
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:GSDMB-NT5C3B (FusionGDB2 ID:34988) |
Fusion Gene Summary for GSDMB-NT5C3B |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: GSDMB-NT5C3B | Fusion gene ID: 34988 | Hgene | Tgene | Gene symbol | GSDMB | NT5C3B | Gene ID | 55876 | 115024 |
| Gene name | gasdermin B | 5'-nucleotidase, cytosolic IIIB | |
| Synonyms | GSDMB-1|GSDML|PP4052|PRO2521 | NT5C3L|cN-IIIB | |
| Cytomap | 17q21.1 | 17q21.2 | |
| Type of gene | protein-coding | protein-coding | |
| Description | gasdermin-Bgasdermin-like proteingasderminB-1 | 7-methylguanosine phosphate-specific 5'-nucleotidase5'-nucleotidase, cytosolic III-like7-methylguanosine nucleotidaseN(7)-methylguanylate 5'-phosphatasecN-III-like proteincytosolic 5'-nucleotidase 3Bcytosolic 5'-nucleotidase III-like protein | |
| Modification date | 20200313 | 20200320 | |
| UniProtAcc | Q8TAX9 | . | |
| Ensembl transtripts involved in fusion gene | ENST00000309481, ENST00000360317, ENST00000394175, ENST00000394179, ENST00000418519, ENST00000520542, | ENST00000435506, ENST00000521789, ENST00000269534, | |
| Fusion gene scores | * DoF score | 2 X 2 X 2=8 | 5 X 5 X 4=100 |
| # samples | 2 | 5 | |
| ** MAII score | log2(2/8*10)=1.32192809488736 | log2(5/100*10)=-1 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: GSDMB [Title/Abstract] AND NT5C3B [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | GSDMB(38066009)-NT5C3B(39983878), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
 Fusion gene breakpoints across GSDMB (5'-gene) Fusion gene breakpoints across GSDMB (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
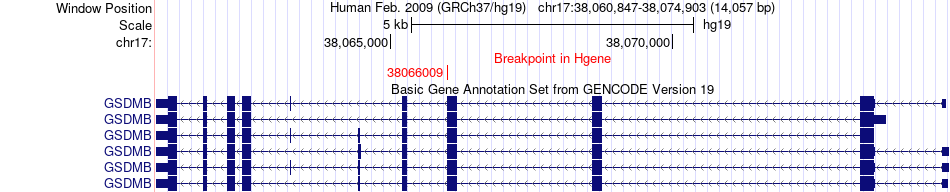 |
 Fusion gene breakpoints across NT5C3B (3'-gene) Fusion gene breakpoints across NT5C3B (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
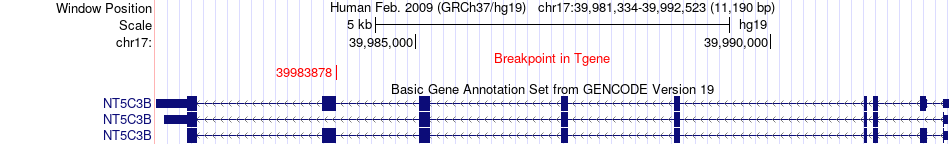 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | STAD | TCGA-VQ-A92D-01A | GSDMB | chr17 | 38066009 | - | NT5C3B | chr17 | 39983878 | - |
Top |
Fusion Gene ORF analysis for GSDMB-NT5C3B |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-intron | ENST00000309481 | ENST00000435506 | GSDMB | chr17 | 38066009 | - | NT5C3B | chr17 | 39983878 | - |
| 5CDS-intron | ENST00000309481 | ENST00000521789 | GSDMB | chr17 | 38066009 | - | NT5C3B | chr17 | 39983878 | - |
| 5CDS-intron | ENST00000360317 | ENST00000435506 | GSDMB | chr17 | 38066009 | - | NT5C3B | chr17 | 39983878 | - |
| 5CDS-intron | ENST00000360317 | ENST00000521789 | GSDMB | chr17 | 38066009 | - | NT5C3B | chr17 | 39983878 | - |
| 5CDS-intron | ENST00000394175 | ENST00000435506 | GSDMB | chr17 | 38066009 | - | NT5C3B | chr17 | 39983878 | - |
| 5CDS-intron | ENST00000394175 | ENST00000521789 | GSDMB | chr17 | 38066009 | - | NT5C3B | chr17 | 39983878 | - |
| 5CDS-intron | ENST00000394179 | ENST00000435506 | GSDMB | chr17 | 38066009 | - | NT5C3B | chr17 | 39983878 | - |
| 5CDS-intron | ENST00000394179 | ENST00000521789 | GSDMB | chr17 | 38066009 | - | NT5C3B | chr17 | 39983878 | - |
| 5CDS-intron | ENST00000418519 | ENST00000435506 | GSDMB | chr17 | 38066009 | - | NT5C3B | chr17 | 39983878 | - |
| 5CDS-intron | ENST00000418519 | ENST00000521789 | GSDMB | chr17 | 38066009 | - | NT5C3B | chr17 | 39983878 | - |
| 5CDS-intron | ENST00000520542 | ENST00000435506 | GSDMB | chr17 | 38066009 | - | NT5C3B | chr17 | 39983878 | - |
| 5CDS-intron | ENST00000520542 | ENST00000521789 | GSDMB | chr17 | 38066009 | - | NT5C3B | chr17 | 39983878 | - |
| In-frame | ENST00000309481 | ENST00000269534 | GSDMB | chr17 | 38066009 | - | NT5C3B | chr17 | 39983878 | - |
| In-frame | ENST00000360317 | ENST00000269534 | GSDMB | chr17 | 38066009 | - | NT5C3B | chr17 | 39983878 | - |
| In-frame | ENST00000394175 | ENST00000269534 | GSDMB | chr17 | 38066009 | - | NT5C3B | chr17 | 39983878 | - |
| In-frame | ENST00000394179 | ENST00000269534 | GSDMB | chr17 | 38066009 | - | NT5C3B | chr17 | 39983878 | - |
| In-frame | ENST00000418519 | ENST00000269534 | GSDMB | chr17 | 38066009 | - | NT5C3B | chr17 | 39983878 | - |
| In-frame | ENST00000520542 | ENST00000269534 | GSDMB | chr17 | 38066009 | - | NT5C3B | chr17 | 39983878 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000360317 | GSDMB | chr17 | 38066009 | - | ENST00000269534 | NT5C3B | chr17 | 39983878 | - | 1352 | 576 | 0 | 911 | 303 |
| ENST00000394175 | GSDMB | chr17 | 38066009 | - | ENST00000269534 | NT5C3B | chr17 | 39983878 | - | 1576 | 800 | 224 | 1135 | 303 |
| ENST00000309481 | GSDMB | chr17 | 38066009 | - | ENST00000269534 | NT5C3B | chr17 | 39983878 | - | 1423 | 647 | 71 | 982 | 303 |
| ENST00000520542 | GSDMB | chr17 | 38066009 | - | ENST00000269534 | NT5C3B | chr17 | 39983878 | - | 1459 | 683 | 107 | 1018 | 303 |
| ENST00000418519 | GSDMB | chr17 | 38066009 | - | ENST00000269534 | NT5C3B | chr17 | 39983878 | - | 1483 | 707 | 131 | 1042 | 303 |
| ENST00000394179 | GSDMB | chr17 | 38066009 | - | ENST00000269534 | NT5C3B | chr17 | 39983878 | - | 1483 | 707 | 131 | 1042 | 303 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000360317 | ENST00000269534 | GSDMB | chr17 | 38066009 | - | NT5C3B | chr17 | 39983878 | - | 0.001893743 | 0.99810624 |
| ENST00000394175 | ENST00000269534 | GSDMB | chr17 | 38066009 | - | NT5C3B | chr17 | 39983878 | - | 0.001587055 | 0.99841297 |
| ENST00000309481 | ENST00000269534 | GSDMB | chr17 | 38066009 | - | NT5C3B | chr17 | 39983878 | - | 0.002023506 | 0.9979765 |
| ENST00000520542 | ENST00000269534 | GSDMB | chr17 | 38066009 | - | NT5C3B | chr17 | 39983878 | - | 0.002123312 | 0.99787664 |
| ENST00000418519 | ENST00000269534 | GSDMB | chr17 | 38066009 | - | NT5C3B | chr17 | 39983878 | - | 0.002043638 | 0.9979564 |
| ENST00000394179 | ENST00000269534 | GSDMB | chr17 | 38066009 | - | NT5C3B | chr17 | 39983878 | - | 0.002043638 | 0.9979564 |
Top |
Fusion Genomic Features for GSDMB-NT5C3B |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
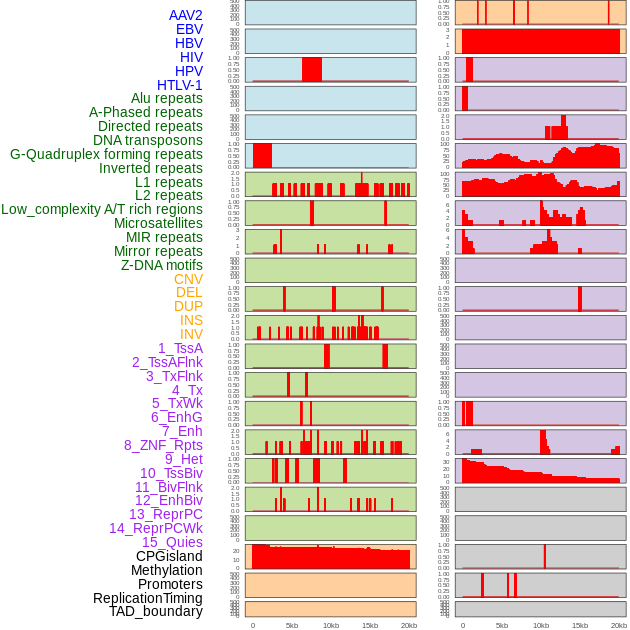 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for GSDMB-NT5C3B |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr17:38066009/chr17:39983878) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| GSDMB | . |
| FUNCTION: [Gasdermin-B]: Precursor of a pore-forming protein that acts as a downstream mediator of granzyme-mediated cell death (PubMed:32299851). This form constitutes the precursor of the pore-forming protein: upon cleavage, the released N-terminal moiety (Gasdermin-B, N-terminal) binds to membranes and forms pores, triggering pyroptosis (PubMed:32299851). {ECO:0000269|PubMed:32299851}.; FUNCTION: [Gasdermin-B, N-terminal]: Pore-forming protein produced by cleavage by granzyme A (GZMA), which causes membrane permeabilization and pyroptosis in target cells of cytotoxic T and natural killer (NK) cells (PubMed:27281216, PubMed:32299851). Key downstream mediator of granzyme-mediated cell death: (1) granzyme A (GZMA), delivered to target cells from cytotoxic T- and NK-cells, (2) specifically cleaves Gasdermin-B to generate this form (PubMed:32299851). After cleavage, moves to the plasma membrane, homooligomerizes within the membrane and forms pores of 10-15 nanometers (nm) of inner diameter, triggering pyroptosis (PubMed:32299851). Binds to membrane inner leaflet lipids, such as phosphatidylinositol 4-phosphate, phosphatidylinositol 5-phosphate, bisphosphorylated phosphatidylinositols, such as phosphatidylinositol (4,5)-bisphosphate, and more weakly to phosphatidic acid (PubMed:28154144). Also binds sufatide, a component of the apical membrane of epithelial cells (PubMed:28154144). {ECO:0000269|PubMed:27281216, ECO:0000269|PubMed:28154144, ECO:0000269|PubMed:32299851}. | FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | GSDMB | chr17:38066009 | chr17:39983878 | ENST00000309481 | - | 4 | 10 | 243_271 | 192 | 404.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | GSDMB | chr17:38066009 | chr17:39983878 | ENST00000360317 | - | 3 | 10 | 243_271 | 192 | 417.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | GSDMB | chr17:38066009 | chr17:39983878 | ENST00000394175 | - | 3 | 8 | 243_271 | 192 | 395.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | GSDMB | chr17:38066009 | chr17:39983878 | ENST00000394179 | - | 4 | 10 | 243_271 | 192 | 412.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | GSDMB | chr17:38066009 | chr17:39983878 | ENST00000418519 | - | 4 | 11 | 243_271 | 192 | 417.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | GSDMB | chr17:38066009 | chr17:39983878 | ENST00000520542 | - | 4 | 10 | 243_271 | 192 | 408.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | GSDMB | chr17:38066009 | chr17:39983878 | ENST00000309481 | - | 4 | 10 | 1_275 | 192 | 404.0 | Region | Triggers pyroptosis |
| Hgene | GSDMB | chr17:38066009 | chr17:39983878 | ENST00000360317 | - | 3 | 10 | 1_275 | 192 | 417.0 | Region | Triggers pyroptosis |
| Hgene | GSDMB | chr17:38066009 | chr17:39983878 | ENST00000394175 | - | 3 | 8 | 1_275 | 192 | 395.0 | Region | Triggers pyroptosis |
| Hgene | GSDMB | chr17:38066009 | chr17:39983878 | ENST00000394179 | - | 4 | 10 | 1_275 | 192 | 412.0 | Region | Triggers pyroptosis |
| Hgene | GSDMB | chr17:38066009 | chr17:39983878 | ENST00000418519 | - | 4 | 11 | 1_275 | 192 | 417.0 | Region | Triggers pyroptosis |
| Hgene | GSDMB | chr17:38066009 | chr17:39983878 | ENST00000520542 | - | 4 | 10 | 1_275 | 192 | 408.0 | Region | Triggers pyroptosis |
| Tgene | NT5C3B | chr17:38066009 | chr17:39983878 | ENST00000269534 | 6 | 9 | 156_157 | 181 | 293.0 | Region | Substrate binding | |
| Tgene | NT5C3B | chr17:38066009 | chr17:39983878 | ENST00000435506 | 6 | 9 | 156_157 | 189 | 301.0 | Region | Substrate binding |
Top |
Fusion Gene Sequence for GSDMB-NT5C3B |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >34988_34988_1_GSDMB-NT5C3B_GSDMB_chr17_38066009_ENST00000309481_NT5C3B_chr17_39983878_ENST00000269534_length(transcript)=1423nt_BP=647nt ATCTGTGGGGATTCTCACAACTTCCATTTCTGGTGAACAGCTGAGGTCAGAGAGGAGTTGGTCCAGGCGCAATGTTCAGCGTATTTGAGG AAATCACAAGAATTGTAGTTAAGGAGATGGATGCTGGAGGGGATATGATTGCCGTTAGAAGCCTTGTTGATGCTGATAGATTCCGCTGCT TCCATCTGGTGGGGGAGAAGAGAACTTTCTTTGGATGCCGGCACTACACAACAGGCCTCACCCTGATGGACATTCTGGACACAGATGGGG ACAAGTGGTTAGATGAACTGGATTCTGGGCTCCAAGGTCAAAAGGCTGAGTTTCAAATTCTGGATAATGTAGACTCAACGGGAGAGTTGA TAGTGAGATTACCCAAAGAAATAACAATTTCAGGCAGTTTCCAGGGCTTCCACCATCAGAAAATCAAGATATCGGAGAACCGGATATCCC AGCAGTATCTGGCTACCCTTGAAAACAGGAAGCTGAAGAGGGAACTACCCTTTTCATTCCGATCAATTAATACGAGAGAAAACCTGTATC TGGTGACAGAAACTCTGGAGACGGTAAAGGAGGAAACCCTGAAAAGCGACCGGCAATATAAATTTTGGAGCCAGATCTCTCAGGGCCATC TCAGCTATAAACACAAGGGTTTTCTCCAGGGATTTAAGGGCCAGCTCATACACACATACAACAAGAACAGCTCTGCGTGTGAGAACTCTG GTTACTTCCAGCAACTTGAGGGCAAAACCAATGTCATCCTGCTGGGAGACTCTATCGGGGACCTCACCATGGCCGATGGGGTTCCTGGTG TGCAGAACATTCTCAAAATTGGCTTCCTGAATGACAAGGTGGAGGAGCGGCGGGAGCGCTACATGGACTCCTATGACATCGTGCTGGAGA AGGACGAGACTCTGGATGTGGTCAACGGGCTACTGCAGCACATCCTGTGCCAGGGGGTCCAGCTGGAGATGCAAGGCCCCTGAAGGCGCA GGCTCCAGCCCGGCCTGCAGGCCGTGGTGAGGAGGGGCGCCTCCCCAGAGTCTGCTCCCCCGTGAACACAGAGCAGAGGCCAGGGTGGCC AGCAGTGGCTGGGTCCTTCCGCGCCCCTCCGTCCTCCTTTCCCTGAGCACCTTCATCACCAGAGGCTTGAAGGAACCCCGCCATGTGGCA GGGCACAGGCACTGTTCCTGGTGAACCTTGGACCACAGCATGTCAGTGCTCTAGGGATTGTCTACTCCAGGGATTTTCTTCAAAATTTTT AAACATGGGAAGTTCAAACAAATATAATGTGTGAAACAGATCAAAATTTTTAAAATGAAAAAAAAGCTGCTCTGATTCAGGGGATGTGGG >34988_34988_1_GSDMB-NT5C3B_GSDMB_chr17_38066009_ENST00000309481_NT5C3B_chr17_39983878_ENST00000269534_length(amino acids)=303AA_BP=192 MFSVFEEITRIVVKEMDAGGDMIAVRSLVDADRFRCFHLVGEKRTFFGCRHYTTGLTLMDILDTDGDKWLDELDSGLQGQKAEFQILDNV DSTGELIVRLPKEITISGSFQGFHHQKIKISENRISQQYLATLENRKLKRELPFSFRSINTRENLYLVTETLETVKEETLKSDRQYKFWS QISQGHLSYKHKGFLQGFKGQLIHTYNKNSSACENSGYFQQLEGKTNVILLGDSIGDLTMADGVPGVQNILKIGFLNDKVEERRERYMDS -------------------------------------------------------------- >34988_34988_2_GSDMB-NT5C3B_GSDMB_chr17_38066009_ENST00000360317_NT5C3B_chr17_39983878_ENST00000269534_length(transcript)=1352nt_BP=576nt ATGTTCAGCGTATTTGAGGAAATCACAAGAATTGTAGTTAAGGAGATGGATGCTGGAGGGGATATGATTGCCGTTAGAAGCCTTGTTGAT GCTGATAGATTCCGCTGCTTCCATCTGGTGGGGGAGAAGAGAACTTTCTTTGGATGCCGGCACTACACAACAGGCCTCACCCTGATGGAC ATTCTGGACACAGATGGGGACAAGTGGTTAGATGAACTGGATTCTGGGCTCCAAGGTCAAAAGGCTGAGTTTCAAATTCTGGATAATGTA GACTCAACGGGAGAGTTGATAGTGAGATTACCCAAAGAAATAACAATTTCAGGCAGTTTCCAGGGCTTCCACCATCAGAAAATCAAGATA TCGGAGAACCGGATATCCCAGCAGTATCTGGCTACCCTTGAAAACAGGAAGCTGAAGAGGGAACTACCCTTTTCATTCCGATCAATTAAT ACGAGAGAAAACCTGTATCTGGTGACAGAAACTCTGGAGACGGTAAAGGAGGAAACCCTGAAAAGCGACCGGCAATATAAATTTTGGAGC CAGATCTCTCAGGGCCATCTCAGCTATAAACACAAGGGTTTTCTCCAGGGATTTAAGGGCCAGCTCATACACACATACAACAAGAACAGC TCTGCGTGTGAGAACTCTGGTTACTTCCAGCAACTTGAGGGCAAAACCAATGTCATCCTGCTGGGAGACTCTATCGGGGACCTCACCATG GCCGATGGGGTTCCTGGTGTGCAGAACATTCTCAAAATTGGCTTCCTGAATGACAAGGTGGAGGAGCGGCGGGAGCGCTACATGGACTCC TATGACATCGTGCTGGAGAAGGACGAGACTCTGGATGTGGTCAACGGGCTACTGCAGCACATCCTGTGCCAGGGGGTCCAGCTGGAGATG CAAGGCCCCTGAAGGCGCAGGCTCCAGCCCGGCCTGCAGGCCGTGGTGAGGAGGGGCGCCTCCCCAGAGTCTGCTCCCCCGTGAACACAG AGCAGAGGCCAGGGTGGCCAGCAGTGGCTGGGTCCTTCCGCGCCCCTCCGTCCTCCTTTCCCTGAGCACCTTCATCACCAGAGGCTTGAA GGAACCCCGCCATGTGGCAGGGCACAGGCACTGTTCCTGGTGAACCTTGGACCACAGCATGTCAGTGCTCTAGGGATTGTCTACTCCAGG GATTTTCTTCAAAATTTTTAAACATGGGAAGTTCAAACAAATATAATGTGTGAAACAGATCAAAATTTTTAAAATGAAAAAAAAGCTGCT CTGATTCAGGGGATGTGGGTGGGGGTAGAACCTGGACCTCTTGGCCCTGGGGGCACATGGGATGCTTCTAGGAACACAGTTTGAGAACCA >34988_34988_2_GSDMB-NT5C3B_GSDMB_chr17_38066009_ENST00000360317_NT5C3B_chr17_39983878_ENST00000269534_length(amino acids)=303AA_BP=192 MFSVFEEITRIVVKEMDAGGDMIAVRSLVDADRFRCFHLVGEKRTFFGCRHYTTGLTLMDILDTDGDKWLDELDSGLQGQKAEFQILDNV DSTGELIVRLPKEITISGSFQGFHHQKIKISENRISQQYLATLENRKLKRELPFSFRSINTRENLYLVTETLETVKEETLKSDRQYKFWS QISQGHLSYKHKGFLQGFKGQLIHTYNKNSSACENSGYFQQLEGKTNVILLGDSIGDLTMADGVPGVQNILKIGFLNDKVEERRERYMDS -------------------------------------------------------------- >34988_34988_3_GSDMB-NT5C3B_GSDMB_chr17_38066009_ENST00000394175_NT5C3B_chr17_39983878_ENST00000269534_length(transcript)=1576nt_BP=800nt GTGGAATGTCATCAGTTAAGGCTATTTTCATTTCTTTTGTGGATCTTCAGTTGCTTCAGGCCATCTGGATGTATACATGCAGGTCACAGG GAATATGATGGCTTAGCTTGGGTTCAGAGGCCTGACACCTCAGGCTGCCAAATGTGGAAGATTTAAATACTTGAACCAATACCCTCCTCC CAAAAACTGAAATTGGCTTCTGTTTCTGAGTTGGTCCAGGCGCAATGTTCAGCGTATTTGAGGAAATCACAAGAATTGTAGTTAAGGAGA TGGATGCTGGAGGGGATATGATTGCCGTTAGAAGCCTTGTTGATGCTGATAGATTCCGCTGCTTCCATCTGGTGGGGGAGAAGAGAACTT TCTTTGGATGCCGGCACTACACAACAGGCCTCACCCTGATGGACATTCTGGACACAGATGGGGACAAGTGGTTAGATGAACTGGATTCTG GGCTCCAAGGTCAAAAGGCTGAGTTTCAAATTCTGGATAATGTAGACTCAACGGGAGAGTTGATAGTGAGATTACCCAAAGAAATAACAA TTTCAGGCAGTTTCCAGGGCTTCCACCATCAGAAAATCAAGATATCGGAGAACCGGATATCCCAGCAGTATCTGGCTACCCTTGAAAACA GGAAGCTGAAGAGGGAACTACCCTTTTCATTCCGATCAATTAATACGAGAGAAAACCTGTATCTGGTGACAGAAACTCTGGAGACGGTAA AGGAGGAAACCCTGAAAAGCGACCGGCAATATAAATTTTGGAGCCAGATCTCTCAGGGCCATCTCAGCTATAAACACAAGGGTTTTCTCC AGGGATTTAAGGGCCAGCTCATACACACATACAACAAGAACAGCTCTGCGTGTGAGAACTCTGGTTACTTCCAGCAACTTGAGGGCAAAA CCAATGTCATCCTGCTGGGAGACTCTATCGGGGACCTCACCATGGCCGATGGGGTTCCTGGTGTGCAGAACATTCTCAAAATTGGCTTCC TGAATGACAAGGTGGAGGAGCGGCGGGAGCGCTACATGGACTCCTATGACATCGTGCTGGAGAAGGACGAGACTCTGGATGTGGTCAACG GGCTACTGCAGCACATCCTGTGCCAGGGGGTCCAGCTGGAGATGCAAGGCCCCTGAAGGCGCAGGCTCCAGCCCGGCCTGCAGGCCGTGG TGAGGAGGGGCGCCTCCCCAGAGTCTGCTCCCCCGTGAACACAGAGCAGAGGCCAGGGTGGCCAGCAGTGGCTGGGTCCTTCCGCGCCCC TCCGTCCTCCTTTCCCTGAGCACCTTCATCACCAGAGGCTTGAAGGAACCCCGCCATGTGGCAGGGCACAGGCACTGTTCCTGGTGAACC TTGGACCACAGCATGTCAGTGCTCTAGGGATTGTCTACTCCAGGGATTTTCTTCAAAATTTTTAAACATGGGAAGTTCAAACAAATATAA TGTGTGAAACAGATCAAAATTTTTAAAATGAAAAAAAAGCTGCTCTGATTCAGGGGATGTGGGTGGGGGTAGAACCTGGACCTCTTGGCC >34988_34988_3_GSDMB-NT5C3B_GSDMB_chr17_38066009_ENST00000394175_NT5C3B_chr17_39983878_ENST00000269534_length(amino acids)=303AA_BP=192 MFSVFEEITRIVVKEMDAGGDMIAVRSLVDADRFRCFHLVGEKRTFFGCRHYTTGLTLMDILDTDGDKWLDELDSGLQGQKAEFQILDNV DSTGELIVRLPKEITISGSFQGFHHQKIKISENRISQQYLATLENRKLKRELPFSFRSINTRENLYLVTETLETVKEETLKSDRQYKFWS QISQGHLSYKHKGFLQGFKGQLIHTYNKNSSACENSGYFQQLEGKTNVILLGDSIGDLTMADGVPGVQNILKIGFLNDKVEERRERYMDS -------------------------------------------------------------- >34988_34988_4_GSDMB-NT5C3B_GSDMB_chr17_38066009_ENST00000394179_NT5C3B_chr17_39983878_ENST00000269534_length(transcript)=1483nt_BP=707nt ACAACTTTCAGTTTCATTATTTTAGCTTCCTGAGATTCAGAGGCCAGGAACTGTGCAGAGATCTGTGGGGATTCTCACAACTTCCATTTC TGGTGAACAGCTGAGGTCAGAGAGGAGTTGGTCCAGGCGCAATGTTCAGCGTATTTGAGGAAATCACAAGAATTGTAGTTAAGGAGATGG ATGCTGGAGGGGATATGATTGCCGTTAGAAGCCTTGTTGATGCTGATAGATTCCGCTGCTTCCATCTGGTGGGGGAGAAGAGAACTTTCT TTGGATGCCGGCACTACACAACAGGCCTCACCCTGATGGACATTCTGGACACAGATGGGGACAAGTGGTTAGATGAACTGGATTCTGGGC TCCAAGGTCAAAAGGCTGAGTTTCAAATTCTGGATAATGTAGACTCAACGGGAGAGTTGATAGTGAGATTACCCAAAGAAATAACAATTT CAGGCAGTTTCCAGGGCTTCCACCATCAGAAAATCAAGATATCGGAGAACCGGATATCCCAGCAGTATCTGGCTACCCTTGAAAACAGGA AGCTGAAGAGGGAACTACCCTTTTCATTCCGATCAATTAATACGAGAGAAAACCTGTATCTGGTGACAGAAACTCTGGAGACGGTAAAGG AGGAAACCCTGAAAAGCGACCGGCAATATAAATTTTGGAGCCAGATCTCTCAGGGCCATCTCAGCTATAAACACAAGGGTTTTCTCCAGG GATTTAAGGGCCAGCTCATACACACATACAACAAGAACAGCTCTGCGTGTGAGAACTCTGGTTACTTCCAGCAACTTGAGGGCAAAACCA ATGTCATCCTGCTGGGAGACTCTATCGGGGACCTCACCATGGCCGATGGGGTTCCTGGTGTGCAGAACATTCTCAAAATTGGCTTCCTGA ATGACAAGGTGGAGGAGCGGCGGGAGCGCTACATGGACTCCTATGACATCGTGCTGGAGAAGGACGAGACTCTGGATGTGGTCAACGGGC TACTGCAGCACATCCTGTGCCAGGGGGTCCAGCTGGAGATGCAAGGCCCCTGAAGGCGCAGGCTCCAGCCCGGCCTGCAGGCCGTGGTGA GGAGGGGCGCCTCCCCAGAGTCTGCTCCCCCGTGAACACAGAGCAGAGGCCAGGGTGGCCAGCAGTGGCTGGGTCCTTCCGCGCCCCTCC GTCCTCCTTTCCCTGAGCACCTTCATCACCAGAGGCTTGAAGGAACCCCGCCATGTGGCAGGGCACAGGCACTGTTCCTGGTGAACCTTG GACCACAGCATGTCAGTGCTCTAGGGATTGTCTACTCCAGGGATTTTCTTCAAAATTTTTAAACATGGGAAGTTCAAACAAATATAATGT GTGAAACAGATCAAAATTTTTAAAATGAAAAAAAAGCTGCTCTGATTCAGGGGATGTGGGTGGGGGTAGAACCTGGACCTCTTGGCCCTG >34988_34988_4_GSDMB-NT5C3B_GSDMB_chr17_38066009_ENST00000394179_NT5C3B_chr17_39983878_ENST00000269534_length(amino acids)=303AA_BP=192 MFSVFEEITRIVVKEMDAGGDMIAVRSLVDADRFRCFHLVGEKRTFFGCRHYTTGLTLMDILDTDGDKWLDELDSGLQGQKAEFQILDNV DSTGELIVRLPKEITISGSFQGFHHQKIKISENRISQQYLATLENRKLKRELPFSFRSINTRENLYLVTETLETVKEETLKSDRQYKFWS QISQGHLSYKHKGFLQGFKGQLIHTYNKNSSACENSGYFQQLEGKTNVILLGDSIGDLTMADGVPGVQNILKIGFLNDKVEERRERYMDS -------------------------------------------------------------- >34988_34988_5_GSDMB-NT5C3B_GSDMB_chr17_38066009_ENST00000418519_NT5C3B_chr17_39983878_ENST00000269534_length(transcript)=1483nt_BP=707nt ACAACTTTCAGTTTCATTATTTTAGCTTCCTGAGATTCAGAGGCCAGGAACTGTGCAGAGATCTGTGGGGATTCTCACAACTTCCATTTC TGGTGAACAGCTGAGGTCAGAGAGGAGTTGGTCCAGGCGCAATGTTCAGCGTATTTGAGGAAATCACAAGAATTGTAGTTAAGGAGATGG ATGCTGGAGGGGATATGATTGCCGTTAGAAGCCTTGTTGATGCTGATAGATTCCGCTGCTTCCATCTGGTGGGGGAGAAGAGAACTTTCT TTGGATGCCGGCACTACACAACAGGCCTCACCCTGATGGACATTCTGGACACAGATGGGGACAAGTGGTTAGATGAACTGGATTCTGGGC TCCAAGGTCAAAAGGCTGAGTTTCAAATTCTGGATAATGTAGACTCAACGGGAGAGTTGATAGTGAGATTACCCAAAGAAATAACAATTT CAGGCAGTTTCCAGGGCTTCCACCATCAGAAAATCAAGATATCGGAGAACCGGATATCCCAGCAGTATCTGGCTACCCTTGAAAACAGGA AGCTGAAGAGGGAACTACCCTTTTCATTCCGATCAATTAATACGAGAGAAAACCTGTATCTGGTGACAGAAACTCTGGAGACGGTAAAGG AGGAAACCCTGAAAAGCGACCGGCAATATAAATTTTGGAGCCAGATCTCTCAGGGCCATCTCAGCTATAAACACAAGGGTTTTCTCCAGG GATTTAAGGGCCAGCTCATACACACATACAACAAGAACAGCTCTGCGTGTGAGAACTCTGGTTACTTCCAGCAACTTGAGGGCAAAACCA ATGTCATCCTGCTGGGAGACTCTATCGGGGACCTCACCATGGCCGATGGGGTTCCTGGTGTGCAGAACATTCTCAAAATTGGCTTCCTGA ATGACAAGGTGGAGGAGCGGCGGGAGCGCTACATGGACTCCTATGACATCGTGCTGGAGAAGGACGAGACTCTGGATGTGGTCAACGGGC TACTGCAGCACATCCTGTGCCAGGGGGTCCAGCTGGAGATGCAAGGCCCCTGAAGGCGCAGGCTCCAGCCCGGCCTGCAGGCCGTGGTGA GGAGGGGCGCCTCCCCAGAGTCTGCTCCCCCGTGAACACAGAGCAGAGGCCAGGGTGGCCAGCAGTGGCTGGGTCCTTCCGCGCCCCTCC GTCCTCCTTTCCCTGAGCACCTTCATCACCAGAGGCTTGAAGGAACCCCGCCATGTGGCAGGGCACAGGCACTGTTCCTGGTGAACCTTG GACCACAGCATGTCAGTGCTCTAGGGATTGTCTACTCCAGGGATTTTCTTCAAAATTTTTAAACATGGGAAGTTCAAACAAATATAATGT GTGAAACAGATCAAAATTTTTAAAATGAAAAAAAAGCTGCTCTGATTCAGGGGATGTGGGTGGGGGTAGAACCTGGACCTCTTGGCCCTG >34988_34988_5_GSDMB-NT5C3B_GSDMB_chr17_38066009_ENST00000418519_NT5C3B_chr17_39983878_ENST00000269534_length(amino acids)=303AA_BP=192 MFSVFEEITRIVVKEMDAGGDMIAVRSLVDADRFRCFHLVGEKRTFFGCRHYTTGLTLMDILDTDGDKWLDELDSGLQGQKAEFQILDNV DSTGELIVRLPKEITISGSFQGFHHQKIKISENRISQQYLATLENRKLKRELPFSFRSINTRENLYLVTETLETVKEETLKSDRQYKFWS QISQGHLSYKHKGFLQGFKGQLIHTYNKNSSACENSGYFQQLEGKTNVILLGDSIGDLTMADGVPGVQNILKIGFLNDKVEERRERYMDS -------------------------------------------------------------- >34988_34988_6_GSDMB-NT5C3B_GSDMB_chr17_38066009_ENST00000520542_NT5C3B_chr17_39983878_ENST00000269534_length(transcript)=1459nt_BP=683nt ATTATTTTAGCTTCCTGAGATTCAGAGGCCAGGAACTGTGCAGAGATCTGTGGGGATTCTCACAACTTCCATTTCTGGTGAACAGCTGAG GTCAGAGAGGAGGCGCAATGTTCAGCGTATTTGAGGAAATCACAAGAATTGTAGTTAAGGAGATGGATGCTGGAGGGGATATGATTGCCG TTAGAAGCCTTGTTGATGCTGATAGATTCCGCTGCTTCCATCTGGTGGGGGAGAAGAGAACTTTCTTTGGATGCCGGCACTACACAACAG GCCTCACCCTGATGGACATTCTGGACACAGATGGGGACAAGTGGTTAGATGAACTGGATTCTGGGCTCCAAGGTCAAAAGGCTGAGTTTC AAATTCTGGATAATGTAGACTCAACGGGAGAGTTGATAGTGAGATTACCCAAAGAAATAACAATTTCAGGCAGTTTCCAGGGCTTCCACC ATCAGAAAATCAAGATATCGGAGAACCGGATATCCCAGCAGTATCTGGCTACCCTTGAAAACAGGAAGCTGAAGAGGGAACTACCCTTTT CATTCCGATCAATTAATACGAGAGAAAACCTGTATCTGGTGACAGAAACTCTGGAGACGGTAAAGGAGGAAACCCTGAAAAGCGACCGGC AATATAAATTTTGGAGCCAGATCTCTCAGGGCCATCTCAGCTATAAACACAAGGGTTTTCTCCAGGGATTTAAGGGCCAGCTCATACACA CATACAACAAGAACAGCTCTGCGTGTGAGAACTCTGGTTACTTCCAGCAACTTGAGGGCAAAACCAATGTCATCCTGCTGGGAGACTCTA TCGGGGACCTCACCATGGCCGATGGGGTTCCTGGTGTGCAGAACATTCTCAAAATTGGCTTCCTGAATGACAAGGTGGAGGAGCGGCGGG AGCGCTACATGGACTCCTATGACATCGTGCTGGAGAAGGACGAGACTCTGGATGTGGTCAACGGGCTACTGCAGCACATCCTGTGCCAGG GGGTCCAGCTGGAGATGCAAGGCCCCTGAAGGCGCAGGCTCCAGCCCGGCCTGCAGGCCGTGGTGAGGAGGGGCGCCTCCCCAGAGTCTG CTCCCCCGTGAACACAGAGCAGAGGCCAGGGTGGCCAGCAGTGGCTGGGTCCTTCCGCGCCCCTCCGTCCTCCTTTCCCTGAGCACCTTC ATCACCAGAGGCTTGAAGGAACCCCGCCATGTGGCAGGGCACAGGCACTGTTCCTGGTGAACCTTGGACCACAGCATGTCAGTGCTCTAG GGATTGTCTACTCCAGGGATTTTCTTCAAAATTTTTAAACATGGGAAGTTCAAACAAATATAATGTGTGAAACAGATCAAAATTTTTAAA ATGAAAAAAAAGCTGCTCTGATTCAGGGGATGTGGGTGGGGGTAGAACCTGGACCTCTTGGCCCTGGGGGCACATGGGATGCTTCTAGGA >34988_34988_6_GSDMB-NT5C3B_GSDMB_chr17_38066009_ENST00000520542_NT5C3B_chr17_39983878_ENST00000269534_length(amino acids)=303AA_BP=192 MFSVFEEITRIVVKEMDAGGDMIAVRSLVDADRFRCFHLVGEKRTFFGCRHYTTGLTLMDILDTDGDKWLDELDSGLQGQKAEFQILDNV DSTGELIVRLPKEITISGSFQGFHHQKIKISENRISQQYLATLENRKLKRELPFSFRSINTRENLYLVTETLETVKEETLKSDRQYKFWS QISQGHLSYKHKGFLQGFKGQLIHTYNKNSSACENSGYFQQLEGKTNVILLGDSIGDLTMADGVPGVQNILKIGFLNDKVEERRERYMDS -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for GSDMB-NT5C3B |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for GSDMB-NT5C3B |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for GSDMB-NT5C3B |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
