
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:GRTP1-COL4A2 (FusionGDB2 ID:34961) |
Fusion Gene Summary for GRTP1-COL4A2 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: GRTP1-COL4A2 | Fusion gene ID: 34961 | Hgene | Tgene | Gene symbol | GRTP1 | COL4A2 | Gene ID | 79774 | 1284 |
| Gene name | growth hormone regulated TBC protein 1 | collagen type IV alpha 2 chain | |
| Synonyms | TBC1D6 | BSVD2|ICH|POREN2 | |
| Cytomap | 13q34 | 13q34 | |
| Type of gene | protein-coding | protein-coding | |
| Description | growth hormone-regulated TBC protein 1TBC1 domain family member 6 | collagen alpha-2(IV) chaincanstatin | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | Q5TC63 | P08572 | |
| Ensembl transtripts involved in fusion gene | ENST00000326039, ENST00000375430, ENST00000375431, ENST00000476439, | ENST00000462309, ENST00000360467, | |
| Fusion gene scores | * DoF score | 8 X 10 X 5=400 | 9 X 10 X 5=450 |
| # samples | 12 | 8 | |
| ** MAII score | log2(12/400*10)=-1.73696559416621 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(8/450*10)=-2.49185309632967 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: GRTP1 [Title/Abstract] AND COL4A2 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | GRTP1(113999178)-COL4A2(111154015), # samples:3 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Tgene | COL4A2 | GO:0016525 | negative regulation of angiogenesis | 10625665 |
 Fusion gene breakpoints across GRTP1 (5'-gene) Fusion gene breakpoints across GRTP1 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
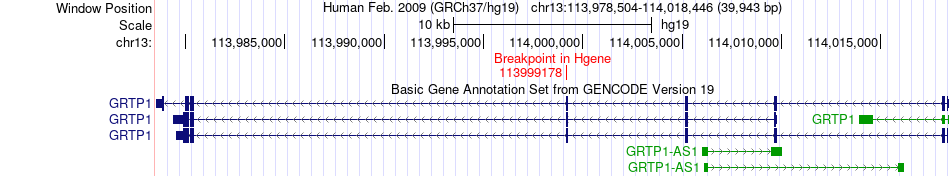 |
 Fusion gene breakpoints across COL4A2 (3'-gene) Fusion gene breakpoints across COL4A2 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
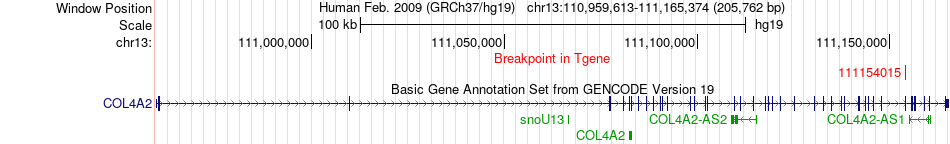 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | LUAD | TCGA-69-7979-01A | GRTP1 | chr13 | 113999178 | - | COL4A2 | chr13 | 111154015 | + |
Top |
Fusion Gene ORF analysis for GRTP1-COL4A2 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-intron | ENST00000326039 | ENST00000462309 | GRTP1 | chr13 | 113999178 | - | COL4A2 | chr13 | 111154015 | + |
| 5CDS-intron | ENST00000375430 | ENST00000462309 | GRTP1 | chr13 | 113999178 | - | COL4A2 | chr13 | 111154015 | + |
| 5CDS-intron | ENST00000375431 | ENST00000462309 | GRTP1 | chr13 | 113999178 | - | COL4A2 | chr13 | 111154015 | + |
| In-frame | ENST00000326039 | ENST00000360467 | GRTP1 | chr13 | 113999178 | - | COL4A2 | chr13 | 111154015 | + |
| In-frame | ENST00000375430 | ENST00000360467 | GRTP1 | chr13 | 113999178 | - | COL4A2 | chr13 | 111154015 | + |
| In-frame | ENST00000375431 | ENST00000360467 | GRTP1 | chr13 | 113999178 | - | COL4A2 | chr13 | 111154015 | + |
| intron-3CDS | ENST00000476439 | ENST00000360467 | GRTP1 | chr13 | 113999178 | - | COL4A2 | chr13 | 111154015 | + |
| intron-intron | ENST00000476439 | ENST00000462309 | GRTP1 | chr13 | 113999178 | - | COL4A2 | chr13 | 111154015 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000375431 | GRTP1 | chr13 | 113999178 | - | ENST00000360467 | COL4A2 | chr13 | 111154015 | + | 2852 | 637 | 42 | 2015 | 657 |
| ENST00000326039 | GRTP1 | chr13 | 113999178 | - | ENST00000360467 | COL4A2 | chr13 | 111154015 | + | 2551 | 336 | 8 | 1714 | 568 |
| ENST00000375430 | GRTP1 | chr13 | 113999178 | - | ENST00000360467 | COL4A2 | chr13 | 111154015 | + | 2824 | 609 | 14 | 1987 | 657 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000375431 | ENST00000360467 | GRTP1 | chr13 | 113999178 | - | COL4A2 | chr13 | 111154015 | + | 0.00778658 | 0.9922134 |
| ENST00000326039 | ENST00000360467 | GRTP1 | chr13 | 113999178 | - | COL4A2 | chr13 | 111154015 | + | 0.006187174 | 0.9938128 |
| ENST00000375430 | ENST00000360467 | GRTP1 | chr13 | 113999178 | - | COL4A2 | chr13 | 111154015 | + | 0.007714164 | 0.99228585 |
Top |
Fusion Genomic Features for GRTP1-COL4A2 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| GRTP1 | chr13 | 113999177 | - | COL4A2 | chr13 | 111154014 | + | 0.00336775 | 0.9966323 |
| GRTP1 | chr13 | 113999177 | - | COL4A2 | chr13 | 111154014 | + | 0.00336775 | 0.9966323 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
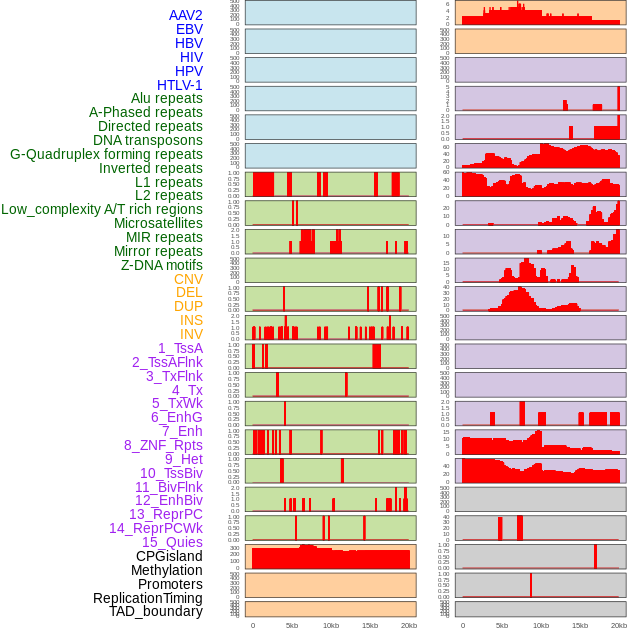 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
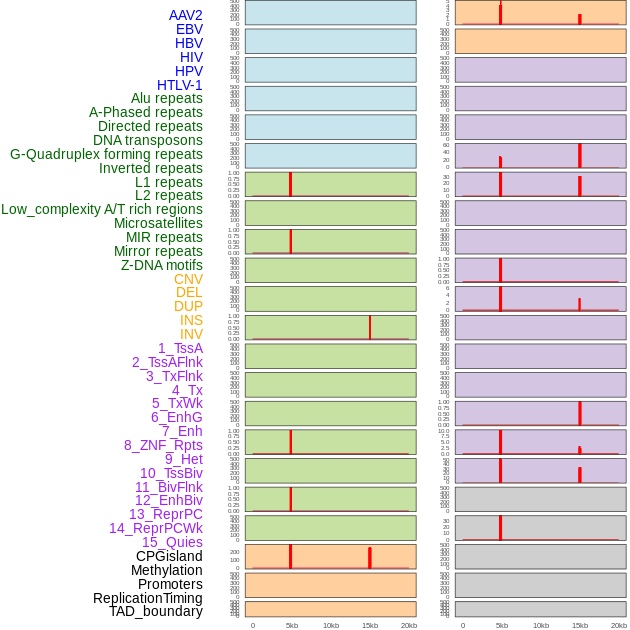 |
Top |
Fusion Protein Features for GRTP1-COL4A2 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr13:113999178/chr13:111154015) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| GRTP1 | COL4A2 |
| FUNCTION: May act as a GTPase-activating protein for Rab family protein(s). | FUNCTION: Type IV collagen is the major structural component of glomerular basement membranes (GBM), forming a 'chicken-wire' meshwork together with laminins, proteoglycans and entactin/nidogen.; FUNCTION: Canstatin, a cleavage product corresponding to the collagen alpha 2(IV) NC1 domain, possesses both anti-angiogenic and anti-tumor cell activity. It inhibits proliferation and migration of endothelial cells, reduces mitochondrial membrane potential, and induces apoptosis. Specifically induces Fas-dependent apoptosis and activates procaspase-8 and -9 activity. Ligand for alphavbeta3 and alphavbeta5 integrins. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | COL4A2 | chr13:113999178 | chr13:111154015 | ENST00000360467 | 39 | 48 | 1489_1712 | 1253 | 1713.0 | Domain | Collagen IV NC1 |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | GRTP1 | chr13:113999178 | chr13:111154015 | ENST00000326039 | - | 3 | 5 | 68_258 | 109 | 267.0 | Domain | Rab-GAP TBC |
| Hgene | GRTP1 | chr13:113999178 | chr13:111154015 | ENST00000375431 | - | 5 | 8 | 68_258 | 187 | 337.0 | Domain | Rab-GAP TBC |
| Tgene | COL4A2 | chr13:113999178 | chr13:111154015 | ENST00000360467 | 39 | 48 | 184_1484 | 1253 | 1713.0 | Region | Note=Triple-helical region |
Top |
Fusion Gene Sequence for GRTP1-COL4A2 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >34961_34961_1_GRTP1-COL4A2_GRTP1_chr13_113999178_ENST00000326039_COL4A2_chr13_111154015_ENST00000360467_length(transcript)=2551nt_BP=336nt GCGTCTGGATGGTGCTGAGTGGGGCCCAGGCGCAGATGGACCAGAATCCCGGCTACTACCACCAGCTTCTCCAGGGAGAGAGAAACCCCA GGCTGGAGGACGCCATCAGGACAGACCTGAACCGGACCTTCCCCGACAACGTGAAGTTCCGGAAGACCACGGACCCCTGCTTACAGAGGA CCCTGTACAATGTGCTGCTGGCATATGGGCACCATAACCAGGGAGTGGGCTACTGCCAGGGAATGAATTTTATAGCAGGATATCTGATTC TTATAACAAATAATGAAGAAGAATCTTTTTGGCTGTTAGATGCTCTTGTTGGAAGAATACTACCAGGGGAACGAGGCCCACCTGGGAGCC CAGGACTTCAGGGGTTCCCTGGTATCACACCCCCTTCCAACATCTCTGGGGCACCTGGTGACAAAGGGGCGCCAGGGATATTTGGCCTGA AAGGTTATCGGGGCCCACCAGGGCCACCAGGTTCTGCTGCTCTTCCTGGAAGCAAAGGTGACACAGGGAACCCAGGAGCTCCAGGAACCC CAGGGACCAAAGGATGGGCCGGGGACTCCGGGCCCCAGGGCAGGCCTGGTGTGTTTGGTCTCCCAGGAGAAAAAGGGCCCAGGGGTGAAC AAGGCTTCATGGGGAACACTGGACCCACTGGGGCGGTGGGCGACAGAGGCCCCAAGGGACCCAAGGGAGACCCAGGATTCCCTGGTGCCC CCGGGACTGTGGGAGCCCCCGGGATTGCAGGAATCCCCCAGAAGATTGCCGTCCAACCAGGGACAGTGGGTCCCCAGGGGAGGCGAGGCC CCCCTGGGGCACCGGGGGAGATGGGGCCCCAGGGCCCCCCCGGAGAACCAGGTTTCCGTGGGGCTCCAGGGAAAGCTGGGCCCCAAGGAA GAGGTGGTGTGTCTGCTGTTCCCGGCTTCCGGGGAGATGAAGGACCCATAGGCCACCAGGGGCCGATTGGCCAAGAAGGTGCACCAGGCC GTCCAGGGAGCCCGGGCCTGCCGGGTATGCCAGGCCGCAGCGTCAGCATCGGCTACCTCCTGGTGAAGCACAGCCAGACGGACCAGGAGC CCATGTGCCCAGTGGGCATGAACAAACTCTGGAGTGGATACAGCCTGCTGTACTTCGAGGGCCAGGAGAAGGCGCACAACCAGGACCTGG GGCTGGCGGGCTCCTGCCTGGCGCGGTTCAGCACCATGCCCTTCCTGTACTGCAACCCTGGTGATGTCTGCTACTATGCCAGCCGGAACG ACAAGTCCTACTGGCTCTCTACCACTGCGCCGCTGCCCATGATGCCCGTGGCCGAGGACGAGATCAAGCCCTACATCAGCCGCTGTTCTG TGTGTGAGGCCCCGGCCATCGCCATCGCGGTCCACAGTCAGGATGTCTCCATCCCACACTGCCCAGCTGGGTGGCGGAGTTTGTGGATCG GATATTCCTTCCTCATGCACACGGCGGCGGGAGACGAAGGCGGTGGCCAATCACTGGTGTCACCGGGCAGCTGTCTAGAGGACTTCCGCG CCACACCATTCATCGAATGCAATGGAGGCCGCGGCACCTGCCACTACTACGCCAACAAGTACAGCTTCTGGCTGACCACCATTCCCGAGC AGAGCTTCCAGGGCTCGCCCTCCGCCGACACGCTCAAGGCCGGCCTCATCCGCACACACATCAGCCGCTGCCAGGTGTGCATGAAGAACC TGTGAGCCGGCGCGTGCCAGGAAGGGCCATTTTGGTGCTTATTCTTAACTTATTACCTCAGGTGCCAACCCAAAAATTGGTTTTATTTTT TTCTTAAAAAAAAAAAAGTCTACCAAAGGAATTTGCATCCAGCAGCAGCACTTAGACCTGCCAGCCACTGTCACCGAGCGGGTGCAAGCA CTCGGGGTCCCTGGAGGGCAAGCCCTGCCCACAGAAAGCCAGGAGCAGCCCTGGCCCCCATCAGCCCTGCTAGACGCACCGCCTGAAGGC ACAGCTAACCACTTCGCACACACCCATGTAACCACTGCACTTTCCAATGCCACAGACAACTCACATTGTTCAACTCCCTTCTCGGGGTGG GACAGACGAGACAACAGCACACAGGCAGCCAGCCGTGGCCAGAGGCTCGAGGGGCTCAGGGCCTCAGGCACCCGTCCCCACACGAGGGCC CCGTGGGTGGGCCTGGCCCTGCTTTCTACGCCAATGTTATGCCAGCTCCATGTTCTCCCAAATACCGTTGATGTGAATTATTTTAAAGGC AAAACCGTGCTCTTTATTTTAAAAAACACTGATAATCACACTGCGGTAGGTCATTCTTTTGCCACATCCCTATAGACCACTGGGTTTGGC AAAACTCAGGCAGAAGTGGAGACCTTTCTAGACATCATTGTCAGCCTTGCTACTTGAAGGTACACCCCATAGGGTCGGAGGTGCTGTCCC CACTGCCCCACGTTGTCCCTGAGATTTAACCCCTCCACTGCTGGGGGTGAGCTGTACTCTTCTGACTGCCCCCTCCTGTGTAACGACTAC >34961_34961_1_GRTP1-COL4A2_GRTP1_chr13_113999178_ENST00000326039_COL4A2_chr13_111154015_ENST00000360467_length(amino acids)=568AA_BP=0 MVLSGAQAQMDQNPGYYHQLLQGERNPRLEDAIRTDLNRTFPDNVKFRKTTDPCLQRTLYNVLLAYGHHNQGVGYCQGMNFIAGYLILIT NNEEESFWLLDALVGRILPGERGPPGSPGLQGFPGITPPSNISGAPGDKGAPGIFGLKGYRGPPGPPGSAALPGSKGDTGNPGAPGTPGT KGWAGDSGPQGRPGVFGLPGEKGPRGEQGFMGNTGPTGAVGDRGPKGPKGDPGFPGAPGTVGAPGIAGIPQKIAVQPGTVGPQGRRGPPG APGEMGPQGPPGEPGFRGAPGKAGPQGRGGVSAVPGFRGDEGPIGHQGPIGQEGAPGRPGSPGLPGMPGRSVSIGYLLVKHSQTDQEPMC PVGMNKLWSGYSLLYFEGQEKAHNQDLGLAGSCLARFSTMPFLYCNPGDVCYYASRNDKSYWLSTTAPLPMMPVAEDEIKPYISRCSVCE APAIAIAVHSQDVSIPHCPAGWRSLWIGYSFLMHTAAGDEGGGQSLVSPGSCLEDFRATPFIECNGGRGTCHYYANKYSFWLTTIPEQSF -------------------------------------------------------------- >34961_34961_2_GRTP1-COL4A2_GRTP1_chr13_113999178_ENST00000375430_COL4A2_chr13_111154015_ENST00000360467_length(transcript)=2824nt_BP=609nt GAACTTGGGACCCGCTGGCCTCGCTCGGTGCGCGCCTCCCTCCCCGCATGCAGCCCGCCGAGCGCTCGCGGGTCCCCAGGATCGACCCGT ACGGATTCGAGCGGCCTGAGGACTTCGACGACGCCGCCTACGAGAAGTTTTTCTCCAGCTACCTGGTCACGCTCACCCGCAGGGCGATCA AATGGTCCCGGCTGCTGCAGGGCGGGGGCGTCCCCAGGAGCCGGACAGTGAAGCGCTATGTCCGGAAAGGGGTCCCGCTGGAGCACCGTG CCCGCGTCTGGATGGTGCTGAGTGGGGCCCAGGCGCAGATGGACCAGAATCCCGGCTACTACCACCAGCTTCTCCAGGGAGAGAGAAACC CCAGGCTGGAGGACGCCATCAGGACAGACCTGAACCGGACCTTCCCCGACAACGTGAAGTTCCGGAAGACCACGGACCCCTGCTTACAGA GGACCCTGTACAATGTGCTGCTGGCATATGGGCACCATAACCAGGGAGTGGGCTACTGCCAGGGAATGAATTTTATAGCAGGATATCTGA TTCTTATAACAAATAATGAAGAAGAATCTTTTTGGCTGTTAGATGCTCTTGTTGGAAGAATACTACCAGGGGAACGAGGCCCACCTGGGA GCCCAGGACTTCAGGGGTTCCCTGGTATCACACCCCCTTCCAACATCTCTGGGGCACCTGGTGACAAAGGGGCGCCAGGGATATTTGGCC TGAAAGGTTATCGGGGCCCACCAGGGCCACCAGGTTCTGCTGCTCTTCCTGGAAGCAAAGGTGACACAGGGAACCCAGGAGCTCCAGGAA CCCCAGGGACCAAAGGATGGGCCGGGGACTCCGGGCCCCAGGGCAGGCCTGGTGTGTTTGGTCTCCCAGGAGAAAAAGGGCCCAGGGGTG AACAAGGCTTCATGGGGAACACTGGACCCACTGGGGCGGTGGGCGACAGAGGCCCCAAGGGACCCAAGGGAGACCCAGGATTCCCTGGTG CCCCCGGGACTGTGGGAGCCCCCGGGATTGCAGGAATCCCCCAGAAGATTGCCGTCCAACCAGGGACAGTGGGTCCCCAGGGGAGGCGAG GCCCCCCTGGGGCACCGGGGGAGATGGGGCCCCAGGGCCCCCCCGGAGAACCAGGTTTCCGTGGGGCTCCAGGGAAAGCTGGGCCCCAAG GAAGAGGTGGTGTGTCTGCTGTTCCCGGCTTCCGGGGAGATGAAGGACCCATAGGCCACCAGGGGCCGATTGGCCAAGAAGGTGCACCAG GCCGTCCAGGGAGCCCGGGCCTGCCGGGTATGCCAGGCCGCAGCGTCAGCATCGGCTACCTCCTGGTGAAGCACAGCCAGACGGACCAGG AGCCCATGTGCCCAGTGGGCATGAACAAACTCTGGAGTGGATACAGCCTGCTGTACTTCGAGGGCCAGGAGAAGGCGCACAACCAGGACC TGGGGCTGGCGGGCTCCTGCCTGGCGCGGTTCAGCACCATGCCCTTCCTGTACTGCAACCCTGGTGATGTCTGCTACTATGCCAGCCGGA ACGACAAGTCCTACTGGCTCTCTACCACTGCGCCGCTGCCCATGATGCCCGTGGCCGAGGACGAGATCAAGCCCTACATCAGCCGCTGTT CTGTGTGTGAGGCCCCGGCCATCGCCATCGCGGTCCACAGTCAGGATGTCTCCATCCCACACTGCCCAGCTGGGTGGCGGAGTTTGTGGA TCGGATATTCCTTCCTCATGCACACGGCGGCGGGAGACGAAGGCGGTGGCCAATCACTGGTGTCACCGGGCAGCTGTCTAGAGGACTTCC GCGCCACACCATTCATCGAATGCAATGGAGGCCGCGGCACCTGCCACTACTACGCCAACAAGTACAGCTTCTGGCTGACCACCATTCCCG AGCAGAGCTTCCAGGGCTCGCCCTCCGCCGACACGCTCAAGGCCGGCCTCATCCGCACACACATCAGCCGCTGCCAGGTGTGCATGAAGA ACCTGTGAGCCGGCGCGTGCCAGGAAGGGCCATTTTGGTGCTTATTCTTAACTTATTACCTCAGGTGCCAACCCAAAAATTGGTTTTATT TTTTTCTTAAAAAAAAAAAAGTCTACCAAAGGAATTTGCATCCAGCAGCAGCACTTAGACCTGCCAGCCACTGTCACCGAGCGGGTGCAA GCACTCGGGGTCCCTGGAGGGCAAGCCCTGCCCACAGAAAGCCAGGAGCAGCCCTGGCCCCCATCAGCCCTGCTAGACGCACCGCCTGAA GGCACAGCTAACCACTTCGCACACACCCATGTAACCACTGCACTTTCCAATGCCACAGACAACTCACATTGTTCAACTCCCTTCTCGGGG TGGGACAGACGAGACAACAGCACACAGGCAGCCAGCCGTGGCCAGAGGCTCGAGGGGCTCAGGGCCTCAGGCACCCGTCCCCACACGAGG GCCCCGTGGGTGGGCCTGGCCCTGCTTTCTACGCCAATGTTATGCCAGCTCCATGTTCTCCCAAATACCGTTGATGTGAATTATTTTAAA GGCAAAACCGTGCTCTTTATTTTAAAAAACACTGATAATCACACTGCGGTAGGTCATTCTTTTGCCACATCCCTATAGACCACTGGGTTT GGCAAAACTCAGGCAGAAGTGGAGACCTTTCTAGACATCATTGTCAGCCTTGCTACTTGAAGGTACACCCCATAGGGTCGGAGGTGCTGT CCCCACTGCCCCACGTTGTCCCTGAGATTTAACCCCTCCACTGCTGGGGGTGAGCTGTACTCTTCTGACTGCCCCCTCCTGTGTAACGAC >34961_34961_2_GRTP1-COL4A2_GRTP1_chr13_113999178_ENST00000375430_COL4A2_chr13_111154015_ENST00000360467_length(amino acids)=657AA_BP=1 MASLGARLPPRMQPAERSRVPRIDPYGFERPEDFDDAAYEKFFSSYLVTLTRRAIKWSRLLQGGGVPRSRTVKRYVRKGVPLEHRARVWM VLSGAQAQMDQNPGYYHQLLQGERNPRLEDAIRTDLNRTFPDNVKFRKTTDPCLQRTLYNVLLAYGHHNQGVGYCQGMNFIAGYLILITN NEEESFWLLDALVGRILPGERGPPGSPGLQGFPGITPPSNISGAPGDKGAPGIFGLKGYRGPPGPPGSAALPGSKGDTGNPGAPGTPGTK GWAGDSGPQGRPGVFGLPGEKGPRGEQGFMGNTGPTGAVGDRGPKGPKGDPGFPGAPGTVGAPGIAGIPQKIAVQPGTVGPQGRRGPPGA PGEMGPQGPPGEPGFRGAPGKAGPQGRGGVSAVPGFRGDEGPIGHQGPIGQEGAPGRPGSPGLPGMPGRSVSIGYLLVKHSQTDQEPMCP VGMNKLWSGYSLLYFEGQEKAHNQDLGLAGSCLARFSTMPFLYCNPGDVCYYASRNDKSYWLSTTAPLPMMPVAEDEIKPYISRCSVCEA PAIAIAVHSQDVSIPHCPAGWRSLWIGYSFLMHTAAGDEGGGQSLVSPGSCLEDFRATPFIECNGGRGTCHYYANKYSFWLTTIPEQSFQ -------------------------------------------------------------- >34961_34961_3_GRTP1-COL4A2_GRTP1_chr13_113999178_ENST00000375431_COL4A2_chr13_111154015_ENST00000360467_length(transcript)=2852nt_BP=637nt GGACTTAAGCGCCCCGGAGCCGGGAGGCGAACTTGGGACCCGCTGGCCTCGCTCGGTGCGCGCCTCCCTCCCCGCATGCAGCCCGCCGAG CGCTCGCGGGTCCCCAGGATCGACCCGTACGGATTCGAGCGGCCTGAGGACTTCGACGACGCCGCCTACGAGAAGTTTTTCTCCAGCTAC CTGGTCACGCTCACCCGCAGGGCGATCAAATGGTCCCGGCTGCTGCAGGGCGGGGGCGTCCCCAGGAGCCGGACAGTGAAGCGCTATGTC CGGAAAGGGGTCCCGCTGGAGCACCGTGCCCGCGTCTGGATGGTGCTGAGTGGGGCCCAGGCGCAGATGGACCAGAATCCCGGCTACTAC CACCAGCTTCTCCAGGGAGAGAGAAACCCCAGGCTGGAGGACGCCATCAGGACAGACCTGAACCGGACCTTCCCCGACAACGTGAAGTTC CGGAAGACCACGGACCCCTGCTTACAGAGGACCCTGTACAATGTGCTGCTGGCATATGGGCACCATAACCAGGGAGTGGGCTACTGCCAG GGAATGAATTTTATAGCAGGATATCTGATTCTTATAACAAATAATGAAGAAGAATCTTTTTGGCTGTTAGATGCTCTTGTTGGAAGAATA CTACCAGGGGAACGAGGCCCACCTGGGAGCCCAGGACTTCAGGGGTTCCCTGGTATCACACCCCCTTCCAACATCTCTGGGGCACCTGGT GACAAAGGGGCGCCAGGGATATTTGGCCTGAAAGGTTATCGGGGCCCACCAGGGCCACCAGGTTCTGCTGCTCTTCCTGGAAGCAAAGGT GACACAGGGAACCCAGGAGCTCCAGGAACCCCAGGGACCAAAGGATGGGCCGGGGACTCCGGGCCCCAGGGCAGGCCTGGTGTGTTTGGT CTCCCAGGAGAAAAAGGGCCCAGGGGTGAACAAGGCTTCATGGGGAACACTGGACCCACTGGGGCGGTGGGCGACAGAGGCCCCAAGGGA CCCAAGGGAGACCCAGGATTCCCTGGTGCCCCCGGGACTGTGGGAGCCCCCGGGATTGCAGGAATCCCCCAGAAGATTGCCGTCCAACCA GGGACAGTGGGTCCCCAGGGGAGGCGAGGCCCCCCTGGGGCACCGGGGGAGATGGGGCCCCAGGGCCCCCCCGGAGAACCAGGTTTCCGT GGGGCTCCAGGGAAAGCTGGGCCCCAAGGAAGAGGTGGTGTGTCTGCTGTTCCCGGCTTCCGGGGAGATGAAGGACCCATAGGCCACCAG GGGCCGATTGGCCAAGAAGGTGCACCAGGCCGTCCAGGGAGCCCGGGCCTGCCGGGTATGCCAGGCCGCAGCGTCAGCATCGGCTACCTC CTGGTGAAGCACAGCCAGACGGACCAGGAGCCCATGTGCCCAGTGGGCATGAACAAACTCTGGAGTGGATACAGCCTGCTGTACTTCGAG GGCCAGGAGAAGGCGCACAACCAGGACCTGGGGCTGGCGGGCTCCTGCCTGGCGCGGTTCAGCACCATGCCCTTCCTGTACTGCAACCCT GGTGATGTCTGCTACTATGCCAGCCGGAACGACAAGTCCTACTGGCTCTCTACCACTGCGCCGCTGCCCATGATGCCCGTGGCCGAGGAC GAGATCAAGCCCTACATCAGCCGCTGTTCTGTGTGTGAGGCCCCGGCCATCGCCATCGCGGTCCACAGTCAGGATGTCTCCATCCCACAC TGCCCAGCTGGGTGGCGGAGTTTGTGGATCGGATATTCCTTCCTCATGCACACGGCGGCGGGAGACGAAGGCGGTGGCCAATCACTGGTG TCACCGGGCAGCTGTCTAGAGGACTTCCGCGCCACACCATTCATCGAATGCAATGGAGGCCGCGGCACCTGCCACTACTACGCCAACAAG TACAGCTTCTGGCTGACCACCATTCCCGAGCAGAGCTTCCAGGGCTCGCCCTCCGCCGACACGCTCAAGGCCGGCCTCATCCGCACACAC ATCAGCCGCTGCCAGGTGTGCATGAAGAACCTGTGAGCCGGCGCGTGCCAGGAAGGGCCATTTTGGTGCTTATTCTTAACTTATTACCTC AGGTGCCAACCCAAAAATTGGTTTTATTTTTTTCTTAAAAAAAAAAAAGTCTACCAAAGGAATTTGCATCCAGCAGCAGCACTTAGACCT GCCAGCCACTGTCACCGAGCGGGTGCAAGCACTCGGGGTCCCTGGAGGGCAAGCCCTGCCCACAGAAAGCCAGGAGCAGCCCTGGCCCCC ATCAGCCCTGCTAGACGCACCGCCTGAAGGCACAGCTAACCACTTCGCACACACCCATGTAACCACTGCACTTTCCAATGCCACAGACAA CTCACATTGTTCAACTCCCTTCTCGGGGTGGGACAGACGAGACAACAGCACACAGGCAGCCAGCCGTGGCCAGAGGCTCGAGGGGCTCAG GGCCTCAGGCACCCGTCCCCACACGAGGGCCCCGTGGGTGGGCCTGGCCCTGCTTTCTACGCCAATGTTATGCCAGCTCCATGTTCTCCC AAATACCGTTGATGTGAATTATTTTAAAGGCAAAACCGTGCTCTTTATTTTAAAAAACACTGATAATCACACTGCGGTAGGTCATTCTTT TGCCACATCCCTATAGACCACTGGGTTTGGCAAAACTCAGGCAGAAGTGGAGACCTTTCTAGACATCATTGTCAGCCTTGCTACTTGAAG GTACACCCCATAGGGTCGGAGGTGCTGTCCCCACTGCCCCACGTTGTCCCTGAGATTTAACCCCTCCACTGCTGGGGGTGAGCTGTACTC >34961_34961_3_GRTP1-COL4A2_GRTP1_chr13_113999178_ENST00000375431_COL4A2_chr13_111154015_ENST00000360467_length(amino acids)=657AA_BP=1 MASLGARLPPRMQPAERSRVPRIDPYGFERPEDFDDAAYEKFFSSYLVTLTRRAIKWSRLLQGGGVPRSRTVKRYVRKGVPLEHRARVWM VLSGAQAQMDQNPGYYHQLLQGERNPRLEDAIRTDLNRTFPDNVKFRKTTDPCLQRTLYNVLLAYGHHNQGVGYCQGMNFIAGYLILITN NEEESFWLLDALVGRILPGERGPPGSPGLQGFPGITPPSNISGAPGDKGAPGIFGLKGYRGPPGPPGSAALPGSKGDTGNPGAPGTPGTK GWAGDSGPQGRPGVFGLPGEKGPRGEQGFMGNTGPTGAVGDRGPKGPKGDPGFPGAPGTVGAPGIAGIPQKIAVQPGTVGPQGRRGPPGA PGEMGPQGPPGEPGFRGAPGKAGPQGRGGVSAVPGFRGDEGPIGHQGPIGQEGAPGRPGSPGLPGMPGRSVSIGYLLVKHSQTDQEPMCP VGMNKLWSGYSLLYFEGQEKAHNQDLGLAGSCLARFSTMPFLYCNPGDVCYYASRNDKSYWLSTTAPLPMMPVAEDEIKPYISRCSVCEA PAIAIAVHSQDVSIPHCPAGWRSLWIGYSFLMHTAAGDEGGGQSLVSPGSCLEDFRATPFIECNGGRGTCHYYANKYSFWLTTIPEQSFQ -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for GRTP1-COL4A2 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for GRTP1-COL4A2 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for GRTP1-COL4A2 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
