTGTTCTATCCCACCCAACAGCTCAGAG ATGAAACTATGGCTTTAAACTACCCTCCAGAGTGTGCAGACTGATGGGACATCAAATTTGCCACCACTAGAGCTGAGAGTCTGAAAGACA GAATGTCACCAGTCCTGCCCAATGCCTTGACAACAGACTGAATTTTAAATGTTCACAACATAAGGAGAATGTATCTCCTCCTATTTATGA AAACCATATGATATTTTGTCTCCTACCTGCTGCTGCTATTATGTAACATCCAGAAGGTTTGCACCCCTCCTATACCATATGTCTGCTTCT GTCCAGGACATGATACTGATGCCATGTTTAGATTCCAGGATCACAAGAATCACCTCAAATTGTTAGGAAGGGACTGCATAAACCAATGAG CTGTATCTGTAATTAATATTCCTATATGTAGCTTTATCCTTAGGAAAATGCTTCTGTTGTAATAGTCCATGGACAATATAAACTGAAAAA TGTCAGTCTGGTTTATATAAGGCAGTATTATTGAGCTCTATTTCCCCACCCCACTATCCTCACTCCCATAAGCTAAGCCTTATGTGAGCC CCTTCAGGGACTCAAGGGTCCAGAAGTCCCTCCCATCTCTACCCCAAAGAATTCCTGAAGCCAGATCCACCCTATCCCTGTACAGAGTAA GTTCTCAATTATTGGCCTGCTAATAGCTGCTAGGGTAGGAAAGCGTGGTTCCAAGAAAGATCCACCCTCAAATGTCAGAGCTATGTTCCC TCCAGCAGTGGTATTAATACTGCCGGTCACCCAGGCTCTGGAGCCAGAGAGACAGACCGGGGTTCAAGCCATGGCTTCGTCATTTGCAAG CTGAGTGACTGTAGGCAGGGAACCTTAACCTCTCTAAGCCACAGCTTCTTCATCTTTAAAATAAGGATAATAATCATTCTTTCCCCTCAG AGCTCTTATGTGGATTAAACGAGATAATGTATATAAAGTACTTTAGCCTGGTACCTAGCACACAATAAGCATTCAATAAATATTAGTTAA >34900_34900_1_GRM3-CASR_GRM3_chr7_86479793_ENST00000361669_CASR_chr3_122003403_ENST00000498619_length(amino acids)=1045AA_BP=769 MAPPLISPRGTETGFMKMLTRLQVLTLALFSKGFLLSLGDHNFLRREIKIEGDLVLGGLFPINEKGTGTEECGRINEDRGIQRLEAMLFA IDEINKDDYLLPGVKLGVHILDTCSRDTYALEQSLEFVRASLTKVDEAEYMCPDGSYAIQENIPLLIAGVIGGSYSSVSIQVANLLRLFQ IPQISYASTSAKLSDKSRYDYFARTVPPDFYQAKAMAEILRFFNWTYVSTVASEGDYGETGIEAFEQEARLRNICIATAEKVGRSNIRKS YDSVIRELLQKPNARVVVLFMRSDDSRELIAAASRANASFTWVASDGWGAQESIIKGSEHVAYGAITLELASQPVRQFDRYFQSLNPYNN HRNPWFRDFWEQKFQCSLQNKRNHRRVCDKHLAIDSSNYEQESKIMFVVNAVYAMAHALHKMQRTLCPNTTKLCDAMKILDGKKLYKDYL LKINFTAPFNPNKDADSIVKFDTFGDGMGRYNVFNFQNVGGKYSYLKVGHWAETLSLDVNSIHWSRNSVPTSQCSDPCAPNEMKNMQPGD VCCWICIPCEPYEYLADEFTCMDCGSGQWPTADLTGCYDLPEDYIRWEDAWAIGPVTIACLGFMCTCMVVTVFIKHNNTPLVKASGRELC YILLFGVGLSYCMTFFFIAKPSPVICALRRLGLGSSFAICYSALLTKTNCIARIFDGVKNGAQRPKFISPSSQVFICLGLILVQIVMVSV WLILEAPGTRRYTLAEKRETVILKCNVKDSSMLISLTYDVILVILCTVYAFKTRKCPENFNEAKFIGFTMYTTCIIWLAFLPIFYVTSSD YRVVTHRLHLNRFSVSGTGTTYSQSIEEVRCSTAAHAFKVAARATLRRSNVSRKRSSSLGGSTGSTPSSSISSKSNSEDPFPQPERQKQQ QPLALTQQEQQQQPLTLPQQQRSQQQPRCKQKVIFGSGTVTFSLSFDEPQKNAMAHRNSTHQNSLEAQKSSDTLTRHEPLLPLQCGETDL -------------------------------------------------------------- >34900_34900_2_GRM3-CASR_GRM3_chr7_86479793_ENST00000536043_CASR_chr3_122003403_ENST00000498619_length(transcript)=4166nt_BP=2227nt GTGCAGCCGGGAGGGGGCAGGGGCAGGGGGCACTGTGACAGGAAGCTGCGCGCACAAGTTGGCCATTTCGAGGGCAAAATAAGTTCTCCC TTGGATTTGGAAAGGACAAAGCCAGTAAGCTACCTCTTTTGTGTCGGATGAGGAGGACCAACCATGAGCCAGAGCCCGGGTGCAGGCTCA CCGCCGCCGCTGCCACCGCGGTCAGCTCCAGTTCCTGCCAGGAGTTGTCGGTGCGAGGTGGCAAACCTGCTGCGGCTCTTCCAGATCCCT CAGATCAGCTACGCATCCACCAGCGCCAAACTCAGTGATAAGTCGCGCTATGATTACTTTGCCAGGACCGTGCCCCCCGACTTCTACCAG GCCAAAGCCATGGCTGAGATCTTGCGCTTCTTCAACTGGACCTACGTGTCCACAGTAGCCTCCGAGGGTGATTACGGGGAGACAGGGATC GAGGCCTTCGAGCAGGAAGCCCGCCTGCGCAACATCTGCATCGCTACGGCGGAGAAGGTGGGCCGCTCCAACATCCGCAAGTCCTACGAC AGCGTGATCCGAGAACTGTTGCAGAAGCCCAACGCGCGCGTCGTGGTCCTCTTCATGCGCAGCGACGACTCGCGGGAGCTCATTGCAGCC GCCAGCCGCGCCAATGCCTCCTTCACCTGGGTGGCCAGCGACGGCTGGGGCGCGCAGGAGAGCATCATCAAGGGCAGCGAGCATGTGGCC TACGGCGCCATCACCCTGGAGCTGGCCTCCCAGCCTGTCCGCCAGTTCGACCGCTACTTCCAGAGCCTCAACCCCTACAACAACCACCGC AACCCCTGGTTCCGGGACTTCTGGGAGCAAAAGTTTCAGTGCAGCCTCCAGAACAAACGCAACCACAGGCGCGTCTGCGACAAGCACCTG GCCATCGACAGCAGCAACTACGAGCAAGAGTCCAAGATCATGTTTGTGGTGAACGCGGTGTATGCCATGGCCCACGCTTTGCACAAAATG CAGCGCACCCTCTGTCCCAACACTACCAAGCTTTGTGATGCTATGAAGATCCTGGATGGGAAGAAGTTGTACAAGGATTACTTGCTGAAA ATCAACTTCACGGCTCCATTCAACCCAAATAAAGATGCAGATAGCATAGTCAAGTTTGACACTTTTGGAGATGGAATGGGGCGATACAAC GTGTTCAATTTCCAAAATGTAGGTGGAAAGTATTCCTACTTGAAAGTTGGTCACTGGGCAGAAACCTTATCGCTAGATGTCAACTCTATC CACTGGTCCCGGAACTCAGTCCCCACTTCCCAGTGCAGCGACCCCTGTGCCCCCAATGAAATGAAGAATATGCAACCAGGGGATGTCTGC TGCTGGATTTGCATCCCCTGTGAACCCTACGAATACCTGGCTGATGAGTTTACCTGTATGGATTGTGGGTCTGGACAGTGGCCCACTGCA GACCTAACTGGATGCTATGACCTTCCTGAGGACTACATCAGGTGGGAAGACGCCTGGGCCATTGGCCCAGTCACCATTGCCTGTCTGGGT TTTATGTGTACATGCATGGTTGTAACTGTTTTTATCAAGCACAACAACACACCCTTGGTCAAAGCATCGGGCCGAGAACTCTGCTACATC TTATTGTTTGGGGTTGGCCTGTCATACTGCATGACATTCTTCTTCATTGCCAAGCCATCACCAGTCATCTGTGCATTGCGCCGACTCGGG CTGGGGAGTTCCTTCGCTATCTGTTACTCAGCCCTGCTGACCAAGACAAACTGCATTGCCCGCATCTTCGATGGGGTCAAGAATGGCGCT CAGAGGCCAAAATTCATCAGCCCCAGTTCTCAGGTTTTCATCTGCCTGGGTCTGATCCTGGTGCAAATTGTGATGGTGTCTGTGTGGCTC ATCCTGGAGGCCCCAGGCACCAGGAGGTATACCCTTGCAGAGAAGCGGGAAACAGTCATCCTAAAATGCAATGTCAAAGATTCCAGCATG TTGATCTCTCTTACCTACGATGTGATCCTGGTGATCTTATGCACTGTGTACGCCTTCAAAACGCGGAAGTGCCCAGAAAATTTCAACGAA GCTAAGTTCATAGGTTTTACCATGTACACCACGTGCATCATCTGGTTGGCCTTCCTCCCTATATTTTATGTGACATCAAGTGACTACAGA GTTGTCACACACAGACTGCACCTCAACAGGTTCAGTGTCAGTGGAACTGGGACCACATACTCTCAGTCCATCGAGGAGGTGCGTTGCAGC ACCGCAGCTCACGCTTTCAAGGTGGCTGCCCGGGCCACGCTGCGCCGCAGCAACGTCTCCCGCAAGCGGTCCAGCAGCCTTGGAGGCTCC ACGGGATCCACCCCCTCCTCCTCCATCAGCAGCAAGAGCAACAGCGAAGACCCATTCCCACAGCCCGAGAGGCAGAAGCAGCAGCAGCCG CTGGCCCTAACCCAGCAAGAGCAGCAGCAGCAGCCCCTGACCCTCCCACAGCAGCAACGATCTCAGCAGCAGCCCAGATGCAAGCAGAAG GTCATCTTTGGCAGCGGCACGGTCACCTTCTCACTGAGCTTTGATGAGCCTCAGAAGAACGCCATGGCCCACAGGAATTCTACGCACCAG AACTCCCTGGAGGCCCAGAAAAGCAGCGATACGCTGACCCGACACGAGCCATTACTCCCGCTGCAGTGCGGGGAAACGGACTTAGATCTG ACCGTCCAGGAAACAGGTCTGCAAGGACCTGTGGGTGGAGACCAGCGGCCAGAGGTGGAGGACCCTGAAGAGTTGTCCCCAGCACTTGTA GTGTCCAGTTCACAGAGCTTTGTCATCAGTGGTGGAGGCAGCACTGTTACAGAAAACGTAGTGAATTCATAAAATGGAAGGAGAAGACTG GGCTAGGGAGAATGCAGAGAGGTTTCTTGGGGTCCCAGGGAAGAGGAATCGCCCCAGACTCCTTTCCTCTGAGGAAGAAGGGATAATAGA CACATCAAATGCCCCGAATTTAGTCACACCATCTTAAATGACAGTGAATTGACCCATGTTCCCTTTAAAATTAAAAAAAAGAAGAGCCTT GTGTTTCTGTGGTTGCATTTGTCAAAGCATTGAGATCTCCACGGTCAGATTTGCTGTTCACCCACATCTAATGTCTCTTCCTCTGTTCTA TCCCACCCAACAGCTCAGAGATGAAACTATGGCTTTAAACTACCCTCCAGAGTGTGCAGACTGATGGGACATCAAATTTGCCACCACTAG AGCTGAGAGTCTGAAAGACAGAATGTCACCAGTCCTGCCCAATGCCTTGACAACAGACTGAATTTTAAATGTTCACAACATAAGGAGAAT GTATCTCCTCCTATTTATGAAAACCATATGATATTTTGTCTCCTACCTGCTGCTGCTATTATGTAACATCCAGAAGGTTTGCACCCCTCC TATACCATATGTCTGCTTCTGTCCAGGACATGATACTGATGCCATGTTTAGATTCCAGGATCACAAGAATCACCTCAAATTGTTAGGAAG GGACTGCATAAACCAATGAGCTGTATCTGTAATTAATATTCCTATATGTAGCTTTATCCTTAGGAAAATGCTTCTGTTGTAATAGTCCAT GGACAATATAAACTGAAAAATGTCAGTCTGGTTTATATAAGGCAGTATTATTGAGCTCTATTTCCCCACCCCACTATCCTCACTCCCATA AGCTAAGCCTTATGTGAGCCCCTTCAGGGACTCAAGGGTCCAGAAGTCCCTCCCATCTCTACCCCAAAGAATTCCTGAAGCCAGATCCAC CCTATCCCTGTACAGAGTAAGTTCTCAATTATTGGCCTGCTAATAGCTGCTAGGGTAGGAAAGCGTGGTTCCAAGAAAGATCCACCCTCA AATGTCAGAGCTATGTTCCCTCCAGCAGTGGTATTAATACTGCCGGTCACCCAGGCTCTGGAGCCAGAGAGACAGACCGGGGTTCAAGCC ATGGCTTCGTCATTTGCAAGCTGAGTGACTGTAGGCAGGGAACCTTAACCTCTCTAAGCCACAGCTTCTTCATCTTTAAAATAAGGATAA TAATCATTCTTTCCCCTCAGAGCTCTTATGTGGATTAAACGAGATAATGTATATAAAGTACTTTAGCCTGGTACCTAGCACACAATAAGC >34900_34900_2_GRM3-CASR_GRM3_chr7_86479793_ENST00000536043_CASR_chr3_122003403_ENST00000498619_length(amino acids)=902AA_BP=626 MSQSPGAGSPPPLPPRSAPVPARSCRCEVANLLRLFQIPQISYASTSAKLSDKSRYDYFARTVPPDFYQAKAMAEILRFFNWTYVSTVAS EGDYGETGIEAFEQEARLRNICIATAEKVGRSNIRKSYDSVIRELLQKPNARVVVLFMRSDDSRELIAAASRANASFTWVASDGWGAQES IIKGSEHVAYGAITLELASQPVRQFDRYFQSLNPYNNHRNPWFRDFWEQKFQCSLQNKRNHRRVCDKHLAIDSSNYEQESKIMFVVNAVY AMAHALHKMQRTLCPNTTKLCDAMKILDGKKLYKDYLLKINFTAPFNPNKDADSIVKFDTFGDGMGRYNVFNFQNVGGKYSYLKVGHWAE TLSLDVNSIHWSRNSVPTSQCSDPCAPNEMKNMQPGDVCCWICIPCEPYEYLADEFTCMDCGSGQWPTADLTGCYDLPEDYIRWEDAWAI GPVTIACLGFMCTCMVVTVFIKHNNTPLVKASGRELCYILLFGVGLSYCMTFFFIAKPSPVICALRRLGLGSSFAICYSALLTKTNCIAR IFDGVKNGAQRPKFISPSSQVFICLGLILVQIVMVSVWLILEAPGTRRYTLAEKRETVILKCNVKDSSMLISLTYDVILVILCTVYAFKT RKCPENFNEAKFIGFTMYTTCIIWLAFLPIFYVTSSDYRVVTHRLHLNRFSVSGTGTTYSQSIEEVRCSTAAHAFKVAARATLRRSNVSR KRSSSLGGSTGSTPSSSISSKSNSEDPFPQPERQKQQQPLALTQQEQQQQPLTLPQQQRSQQQPRCKQKVIFGSGTVTFSLSFDEPQKNA MAHRNSTHQNSLEAQKSSDTLTRHEPLLPLQCGETDLDLTVQETGLQGPVGGDQRPEVEDPEELSPALVVSSSQSFVISGGGSTVTENVV -------------------------------------------------------------- >34900_34900_3_GRM3-CASR_GRM3_chr7_86479793_ENST00000546348_CASR_chr3_122003403_ENST00000498619_length(transcript)=3556nt_BP=1617nt AATTAGATGCGACGGCTTCAGCCTGGTCAAGGTGAAGGAAAGTTGCTTCCGCGCCTAGGAAGTGGGTTTGCCTGATAAGAGAAGGAGGAG GGGACTCGGCTGGGAAGAGCTCCCCTCCCCTCCGCGGAAGACCACTGGGTCCCCTCTTTCCCCAACCTCCTCCCTCTCTTCTACTCCACC CCTCCGTTTTCCCACTCCCCACTGACTCGGATGCCTGGATGTTCTGCCACCGGGCAGTGGTCCAGCGTGCAGCCGGGAGGGGGCAGGGGC AGGGGGCACTGTGACAGGAAGCTGCGCGCACAAGTTGGCCATTTCGAGGGCAAAATAAGTTCTCCCTTGGATTTGGAAAGGACAAAGCCA GTAAGCTACCTCTTTTGTGTCGGATGAGGAGGACCAACCATGAGCCAGAGCCCGGGTGCAGGCTCACCGCCGCCGCTGCCACCGCGGTCA GCTCCAGTTCCTGCCAGGAGTTGTCGGTGCGAGCTCCATTCAACCCAAATAAAGATGCAGATAGCATAGTCAAGTTTGACACTTTTGGAG ATGGAATGGGGCGATACAACGTGTTCAATTTCCAAAATGTAGGTGGAAAGTATTCCTACTTGAAAGTTGGTCACTGGGCAGAAACCTTAT CGCTAGATGTCAACTCTATCCACTGGTCCCGGAACTCAGTCCCCACTTCCCAGTGCAGCGACCCCTGTGCCCCCAATGAAATGAAGAATA TGCAACCAGGGGATGTCTGCTGCTGGATTTGCATCCCCTGTGAACCCTACGAATACCTGGCTGATGAGTTTACCTGTATGGATTGTGGGT CTGGACAGTGGCCCACTGCAGACCTAACTGGATGCTATGACCTTCCTGAGGACTACATCAGGTGGGAAGACGCCTGGGCCATTGGCCCAG TCACCATTGCCTGTCTGGGTTTTATGTGTACATGCATGGTTGTAACTGTTTTTATCAAGCACAACAACACACCCTTGGTCAAAGCATCGG GCCGAGAACTCTGCTACATCTTATTGTTTGGGGTTGGCCTGTCATACTGCATGACATTCTTCTTCATTGCCAAGCCATCACCAGTCATCT GTGCATTGCGCCGACTCGGGCTGGGGAGTTCCTTCGCTATCTGTTACTCAGCCCTGCTGACCAAGACAAACTGCATTGCCCGCATCTTCG ATGGGGTCAAGAATGGCGCTCAGAGGCCAAAATTCATCAGCCCCAGTTCTCAGGTTTTCATCTGCCTGGGTCTGATCCTGGTGCAAATTG TGATGGTGTCTGTGTGGCTCATCCTGGAGGCCCCAGGCACCAGGAGGTATACCCTTGCAGAGAAGCGGGAAACAGTCATCCTAAAATGCA ATGTCAAAGATTCCAGCATGTTGATCTCTCTTACCTACGATGTGATCCTGGTGATCTTATGCACTGTGTACGCCTTCAAAACGCGGAAGT GCCCAGAAAATTTCAACGAAGCTAAGTTCATAGGTTTTACCATGTACACCACGTGCATCATCTGGTTGGCCTTCCTCCCTATATTTTATG TGACATCAAGTGACTACAGAGTTGTCACACACAGACTGCACCTCAACAGGTTCAGTGTCAGTGGAACTGGGACCACATACTCTCAGTCCA TCGAGGAGGTGCGTTGCAGCACCGCAGCTCACGCTTTCAAGGTGGCTGCCCGGGCCACGCTGCGCCGCAGCAACGTCTCCCGCAAGCGGT CCAGCAGCCTTGGAGGCTCCACGGGATCCACCCCCTCCTCCTCCATCAGCAGCAAGAGCAACAGCGAAGACCCATTCCCACAGCCCGAGA GGCAGAAGCAGCAGCAGCCGCTGGCCCTAACCCAGCAAGAGCAGCAGCAGCAGCCCCTGACCCTCCCACAGCAGCAACGATCTCAGCAGC AGCCCAGATGCAAGCAGAAGGTCATCTTTGGCAGCGGCACGGTCACCTTCTCACTGAGCTTTGATGAGCCTCAGAAGAACGCCATGGCCC ACAGGAATTCTACGCACCAGAACTCCCTGGAGGCCCAGAAAAGCAGCGATACGCTGACCCGACACGAGCCATTACTCCCGCTGCAGTGCG GGGAAACGGACTTAGATCTGACCGTCCAGGAAACAGGTCTGCAAGGACCTGTGGGTGGAGACCAGCGGCCAGAGGTGGAGGACCCTGAAG AGTTGTCCCCAGCACTTGTAGTGTCCAGTTCACAGAGCTTTGTCATCAGTGGTGGAGGCAGCACTGTTACAGAAAACGTAGTGAATTCAT AAAATGGAAGGAGAAGACTGGGCTAGGGAGAATGCAGAGAGGTTTCTTGGGGTCCCAGGGAAGAGGAATCGCCCCAGACTCCTTTCCTCT GAGGAAGAAGGGATAATAGACACATCAAATGCCCCGAATTTAGTCACACCATCTTAAATGACAGTGAATTGACCCATGTTCCCTTTAAAA TTAAAAAAAAGAAGAGCCTTGTGTTTCTGTGGTTGCATTTGTCAAAGCATTGAGATCTCCACGGTCAGATTTGCTGTTCACCCACATCTA ATGTCTCTTCCTCTGTTCTATCCCACCCAACAGCTCAGAGATGAAACTATGGCTTTAAACTACCCTCCAGAGTGTGCAGACTGATGGGAC ATCAAATTTGCCACCACTAGAGCTGAGAGTCTGAAAGACAGAATGTCACCAGTCCTGCCCAATGCCTTGACAACAGACTGAATTTTAAAT GTTCACAACATAAGGAGAATGTATCTCCTCCTATTTATGAAAACCATATGATATTTTGTCTCCTACCTGCTGCTGCTATTATGTAACATC CAGAAGGTTTGCACCCCTCCTATACCATATGTCTGCTTCTGTCCAGGACATGATACTGATGCCATGTTTAGATTCCAGGATCACAAGAAT CACCTCAAATTGTTAGGAAGGGACTGCATAAACCAATGAGCTGTATCTGTAATTAATATTCCTATATGTAGCTTTATCCTTAGGAAAATG CTTCTGTTGTAATAGTCCATGGACAATATAAACTGAAAAATGTCAGTCTGGTTTATATAAGGCAGTATTATTGAGCTCTATTTCCCCACC CCACTATCCTCACTCCCATAAGCTAAGCCTTATGTGAGCCCCTTCAGGGACTCAAGGGTCCAGAAGTCCCTCCCATCTCTACCCCAAAGA ATTCCTGAAGCCAGATCCACCCTATCCCTGTACAGAGTAAGTTCTCAATTATTGGCCTGCTAATAGCTGCTAGGGTAGGAAAGCGTGGTT CCAAGAAAGATCCACCCTCAAATGTCAGAGCTATGTTCCCTCCAGCAGTGGTATTAATACTGCCGGTCACCCAGGCTCTGGAGCCAGAGA GACAGACCGGGGTTCAAGCCATGGCTTCGTCATTTGCAAGCTGAGTGACTGTAGGCAGGGAACCTTAACCTCTCTAAGCCACAGCTTCTT CATCTTTAAAATAAGGATAATAATCATTCTTTCCCCTCAGAGCTCTTATGTGGATTAAACGAGATAATGTATATAAAGTACTTTAGCCTG >34900_34900_3_GRM3-CASR_GRM3_chr7_86479793_ENST00000546348_CASR_chr3_122003403_ENST00000498619_length(amino acids)=625AA_BP=349 MCRMRRTNHEPEPGCRLTAAAATAVSSSSCQELSVRAPFNPNKDADSIVKFDTFGDGMGRYNVFNFQNVGGKYSYLKVGHWAETLSLDVN SIHWSRNSVPTSQCSDPCAPNEMKNMQPGDVCCWICIPCEPYEYLADEFTCMDCGSGQWPTADLTGCYDLPEDYIRWEDAWAIGPVTIAC LGFMCTCMVVTVFIKHNNTPLVKASGRELCYILLFGVGLSYCMTFFFIAKPSPVICALRRLGLGSSFAICYSALLTKTNCIARIFDGVKN GAQRPKFISPSSQVFICLGLILVQIVMVSVWLILEAPGTRRYTLAEKRETVILKCNVKDSSMLISLTYDVILVILCTVYAFKTRKCPENF NEAKFIGFTMYTTCIIWLAFLPIFYVTSSDYRVVTHRLHLNRFSVSGTGTTYSQSIEEVRCSTAAHAFKVAARATLRRSNVSRKRSSSLG GSTGSTPSSSISSKSNSEDPFPQPERQKQQQPLALTQQEQQQQPLTLPQQQRSQQQPRCKQKVIFGSGTVTFSLSFDEPQKNAMAHRNST -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for GRM3-CASR |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
| Tgene | CASR | chr7:86479793 | chr3:122003403 | ENST00000296154 | 0 | 6 | 880_900 | 0 | 1079.0 | RNF19A | |
| Tgene | CASR | chr7:86479793 | chr3:122003403 | ENST00000490131 | 0 | 7 | 880_900 | 0 | 1079.0 | RNF19A |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for GRM3-CASR |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for GRM3-CASR |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
