
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:GSE1-NOTCH3 (FusionGDB2 ID:35011) |
Fusion Gene Summary for GSE1-NOTCH3 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: GSE1-NOTCH3 | Fusion gene ID: 35011 | Hgene | Tgene | Gene symbol | GSE1 | NOTCH3 | Gene ID | 23199 | 4854 |
| Gene name | Gse1 coiled-coil protein | notch receptor 3 | |
| Synonyms | CRHSP24|KIAA0182 | CADASIL|CADASIL1|CASIL|IMF2|LMNS | |
| Cytomap | 16q24.1 | 19p13.12 | |
| Type of gene | protein-coding | protein-coding | |
| Description | genetic suppressor element 1CTC-786C10.1Gse1 coiled-coil protein homolog | neurogenic locus notch homolog protein 3Notch homolog 3notch 3 | |
| Modification date | 20200327 | 20200329 | |
| UniProtAcc | Q14687 | Q9UM47 | |
| Ensembl transtripts involved in fusion gene | ENST00000253458, ENST00000393243, ENST00000405402, ENST00000471070, | ENST00000263388, | |
| Fusion gene scores | * DoF score | 23 X 18 X 8=3312 | 4 X 5 X 5=100 |
| # samples | 23 | 6 | |
| ** MAII score | log2(23/3312*10)=-3.84799690655495 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(6/100*10)=-0.736965594166206 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: GSE1 [Title/Abstract] AND NOTCH3 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | GSE1(85647004)-NOTCH3(15276902), # samples:2 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
 Fusion gene breakpoints across GSE1 (5'-gene) Fusion gene breakpoints across GSE1 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
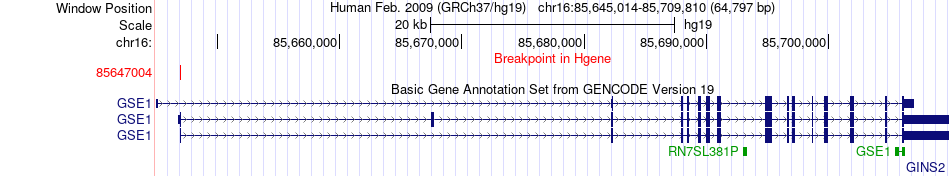 |
 Fusion gene breakpoints across NOTCH3 (3'-gene) Fusion gene breakpoints across NOTCH3 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
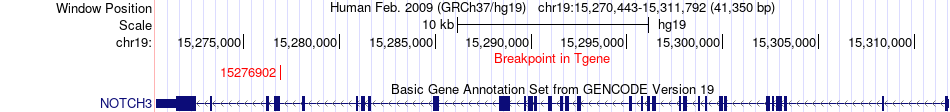 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | LUAD | TCGA-05-4427-01A | GSE1 | chr16 | 85647004 | + | NOTCH3 | chr19 | 15276902 | - |
Top |
Fusion Gene ORF analysis for GSE1-NOTCH3 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| In-frame | ENST00000253458 | ENST00000263388 | GSE1 | chr16 | 85647004 | + | NOTCH3 | chr19 | 15276902 | - |
| In-frame | ENST00000393243 | ENST00000263388 | GSE1 | chr16 | 85647004 | + | NOTCH3 | chr19 | 15276902 | - |
| intron-3CDS | ENST00000405402 | ENST00000263388 | GSE1 | chr16 | 85647004 | + | NOTCH3 | chr19 | 15276902 | - |
| intron-3CDS | ENST00000471070 | ENST00000263388 | GSE1 | chr16 | 85647004 | + | NOTCH3 | chr19 | 15276902 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000253458 | GSE1 | chr16 | 85647004 | + | ENST00000263388 | NOTCH3 | chr19 | 15276902 | - | 2816 | 183 | 161 | 1786 | 541 |
| ENST00000393243 | GSE1 | chr16 | 85647004 | + | ENST00000263388 | NOTCH3 | chr19 | 15276902 | - | 2680 | 47 | 25 | 1650 | 541 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000253458 | ENST00000263388 | GSE1 | chr16 | 85647004 | + | NOTCH3 | chr19 | 15276902 | - | 0.030220913 | 0.96977913 |
| ENST00000393243 | ENST00000263388 | GSE1 | chr16 | 85647004 | + | NOTCH3 | chr19 | 15276902 | - | 0.028835285 | 0.9711647 |
Top |
Fusion Genomic Features for GSE1-NOTCH3 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
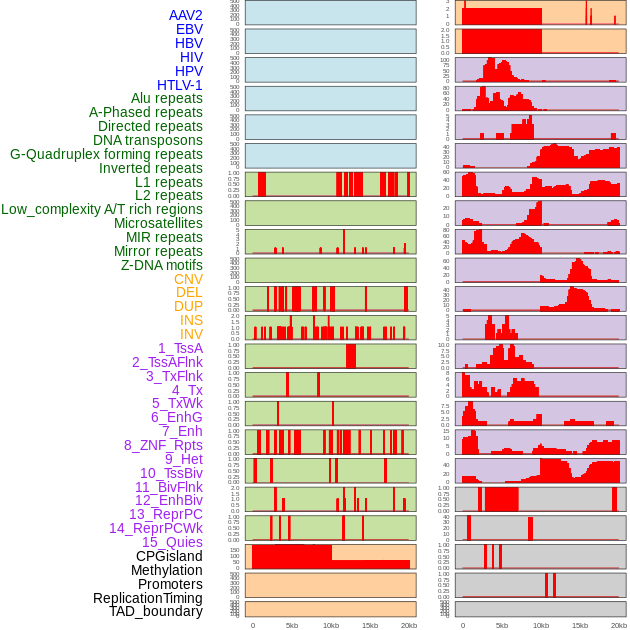 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for GSE1-NOTCH3 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr16:85647004/chr19:15276902) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| GSE1 | NOTCH3 |
| FUNCTION: Functions as a receptor for membrane-bound ligands Jagged1, Jagged2 and Delta1 to regulate cell-fate determination (PubMed:15350543). Upon ligand activation through the released notch intracellular domain (NICD) it forms a transcriptional activator complex with RBPJ/RBPSUH and activates genes of the enhancer of split locus. Affects the implementation of differentiation, proliferation and apoptotic programs (By similarity). {ECO:0000250|UniProtKB:Q9R172, ECO:0000269|PubMed:15350543}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | NOTCH3 | chr16:85647004 | chr19:15276902 | ENST00000263388 | 28 | 33 | 1838_1867 | 1787 | 2322.0 | Repeat | Note=ANK 1 | |
| Tgene | NOTCH3 | chr16:85647004 | chr19:15276902 | ENST00000263388 | 28 | 33 | 1871_1901 | 1787 | 2322.0 | Repeat | Note=ANK 2 | |
| Tgene | NOTCH3 | chr16:85647004 | chr19:15276902 | ENST00000263388 | 28 | 33 | 1905_1934 | 1787 | 2322.0 | Repeat | Note=ANK 3 | |
| Tgene | NOTCH3 | chr16:85647004 | chr19:15276902 | ENST00000263388 | 28 | 33 | 1938_1967 | 1787 | 2322.0 | Repeat | Note=ANK 4 | |
| Tgene | NOTCH3 | chr16:85647004 | chr19:15276902 | ENST00000263388 | 28 | 33 | 1971_2000 | 1787 | 2322.0 | Repeat | Note=ANK 5 |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | GSE1 | chr16:85647004 | chr19:15276902 | ENST00000253458 | + | 1 | 16 | 1127_1201 | 2 | 1218.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | GSE1 | chr16:85647004 | chr19:15276902 | ENST00000253458 | + | 1 | 16 | 321_403 | 2 | 1218.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | GSE1 | chr16:85647004 | chr19:15276902 | ENST00000393243 | + | 1 | 15 | 1127_1201 | 2 | 1145.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | GSE1 | chr16:85647004 | chr19:15276902 | ENST00000393243 | + | 1 | 15 | 321_403 | 2 | 1145.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | GSE1 | chr16:85647004 | chr19:15276902 | ENST00000405402 | + | 1 | 15 | 1127_1201 | 0 | 1114.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | GSE1 | chr16:85647004 | chr19:15276902 | ENST00000405402 | + | 1 | 15 | 321_403 | 0 | 1114.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | GSE1 | chr16:85647004 | chr19:15276902 | ENST00000253458 | + | 1 | 16 | 1102_1107 | 2 | 1218.0 | Compositional bias | Note=Poly-Glu |
| Hgene | GSE1 | chr16:85647004 | chr19:15276902 | ENST00000253458 | + | 1 | 16 | 532_722 | 2 | 1218.0 | Compositional bias | Note=Pro-rich |
| Hgene | GSE1 | chr16:85647004 | chr19:15276902 | ENST00000253458 | + | 1 | 16 | 745_748 | 2 | 1218.0 | Compositional bias | Note=Poly-Arg |
| Hgene | GSE1 | chr16:85647004 | chr19:15276902 | ENST00000393243 | + | 1 | 15 | 1102_1107 | 2 | 1145.0 | Compositional bias | Note=Poly-Glu |
| Hgene | GSE1 | chr16:85647004 | chr19:15276902 | ENST00000393243 | + | 1 | 15 | 532_722 | 2 | 1145.0 | Compositional bias | Note=Pro-rich |
| Hgene | GSE1 | chr16:85647004 | chr19:15276902 | ENST00000393243 | + | 1 | 15 | 745_748 | 2 | 1145.0 | Compositional bias | Note=Poly-Arg |
| Hgene | GSE1 | chr16:85647004 | chr19:15276902 | ENST00000405402 | + | 1 | 15 | 1102_1107 | 0 | 1114.0 | Compositional bias | Note=Poly-Glu |
| Hgene | GSE1 | chr16:85647004 | chr19:15276902 | ENST00000405402 | + | 1 | 15 | 532_722 | 0 | 1114.0 | Compositional bias | Note=Pro-rich |
| Hgene | GSE1 | chr16:85647004 | chr19:15276902 | ENST00000405402 | + | 1 | 15 | 745_748 | 0 | 1114.0 | Compositional bias | Note=Poly-Arg |
| Tgene | NOTCH3 | chr16:85647004 | chr19:15276902 | ENST00000263388 | 28 | 33 | 1000_1034 | 1787 | 2322.0 | Domain | EGF-like 26 | |
| Tgene | NOTCH3 | chr16:85647004 | chr19:15276902 | ENST00000263388 | 28 | 33 | 1036_1082 | 1787 | 2322.0 | Domain | EGF-like 27 | |
| Tgene | NOTCH3 | chr16:85647004 | chr19:15276902 | ENST00000263388 | 28 | 33 | 1084_1120 | 1787 | 2322.0 | Domain | EGF-like 28 | |
| Tgene | NOTCH3 | chr16:85647004 | chr19:15276902 | ENST00000263388 | 28 | 33 | 1122_1158 | 1787 | 2322.0 | Domain | EGF-like 29%3B calcium-binding | |
| Tgene | NOTCH3 | chr16:85647004 | chr19:15276902 | ENST00000263388 | 28 | 33 | 1160_1203 | 1787 | 2322.0 | Domain | EGF-like 30%3B calcium-binding | |
| Tgene | NOTCH3 | chr16:85647004 | chr19:15276902 | ENST00000263388 | 28 | 33 | 119_156 | 1787 | 2322.0 | Domain | EGF-like 3 | |
| Tgene | NOTCH3 | chr16:85647004 | chr19:15276902 | ENST00000263388 | 28 | 33 | 1205_1244 | 1787 | 2322.0 | Domain | EGF-like 31 | |
| Tgene | NOTCH3 | chr16:85647004 | chr19:15276902 | ENST00000263388 | 28 | 33 | 1246_1287 | 1787 | 2322.0 | Domain | EGF-like 32 | |
| Tgene | NOTCH3 | chr16:85647004 | chr19:15276902 | ENST00000263388 | 28 | 33 | 1289_1325 | 1787 | 2322.0 | Domain | EGF-like 33 | |
| Tgene | NOTCH3 | chr16:85647004 | chr19:15276902 | ENST00000263388 | 28 | 33 | 1335_1373 | 1787 | 2322.0 | Domain | EGF-like 34 | |
| Tgene | NOTCH3 | chr16:85647004 | chr19:15276902 | ENST00000263388 | 28 | 33 | 158_195 | 1787 | 2322.0 | Domain | EGF-like 4%3B calcium-binding | |
| Tgene | NOTCH3 | chr16:85647004 | chr19:15276902 | ENST00000263388 | 28 | 33 | 197_234 | 1787 | 2322.0 | Domain | EGF-like 5 | |
| Tgene | NOTCH3 | chr16:85647004 | chr19:15276902 | ENST00000263388 | 28 | 33 | 236_272 | 1787 | 2322.0 | Domain | EGF-like 6%3B calcium-binding | |
| Tgene | NOTCH3 | chr16:85647004 | chr19:15276902 | ENST00000263388 | 28 | 33 | 274_312 | 1787 | 2322.0 | Domain | EGF-like 7 | |
| Tgene | NOTCH3 | chr16:85647004 | chr19:15276902 | ENST00000263388 | 28 | 33 | 314_350 | 1787 | 2322.0 | Domain | EGF-like 8%3B calcium-binding | |
| Tgene | NOTCH3 | chr16:85647004 | chr19:15276902 | ENST00000263388 | 28 | 33 | 351_389 | 1787 | 2322.0 | Domain | EGF-like 9 | |
| Tgene | NOTCH3 | chr16:85647004 | chr19:15276902 | ENST00000263388 | 28 | 33 | 391_429 | 1787 | 2322.0 | Domain | EGF-like 10%3B calcium-binding | |
| Tgene | NOTCH3 | chr16:85647004 | chr19:15276902 | ENST00000263388 | 28 | 33 | 40_77 | 1787 | 2322.0 | Domain | EGF-like 1 | |
| Tgene | NOTCH3 | chr16:85647004 | chr19:15276902 | ENST00000263388 | 28 | 33 | 431_467 | 1787 | 2322.0 | Domain | EGF-like 11%3B calcium-binding | |
| Tgene | NOTCH3 | chr16:85647004 | chr19:15276902 | ENST00000263388 | 28 | 33 | 469_505 | 1787 | 2322.0 | Domain | EGF-like 12%3B calcium-binding | |
| Tgene | NOTCH3 | chr16:85647004 | chr19:15276902 | ENST00000263388 | 28 | 33 | 507_543 | 1787 | 2322.0 | Domain | EGF-like 13%3B calcium-binding | |
| Tgene | NOTCH3 | chr16:85647004 | chr19:15276902 | ENST00000263388 | 28 | 33 | 545_580 | 1787 | 2322.0 | Domain | EGF-like 14%3B calcium-binding | |
| Tgene | NOTCH3 | chr16:85647004 | chr19:15276902 | ENST00000263388 | 28 | 33 | 582_618 | 1787 | 2322.0 | Domain | EGF-like 15%3B calcium-binding | |
| Tgene | NOTCH3 | chr16:85647004 | chr19:15276902 | ENST00000263388 | 28 | 33 | 620_655 | 1787 | 2322.0 | Domain | EGF-like 16%3B calcium-binding | |
| Tgene | NOTCH3 | chr16:85647004 | chr19:15276902 | ENST00000263388 | 28 | 33 | 657_693 | 1787 | 2322.0 | Domain | EGF-like 17%3B calcium-binding | |
| Tgene | NOTCH3 | chr16:85647004 | chr19:15276902 | ENST00000263388 | 28 | 33 | 695_730 | 1787 | 2322.0 | Domain | EGF-like 18 | |
| Tgene | NOTCH3 | chr16:85647004 | chr19:15276902 | ENST00000263388 | 28 | 33 | 734_770 | 1787 | 2322.0 | Domain | EGF-like 19 | |
| Tgene | NOTCH3 | chr16:85647004 | chr19:15276902 | ENST00000263388 | 28 | 33 | 771_808 | 1787 | 2322.0 | Domain | EGF-like 20 | |
| Tgene | NOTCH3 | chr16:85647004 | chr19:15276902 | ENST00000263388 | 28 | 33 | 78_118 | 1787 | 2322.0 | Domain | EGF-like 2 | |
| Tgene | NOTCH3 | chr16:85647004 | chr19:15276902 | ENST00000263388 | 28 | 33 | 810_847 | 1787 | 2322.0 | Domain | EGF-like 21%3B calcium-binding | |
| Tgene | NOTCH3 | chr16:85647004 | chr19:15276902 | ENST00000263388 | 28 | 33 | 849_885 | 1787 | 2322.0 | Domain | EGF-like 22%3B calcium-binding | |
| Tgene | NOTCH3 | chr16:85647004 | chr19:15276902 | ENST00000263388 | 28 | 33 | 887_922 | 1787 | 2322.0 | Domain | EGF-like 23%3B calcium-binding | |
| Tgene | NOTCH3 | chr16:85647004 | chr19:15276902 | ENST00000263388 | 28 | 33 | 924_960 | 1787 | 2322.0 | Domain | EGF-like 24 | |
| Tgene | NOTCH3 | chr16:85647004 | chr19:15276902 | ENST00000263388 | 28 | 33 | 962_998 | 1787 | 2322.0 | Domain | EGF-like 25 | |
| Tgene | NOTCH3 | chr16:85647004 | chr19:15276902 | ENST00000263388 | 28 | 33 | 1387_1427 | 1787 | 2322.0 | Repeat | Note=LNR 1 | |
| Tgene | NOTCH3 | chr16:85647004 | chr19:15276902 | ENST00000263388 | 28 | 33 | 1428_1458 | 1787 | 2322.0 | Repeat | Note=LNR 2 | |
| Tgene | NOTCH3 | chr16:85647004 | chr19:15276902 | ENST00000263388 | 28 | 33 | 1467_1505 | 1787 | 2322.0 | Repeat | Note=LNR 3 | |
| Tgene | NOTCH3 | chr16:85647004 | chr19:15276902 | ENST00000263388 | 28 | 33 | 1665_2321 | 1787 | 2322.0 | Topological domain | Cytoplasmic | |
| Tgene | NOTCH3 | chr16:85647004 | chr19:15276902 | ENST00000263388 | 28 | 33 | 40_1643 | 1787 | 2322.0 | Topological domain | Extracellular | |
| Tgene | NOTCH3 | chr16:85647004 | chr19:15276902 | ENST00000263388 | 28 | 33 | 1644_1664 | 1787 | 2322.0 | Transmembrane | Helical |
Top |
Fusion Gene Sequence for GSE1-NOTCH3 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >35011_35011_1_GSE1-NOTCH3_GSE1_chr16_85647004_ENST00000253458_NOTCH3_chr19_15276902_ENST00000263388_length(transcript)=2816nt_BP=183nt TCCTCGGCGGCGACAGCAGCAGGTGTTTCTTGGCCGGGGCCCCGGAAGCTCCACCTCCCCAGCGGGCCCGAGCTGCCGCCGCCGAGCAGC CCCGGGTGAGATAAGCAGTTTAGACAAACACTGGGCGACGGTGGCTCCAGCATGTATCAGCCGAGGTGGAGCTGCGGGGCCCTGGCATGA AAGATGGCTTCACCCCGCTAATGCTGGCTTCCTTCTGTGGGGGGGCTCTGGAGCCAATGCCAACTGAAGAGGATGAGGCAGATGACACAT CAGCTAGCATCATCTCCGACCTGATCTGCCAGGGGGCTCAGCTTGGGGCACGGACTGACCGTACTGGCGAGACTGCTTTGCACCTGGCTG CCCGTTATGCCCGTGCTGATGCAGCCAAGCGGCTGCTGGATGCTGGGGCAGACACCAATGCCCAGGACCACTCAGGCCGCACTCCCCTGC ACACAGCTGTCACAGCCGATGCCCAGGGTGTCTTCCAGATTCTCATCCGAAACCGCTCTACAGACTTGGATGCCCGCATGGCAGATGGCT CAACGGCACTGATCCTGGCGGCCCGCCTGGCAGTAGAGGGCATGGTGGAAGAGCTCATCGCCAGCCATGCTGATGTCAATGCTGTGGATG AGCTTGGGAAATCAGCCTTACACTGGGCTGCGGCTGTGAACAACGTGGAAGCCACTTTGGCCCTGCTCAAAAATGGAGCCAATAAGGACA TGCAGGATAGCAAGGAGGAGACCCCCCTATTCCTGGCCGCCCGCGAGGGCAGCTATGAGGCTGCCAAGCTGCTGTTGGACCACTTTGCCA ACCGTGAGATCACCGACCACCTGGACAGGCTGCCGCGGGACGTAGCCCAGGAGAGACTGCACCAGGACATCGTGCGCTTGCTGGATCAAC CCAGTGGGCCCCGCAGCCCCCCCGGTCCCCACGGCCTGGGGCCTCTGCTCTGTCCTCCAGGGGCCTTCCTCCCTGGCCTCAAAGCGGCAC AGTCGGGGTCCAAGAAGAGCAGGAGGCCCCCCGGGAAGGCGGGGCTGGGGCCGCAGGGGCCCCGGGGGCGGGGCAAGAAGCTGACGCTGG CCTGCCCGGGCCCCCTGGCTGACAGCTCGGTCACGCTGTCGCCCGTGGACTCGCTGGACTCCCCGCGGCCTTTCGGTGGGCCCCCTGCTT CCCCTGGTGGCTTCCCCCTTGAGGGGCCCTATGCAGCTGCCACTGCCACTGCAGTGTCTCTGGCACAGCTTGGTGGCCCAGGCCGGGCGG GTCTAGGGCGCCAGCCCCCTGGAGGATGTGTACTCAGCCTGGGCCTGCTGAACCCTGTGGCTGTGCCCCTCGATTGGGCCCGGCTGCCCC CACCTGCCCCTCCAGGCCCCTCGTTCCTGCTGCCACTGGCGCCGGGACCCCAGCTGCTCAACCCAGGGACCCCCGTCTCCCCGCAGGAGC GGCCCCCGCCTTACCTGGCAGTCCCAGGACATGGCGAGGAGTACCCGGCGGCTGGGGCACACAGCAGCCCCCCAAAGGCCCGCTTCCTGC GGGTTCCCAGTGAGCACCCTTACCTGACCCCATCCCCCGAATCCCCTGAGCACTGGGCCAGCCCCTCACCTCCCTCCCTCTCAGACTGGT CCGAATCCACGCCTAGCCCAGCCACTGCCACTGGGGCCATGGCCACCACCACTGGGGCACTGCCTGCCCAGCCACTTCCCTTGTCTGTTC CCAGCTCCCTTGCTCAGGCCCAGACCCAGCTGGGGCCCCAGCCGGAAGTTACCCCCAAGAGGCAAGTGTTGGCCTGAGACGCTCGTCAGT TCTTAGATCTTGGGGGCCTAAAGAGACCCCCGTCCTGCCTCCTTTCTTTCTCTGTCTCTTCCTTCCTTTTAGTCTTTTTCATCCTCTTCT CTTTCCACCAACCCTCCTGCATCCTTGCCTTGCAGCGTGACCGAGATAGGTCATCAGCCCAGGGCTTCAGTCTTCCTTTATTTATAATGG GTGGGGGCTACCACCCACCCTCTCAGTCTTGTGAAGAGTCTGGGACCTCCTTCTTCCCCACTTCTCTCTTCCCTCATTCCTTTCTCTCTC CTTCTGGCCTCTCATTTCCTTACACTCTGACATGAATGAATTATTATTATTTTTATTTTTCTTTTTTTTTTTACATTTTGTATAGAAACA AATTCATTTAAACAAACTTATTATTATTATTTTTTACAAAATATATATATGGAGATGCTCCCTCCCCCTGTGAACCCCCCAGTGCCCCCG TGGGGCTGAGTCTGTGGGCCCATTCGGCCAAGCTGGATTCTGTGTACCTAGTACACAGGCATGACTGGGATCCCGTGTACCGAGTACACG ACCCAGGTATGTACCAAGTAGGCACCCTTGGGCGCACCCACTGGGGCCAGGGGTCGGGGGAGTGTTGGGAGCCTCCTCCCCACCCCACCT CCCTCACTTCACTGCATTCCAGATGGGACATGTTCCATAGCCTTGCTGGGGAAGGGCCCACTGCCAACTCCCTCTGCCCCAGCCCCACCC TTGGCCATCTCCCTTTGGGAACTAGGGGGCTGCTGGTGGGAAATGGGAGCCAGGGCAGATGTATGCATTCCTTTGTGTCCCTGTAAATGT GGGACTACAAGAAGAGGAGCTGCCTGAGTGGTACTTTCTCTTCCTGGTAATCCTCTGGCCCAGCCTCATGGCAGAATAGAGGTATTTTTA GGCTATTTTTGTAATATGGCTTCTGGTCAAAATCCCTGTGTAGCTGAATTCCCAAGCCCTGCATTGTACAGCCCCCCACTCCCCTCACCA >35011_35011_1_GSE1-NOTCH3_GSE1_chr16_85647004_ENST00000253458_NOTCH3_chr19_15276902_ENST00000263388_length(amino acids)=541AA_BP=7 MRGPGMKDGFTPLMLASFCGGALEPMPTEEDEADDTSASIISDLICQGAQLGARTDRTGETALHLAARYARADAAKRLLDAGADTNAQDH SGRTPLHTAVTADAQGVFQILIRNRSTDLDARMADGSTALILAARLAVEGMVEELIASHADVNAVDELGKSALHWAAAVNNVEATLALLK NGANKDMQDSKEETPLFLAAREGSYEAAKLLLDHFANREITDHLDRLPRDVAQERLHQDIVRLLDQPSGPRSPPGPHGLGPLLCPPGAFL PGLKAAQSGSKKSRRPPGKAGLGPQGPRGRGKKLTLACPGPLADSSVTLSPVDSLDSPRPFGGPPASPGGFPLEGPYAAATATAVSLAQL GGPGRAGLGRQPPGGCVLSLGLLNPVAVPLDWARLPPPAPPGPSFLLPLAPGPQLLNPGTPVSPQERPPPYLAVPGHGEEYPAAGAHSSP PKARFLRVPSEHPYLTPSPESPEHWASPSPPSLSDWSESTPSPATATGAMATTTGALPAQPLPLSVPSSLAQAQTQLGPQPEVTPKRQVL -------------------------------------------------------------- >35011_35011_2_GSE1-NOTCH3_GSE1_chr16_85647004_ENST00000393243_NOTCH3_chr19_15276902_ENST00000263388_length(transcript)=2680nt_BP=47nt CCAGCATGTATCAGCCGAGGTGGAGCTGCGGGGCCCTGGCATGAAAGATGGCTTCACCCCGCTAATGCTGGCTTCCTTCTGTGGGGGGGC TCTGGAGCCAATGCCAACTGAAGAGGATGAGGCAGATGACACATCAGCTAGCATCATCTCCGACCTGATCTGCCAGGGGGCTCAGCTTGG GGCACGGACTGACCGTACTGGCGAGACTGCTTTGCACCTGGCTGCCCGTTATGCCCGTGCTGATGCAGCCAAGCGGCTGCTGGATGCTGG GGCAGACACCAATGCCCAGGACCACTCAGGCCGCACTCCCCTGCACACAGCTGTCACAGCCGATGCCCAGGGTGTCTTCCAGATTCTCAT CCGAAACCGCTCTACAGACTTGGATGCCCGCATGGCAGATGGCTCAACGGCACTGATCCTGGCGGCCCGCCTGGCAGTAGAGGGCATGGT GGAAGAGCTCATCGCCAGCCATGCTGATGTCAATGCTGTGGATGAGCTTGGGAAATCAGCCTTACACTGGGCTGCGGCTGTGAACAACGT GGAAGCCACTTTGGCCCTGCTCAAAAATGGAGCCAATAAGGACATGCAGGATAGCAAGGAGGAGACCCCCCTATTCCTGGCCGCCCGCGA GGGCAGCTATGAGGCTGCCAAGCTGCTGTTGGACCACTTTGCCAACCGTGAGATCACCGACCACCTGGACAGGCTGCCGCGGGACGTAGC CCAGGAGAGACTGCACCAGGACATCGTGCGCTTGCTGGATCAACCCAGTGGGCCCCGCAGCCCCCCCGGTCCCCACGGCCTGGGGCCTCT GCTCTGTCCTCCAGGGGCCTTCCTCCCTGGCCTCAAAGCGGCACAGTCGGGGTCCAAGAAGAGCAGGAGGCCCCCCGGGAAGGCGGGGCT GGGGCCGCAGGGGCCCCGGGGGCGGGGCAAGAAGCTGACGCTGGCCTGCCCGGGCCCCCTGGCTGACAGCTCGGTCACGCTGTCGCCCGT GGACTCGCTGGACTCCCCGCGGCCTTTCGGTGGGCCCCCTGCTTCCCCTGGTGGCTTCCCCCTTGAGGGGCCCTATGCAGCTGCCACTGC CACTGCAGTGTCTCTGGCACAGCTTGGTGGCCCAGGCCGGGCGGGTCTAGGGCGCCAGCCCCCTGGAGGATGTGTACTCAGCCTGGGCCT GCTGAACCCTGTGGCTGTGCCCCTCGATTGGGCCCGGCTGCCCCCACCTGCCCCTCCAGGCCCCTCGTTCCTGCTGCCACTGGCGCCGGG ACCCCAGCTGCTCAACCCAGGGACCCCCGTCTCCCCGCAGGAGCGGCCCCCGCCTTACCTGGCAGTCCCAGGACATGGCGAGGAGTACCC GGCGGCTGGGGCACACAGCAGCCCCCCAAAGGCCCGCTTCCTGCGGGTTCCCAGTGAGCACCCTTACCTGACCCCATCCCCCGAATCCCC TGAGCACTGGGCCAGCCCCTCACCTCCCTCCCTCTCAGACTGGTCCGAATCCACGCCTAGCCCAGCCACTGCCACTGGGGCCATGGCCAC CACCACTGGGGCACTGCCTGCCCAGCCACTTCCCTTGTCTGTTCCCAGCTCCCTTGCTCAGGCCCAGACCCAGCTGGGGCCCCAGCCGGA AGTTACCCCCAAGAGGCAAGTGTTGGCCTGAGACGCTCGTCAGTTCTTAGATCTTGGGGGCCTAAAGAGACCCCCGTCCTGCCTCCTTTC TTTCTCTGTCTCTTCCTTCCTTTTAGTCTTTTTCATCCTCTTCTCTTTCCACCAACCCTCCTGCATCCTTGCCTTGCAGCGTGACCGAGA TAGGTCATCAGCCCAGGGCTTCAGTCTTCCTTTATTTATAATGGGTGGGGGCTACCACCCACCCTCTCAGTCTTGTGAAGAGTCTGGGAC CTCCTTCTTCCCCACTTCTCTCTTCCCTCATTCCTTTCTCTCTCCTTCTGGCCTCTCATTTCCTTACACTCTGACATGAATGAATTATTA TTATTTTTATTTTTCTTTTTTTTTTTACATTTTGTATAGAAACAAATTCATTTAAACAAACTTATTATTATTATTTTTTACAAAATATAT ATATGGAGATGCTCCCTCCCCCTGTGAACCCCCCAGTGCCCCCGTGGGGCTGAGTCTGTGGGCCCATTCGGCCAAGCTGGATTCTGTGTA CCTAGTACACAGGCATGACTGGGATCCCGTGTACCGAGTACACGACCCAGGTATGTACCAAGTAGGCACCCTTGGGCGCACCCACTGGGG CCAGGGGTCGGGGGAGTGTTGGGAGCCTCCTCCCCACCCCACCTCCCTCACTTCACTGCATTCCAGATGGGACATGTTCCATAGCCTTGC TGGGGAAGGGCCCACTGCCAACTCCCTCTGCCCCAGCCCCACCCTTGGCCATCTCCCTTTGGGAACTAGGGGGCTGCTGGTGGGAAATGG GAGCCAGGGCAGATGTATGCATTCCTTTGTGTCCCTGTAAATGTGGGACTACAAGAAGAGGAGCTGCCTGAGTGGTACTTTCTCTTCCTG GTAATCCTCTGGCCCAGCCTCATGGCAGAATAGAGGTATTTTTAGGCTATTTTTGTAATATGGCTTCTGGTCAAAATCCCTGTGTAGCTG >35011_35011_2_GSE1-NOTCH3_GSE1_chr16_85647004_ENST00000393243_NOTCH3_chr19_15276902_ENST00000263388_length(amino acids)=541AA_BP=7 MRGPGMKDGFTPLMLASFCGGALEPMPTEEDEADDTSASIISDLICQGAQLGARTDRTGETALHLAARYARADAAKRLLDAGADTNAQDH SGRTPLHTAVTADAQGVFQILIRNRSTDLDARMADGSTALILAARLAVEGMVEELIASHADVNAVDELGKSALHWAAAVNNVEATLALLK NGANKDMQDSKEETPLFLAAREGSYEAAKLLLDHFANREITDHLDRLPRDVAQERLHQDIVRLLDQPSGPRSPPGPHGLGPLLCPPGAFL PGLKAAQSGSKKSRRPPGKAGLGPQGPRGRGKKLTLACPGPLADSSVTLSPVDSLDSPRPFGGPPASPGGFPLEGPYAAATATAVSLAQL GGPGRAGLGRQPPGGCVLSLGLLNPVAVPLDWARLPPPAPPGPSFLLPLAPGPQLLNPGTPVSPQERPPPYLAVPGHGEEYPAAGAHSSP PKARFLRVPSEHPYLTPSPESPEHWASPSPPSLSDWSESTPSPATATGAMATTTGALPAQPLPLSVPSSLAQAQTQLGPQPEVTPKRQVL -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for GSE1-NOTCH3 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for GSE1-NOTCH3 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for GSE1-NOTCH3 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
