
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:GUK1-TARBP1 (FusionGDB2 ID:35368) |
Fusion Gene Summary for GUK1-TARBP1 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: GUK1-TARBP1 | Fusion gene ID: 35368 | Hgene | Tgene | Gene symbol | GUK1 | TARBP1 | Gene ID | 2987 | 6894 |
| Gene name | guanylate kinase 1 | TAR (HIV-1) RNA binding protein 1 | |
| Synonyms | GMK | TRM3|TRMT3|TRP-185|TRP185 | |
| Cytomap | 1q42.13 | 1q42.2 | |
| Type of gene | protein-coding | protein-coding | |
| Description | guanylate kinaseATP:GMP phosphotransferaseGMP kinase | probable methyltransferase TARBP1TAR (HIV) RNA-binding protein 1TAR RNA loop binding proteinTAR RNA-binding protein 1TAR RNA-binding protein of 185 kDatRNA methyltransferase 3 homolog | |
| Modification date | 20200320 | 20200313 | |
| UniProtAcc | Q16774 | . | |
| Ensembl transtripts involved in fusion gene | ENST00000366723, ENST00000366728, ENST00000391865, ENST00000312726, ENST00000366721, ENST00000366722, ENST00000366726, ENST00000366716, ENST00000366718, ENST00000366730, ENST00000470040, | ENST00000483404, ENST00000040877, | |
| Fusion gene scores | * DoF score | 14 X 13 X 6=1092 | 3 X 3 X 3=27 |
| # samples | 16 | 3 | |
| ** MAII score | log2(16/1092*10)=-2.77082904603249 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(3/27*10)=0.15200309344505 effective Gene in Pan-Cancer Fusion Genes (eGinPCFGs). DoF>8 and MAII>0 | |
| Context | PubMed: GUK1 [Title/Abstract] AND TARBP1 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | GUK1(228328064)-TARBP1(234603396), # samples:2 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | GUK1 | GO:0006163 | purine nucleotide metabolic process | 8663313 |
| Hgene | GUK1 | GO:0006805 | xenobiotic metabolic process | 6306664 |
 Fusion gene breakpoints across GUK1 (5'-gene) Fusion gene breakpoints across GUK1 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
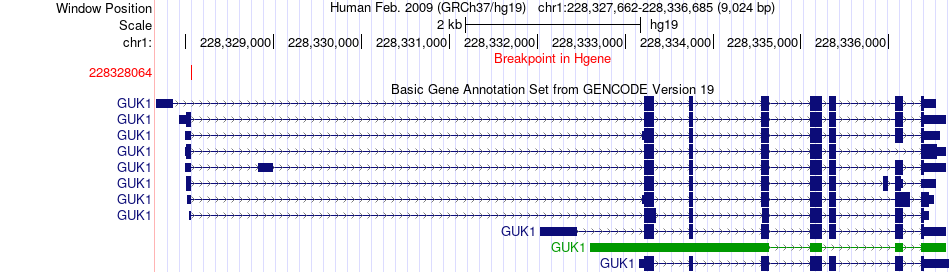 |
 Fusion gene breakpoints across TARBP1 (3'-gene) Fusion gene breakpoints across TARBP1 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
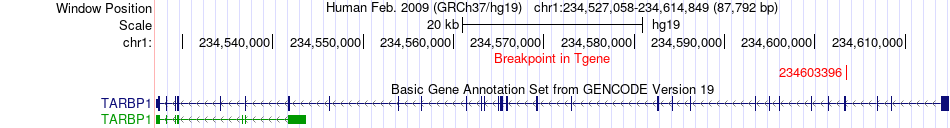 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | BRCA | TCGA-GM-A3XL-01A | GUK1 | chr1 | 228328064 | + | TARBP1 | chr1 | 234603396 | - |
Top |
Fusion Gene ORF analysis for GUK1-TARBP1 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-intron | ENST00000366723 | ENST00000483404 | GUK1 | chr1 | 228328064 | + | TARBP1 | chr1 | 234603396 | - |
| 5CDS-intron | ENST00000366728 | ENST00000483404 | GUK1 | chr1 | 228328064 | + | TARBP1 | chr1 | 234603396 | - |
| 5CDS-intron | ENST00000391865 | ENST00000483404 | GUK1 | chr1 | 228328064 | + | TARBP1 | chr1 | 234603396 | - |
| 5UTR-3CDS | ENST00000312726 | ENST00000040877 | GUK1 | chr1 | 228328064 | + | TARBP1 | chr1 | 234603396 | - |
| 5UTR-3CDS | ENST00000366721 | ENST00000040877 | GUK1 | chr1 | 228328064 | + | TARBP1 | chr1 | 234603396 | - |
| 5UTR-3CDS | ENST00000366722 | ENST00000040877 | GUK1 | chr1 | 228328064 | + | TARBP1 | chr1 | 234603396 | - |
| 5UTR-3CDS | ENST00000366726 | ENST00000040877 | GUK1 | chr1 | 228328064 | + | TARBP1 | chr1 | 234603396 | - |
| 5UTR-intron | ENST00000312726 | ENST00000483404 | GUK1 | chr1 | 228328064 | + | TARBP1 | chr1 | 234603396 | - |
| 5UTR-intron | ENST00000366721 | ENST00000483404 | GUK1 | chr1 | 228328064 | + | TARBP1 | chr1 | 234603396 | - |
| 5UTR-intron | ENST00000366722 | ENST00000483404 | GUK1 | chr1 | 228328064 | + | TARBP1 | chr1 | 234603396 | - |
| 5UTR-intron | ENST00000366726 | ENST00000483404 | GUK1 | chr1 | 228328064 | + | TARBP1 | chr1 | 234603396 | - |
| In-frame | ENST00000366723 | ENST00000040877 | GUK1 | chr1 | 228328064 | + | TARBP1 | chr1 | 234603396 | - |
| In-frame | ENST00000366728 | ENST00000040877 | GUK1 | chr1 | 228328064 | + | TARBP1 | chr1 | 234603396 | - |
| In-frame | ENST00000391865 | ENST00000040877 | GUK1 | chr1 | 228328064 | + | TARBP1 | chr1 | 234603396 | - |
| intron-3CDS | ENST00000366716 | ENST00000040877 | GUK1 | chr1 | 228328064 | + | TARBP1 | chr1 | 234603396 | - |
| intron-3CDS | ENST00000366718 | ENST00000040877 | GUK1 | chr1 | 228328064 | + | TARBP1 | chr1 | 234603396 | - |
| intron-3CDS | ENST00000366730 | ENST00000040877 | GUK1 | chr1 | 228328064 | + | TARBP1 | chr1 | 234603396 | - |
| intron-3CDS | ENST00000470040 | ENST00000040877 | GUK1 | chr1 | 228328064 | + | TARBP1 | chr1 | 234603396 | - |
| intron-intron | ENST00000366716 | ENST00000483404 | GUK1 | chr1 | 228328064 | + | TARBP1 | chr1 | 234603396 | - |
| intron-intron | ENST00000366718 | ENST00000483404 | GUK1 | chr1 | 228328064 | + | TARBP1 | chr1 | 234603396 | - |
| intron-intron | ENST00000366730 | ENST00000483404 | GUK1 | chr1 | 228328064 | + | TARBP1 | chr1 | 234603396 | - |
| intron-intron | ENST00000470040 | ENST00000483404 | GUK1 | chr1 | 228328064 | + | TARBP1 | chr1 | 234603396 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000391865 | GUK1 | chr1 | 228328064 | + | ENST00000040877 | TARBP1 | chr1 | 234603396 | - | 4167 | 136 | 36 | 3902 | 1288 |
| ENST00000366728 | GUK1 | chr1 | 228328064 | + | ENST00000040877 | TARBP1 | chr1 | 234603396 | - | 4105 | 74 | 13 | 3840 | 1275 |
| ENST00000366723 | GUK1 | chr1 | 228328064 | + | ENST00000040877 | TARBP1 | chr1 | 234603396 | - | 4093 | 62 | 1 | 3828 | 1275 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000391865 | ENST00000040877 | GUK1 | chr1 | 228328064 | + | TARBP1 | chr1 | 234603396 | - | 0.000210131 | 0.9997899 |
| ENST00000366728 | ENST00000040877 | GUK1 | chr1 | 228328064 | + | TARBP1 | chr1 | 234603396 | - | 0.000205391 | 0.99979466 |
| ENST00000366723 | ENST00000040877 | GUK1 | chr1 | 228328064 | + | TARBP1 | chr1 | 234603396 | - | 0.000202791 | 0.9997973 |
Top |
Fusion Genomic Features for GUK1-TARBP1 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
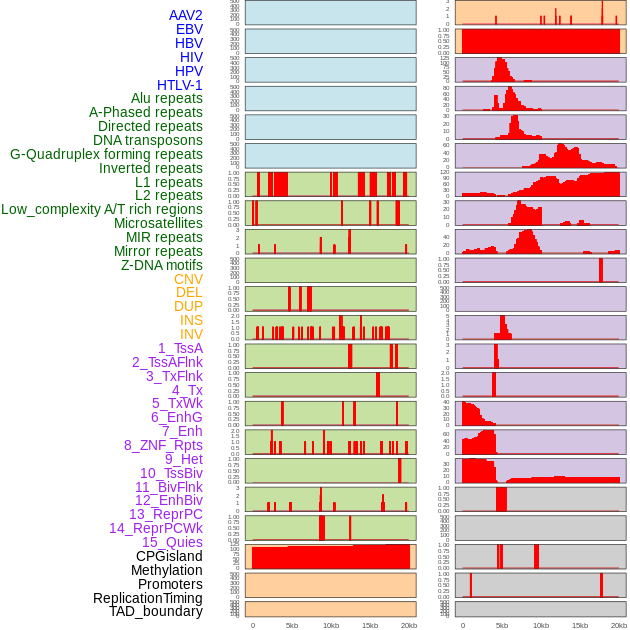 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for GUK1-TARBP1 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr1:228328064/chr1:234603396) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| GUK1 | . |
| FUNCTION: Catalyzes the phosphorylation of GMP to GDP. Essential enzyme for recycling GMP and indirectly, cyclic GMP (cGMP) (PubMed:31201273). Involved in the cGMP metabolism in photoreceptors (By similarity). It may also have a role in the survival and growth progression of some tumors (PubMed:31201273). In addition to its physiological role, GUK1 is essential for convert prodrugs used for the treatment of cancers and viral infections into their pharmacologically active metabolites, most notably acyclovir, ganciclovir, and 6-thioguanine and its closely related analog 6-mercaptopurine (PubMed:197968, PubMed:6248551, PubMed:6306664). {ECO:0000250|UniProtKB:P46195, ECO:0000269|PubMed:197968, ECO:0000269|PubMed:31201273, ECO:0000269|PubMed:6248551, ECO:0000269|PubMed:6306664}. | FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | GUK1 | chr1:228328064 | chr1:234603396 | ENST00000366728 | + | 1 | 7 | 14_19 | 20 | 242.0 | Nucleotide binding | ATP |
| Hgene | GUK1 | chr1:228328064 | chr1:234603396 | ENST00000391865 | + | 1 | 8 | 14_19 | 20 | 219.0 | Nucleotide binding | ATP |
| Tgene | TARBP1 | chr1:228328064 | chr1:234603396 | ENST00000040877 | 2 | 30 | 1543_1545 | 366 | 1622.0 | Region | S-adenosyl-L-methionine binding | |
| Tgene | TARBP1 | chr1:228328064 | chr1:234603396 | ENST00000040877 | 2 | 30 | 1586_1595 | 366 | 1622.0 | Region | S-adenosyl-L-methionine binding |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | GUK1 | chr1:228328064 | chr1:234603396 | ENST00000312726 | + | 1 | 9 | 4_186 | 0 | 198.0 | Domain | Guanylate kinase-like |
| Hgene | GUK1 | chr1:228328064 | chr1:234603396 | ENST00000366716 | + | 1 | 7 | 4_186 | 0 | 198.0 | Domain | Guanylate kinase-like |
| Hgene | GUK1 | chr1:228328064 | chr1:234603396 | ENST00000366718 | + | 1 | 8 | 4_186 | 0 | 198.0 | Domain | Guanylate kinase-like |
| Hgene | GUK1 | chr1:228328064 | chr1:234603396 | ENST00000366726 | + | 1 | 8 | 4_186 | 0 | 198.0 | Domain | Guanylate kinase-like |
| Hgene | GUK1 | chr1:228328064 | chr1:234603396 | ENST00000366728 | + | 1 | 7 | 4_186 | 20 | 242.0 | Domain | Guanylate kinase-like |
| Hgene | GUK1 | chr1:228328064 | chr1:234603396 | ENST00000366730 | + | 1 | 8 | 4_186 | 0 | 198.0 | Domain | Guanylate kinase-like |
| Hgene | GUK1 | chr1:228328064 | chr1:234603396 | ENST00000391865 | + | 1 | 8 | 4_186 | 20 | 219.0 | Domain | Guanylate kinase-like |
| Hgene | GUK1 | chr1:228328064 | chr1:234603396 | ENST00000312726 | + | 1 | 9 | 14_19 | 0 | 198.0 | Nucleotide binding | ATP |
| Hgene | GUK1 | chr1:228328064 | chr1:234603396 | ENST00000312726 | + | 1 | 9 | 171_172 | 0 | 198.0 | Nucleotide binding | ATP |
| Hgene | GUK1 | chr1:228328064 | chr1:234603396 | ENST00000312726 | + | 1 | 9 | 37_51 | 0 | 198.0 | Nucleotide binding | Substrate binding |
| Hgene | GUK1 | chr1:228328064 | chr1:234603396 | ENST00000366716 | + | 1 | 7 | 14_19 | 0 | 198.0 | Nucleotide binding | ATP |
| Hgene | GUK1 | chr1:228328064 | chr1:234603396 | ENST00000366716 | + | 1 | 7 | 171_172 | 0 | 198.0 | Nucleotide binding | ATP |
| Hgene | GUK1 | chr1:228328064 | chr1:234603396 | ENST00000366716 | + | 1 | 7 | 37_51 | 0 | 198.0 | Nucleotide binding | Substrate binding |
| Hgene | GUK1 | chr1:228328064 | chr1:234603396 | ENST00000366718 | + | 1 | 8 | 14_19 | 0 | 198.0 | Nucleotide binding | ATP |
| Hgene | GUK1 | chr1:228328064 | chr1:234603396 | ENST00000366718 | + | 1 | 8 | 171_172 | 0 | 198.0 | Nucleotide binding | ATP |
| Hgene | GUK1 | chr1:228328064 | chr1:234603396 | ENST00000366718 | + | 1 | 8 | 37_51 | 0 | 198.0 | Nucleotide binding | Substrate binding |
| Hgene | GUK1 | chr1:228328064 | chr1:234603396 | ENST00000366726 | + | 1 | 8 | 14_19 | 0 | 198.0 | Nucleotide binding | ATP |
| Hgene | GUK1 | chr1:228328064 | chr1:234603396 | ENST00000366726 | + | 1 | 8 | 171_172 | 0 | 198.0 | Nucleotide binding | ATP |
| Hgene | GUK1 | chr1:228328064 | chr1:234603396 | ENST00000366726 | + | 1 | 8 | 37_51 | 0 | 198.0 | Nucleotide binding | Substrate binding |
| Hgene | GUK1 | chr1:228328064 | chr1:234603396 | ENST00000366728 | + | 1 | 7 | 171_172 | 20 | 242.0 | Nucleotide binding | ATP |
| Hgene | GUK1 | chr1:228328064 | chr1:234603396 | ENST00000366728 | + | 1 | 7 | 37_51 | 20 | 242.0 | Nucleotide binding | Substrate binding |
| Hgene | GUK1 | chr1:228328064 | chr1:234603396 | ENST00000366730 | + | 1 | 8 | 14_19 | 0 | 198.0 | Nucleotide binding | ATP |
| Hgene | GUK1 | chr1:228328064 | chr1:234603396 | ENST00000366730 | + | 1 | 8 | 171_172 | 0 | 198.0 | Nucleotide binding | ATP |
| Hgene | GUK1 | chr1:228328064 | chr1:234603396 | ENST00000366730 | + | 1 | 8 | 37_51 | 0 | 198.0 | Nucleotide binding | Substrate binding |
| Hgene | GUK1 | chr1:228328064 | chr1:234603396 | ENST00000391865 | + | 1 | 8 | 171_172 | 20 | 219.0 | Nucleotide binding | ATP |
| Hgene | GUK1 | chr1:228328064 | chr1:234603396 | ENST00000391865 | + | 1 | 8 | 37_51 | 20 | 219.0 | Nucleotide binding | Substrate binding |
| Tgene | TARBP1 | chr1:228328064 | chr1:234603396 | ENST00000040877 | 2 | 30 | 99_219 | 366 | 1622.0 | Compositional bias | Note=Ala-rich |
Top |
Fusion Gene Sequence for GUK1-TARBP1 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >35368_35368_1_GUK1-TARBP1_GUK1_chr1_228328064_ENST00000366723_TARBP1_chr1_234603396_ENST00000040877_length(transcript)=4093nt_BP=62nt GATGCTGCGGCGCCCGCTGGCCGGGCTGGCTGCGGCCGCCCTGGGCCGGGCCCCACCGGACGGATGTTGGCTCTTTCACCCATCCTGGCA TATGTGTATTTATAAAAGAATGTTTGAAAGTGAAAACAAAATCCTGTCCAAAGAAGGTGTTATCCATTTTTTGGAGCTGTATGAAACAAA GATTCTTCCATTTTCACCAGAATTTTCTGAGTTTATTATTGGACCATTAATGGATGCGCTTTCAGAGAGCTCTCTGTATAGCAGGTCCCC AGGCCAGCCAATAGGAAGCTGTTCTCCATTGGGACTGAAATTACAGAAGTTTTTAGTCACTTATATTTCTCTTCTTCCAGAAGAAATAAA GAGTAGCTTCCTATTGAAGTTTATTCGGAAGATGACAAGTAGGCATTGGTGTGCTGTTCCCATTTTGTTTCTATCTAAGGCTTTGGCAAA TGTCCCAAGACATAAGGCCCTGGGTATAGATGGGCTTCTTGCTCTCAGGGATGTTATTCATTGCACTATGATCACACATCAGATTCTCCT GAGAGGGGCAGCCCAATGCTACCTTCTTCAAACAGCTATGAATTTGCTAGATGTGGAGAAAGTGTCACTTTCTGATGTCTCAACTTTTCT CATGTCTCTGAGACAAGAGGAATCCTTAGGACGAGGAACTTCATTGTGGACAGAGCTGTGTGACTGGCTACGTGTTAATGAAAGCTATTT TAAGCCATCCCCTACGTGTAGCTCCATTGGACTTCACAAGACATCTTTAAATGCTTATGTAAAGAGCATTGTTCAAGAGTATGTTAAGTC ATCTGCTTGGGAAACAGGAGAAAACTGCTTTATGCCTGATTGGTTTGAAGCCAAGCTTGTTTCTCTGATGGTCTTGCTGGCTGTGGATGT GGAAGGAATGAAGACTCAGTATAGCGGAAAGCAGAGAACAGAGAATGTATTGCGGATATTCTTAGACCCTCTTCTGGATGTGCTTATGAA GTTTAGTACCAATGCCTACATGCCCTTGCTGAAGACTGACAGATGCCTCCAGCTGCTGTTGAAGCTGTTGAACACATGCAGGTTGAAAGG TTCCAGTGCCCAAGATGATGAGGTGTCTACTGTTCTTCAGAACTTTTTCATGTCTACTACAGAGAGCATTTCTGAATTTATTCTCAGAAG ACTTACTATGAATGAGCTAAATAGTGTTTCAGATCTGGATCGTTGCCATTTATACCTGATGGTGTTAACTGAGCTTATAAATCTGCATTT GAAGGTTGGGTGGAAAAGGGGTAACCCTATCTGGAGAGTTATTTCTCTTTTGAAAAATGCATCCATTCAGCATCTTCAAGAGATGGACAG TGGACAGGAGCCAACAGTTGGAAGTCAGATTCAGAGAGTAGTGAGCATGGCTGCCTTGGCCATGGTGTGTGAGGCCATAGACCAGAAGCC TGAGCTGCAGCTGGACTCTCTCCATGCTGGGCCCCTGGAAAGCTTCCTTTCCTCTCTTCAGCTCAATCAGACGCTGCAGAAGCCCCACGC AGAGGAGCAGAGCAGTTATGCTCACCCCTTGGAGTGCAGCAGTGTTTTGGAAGAATCGTCATCTTCCCAAGGATGGGGAAAAATAGTTGC ACAATATATTCATGATCAATGGGTGTGCCTCTCTTTCCTGTTGAAAAAATATCACACCCTTATACCAACCACAGGGAGTGAAATTCTGGA ACCGTTTCTACCTGCCGTTCAGATGCCAATAAGGACTTTGCAGTCTGCACTAGAAGCCCTCACAGTTCTTTCTTCTGATCAAGTTTTACC AGTGTTCCATTGCTTGAAAGTGTTGGTTCCCAAGCTTCTGACTTCCTCTGAATCACTCTGCATAGAGTCTTTTGACATGGCGTGGAAAAT TATATCTTCTTTAAGCAACACTCAGCTGATATTCTGGGCTAATTTAAAAGCTTTTGTTCAGTTTGTTTTTGATAACAAAGTTCTTACCAT TGCTGCCAAAATCAAGGGCCAGGCATATTTCAAAATAAAAGAGATTATGTACAAGATAATTGAAATGTCTGCTATAAAGACTGGAGTCTT CAATACACTGATAAGTTACTGCTGTCAGTCTTGGATAGTGTCTGCTTCAAATGTGTCCCAAGGATCTTTATCAAGTGCTAAAAATTATAG CGAACTTATCCTTGAGGCTTGTATATTTGGAACTGTGTTTAGGCGTGATCAAAGACTTGTTCAGGATGTACAGACCTTCATAGAAAACCT TGGACATGACTGTGCGGCAAATATTGTTATGGAAAATACTAAGAGAGAAGACCATTATGTGAGAATTTGTGCTGTCAAATTCCTGTGTTT ATTAGATGGCTCCAATATGTCCCACAAGTTGTTTATTGAGGATCTTGCAATCAAGCTATTAGATAAAGATGAATTAGTGTCCAAGTCCAA AAAACGCTACTATGTGAATTCTCTACAGCACAGAGTGAAAAACCGAGTGTGGCAGACTCTGCTGGTACTTTTCCCTAGACTTGACCAGAA TTTCTTGAATGGAATTATTGACAGGATTTTCCAGGCTGGTTTCACCAACAATCAAGCATCCATAAAATATTTTATAGAATGGATTATTAT ATTGATTCTTCATAAATTCCCTCAATTTCTTCCAAAGTTCTGGGATTGTTTTTCTTATGGTGAAGAAAATCTTAAAACAAGCATTTGTAC GTTTTTAGCAGTTTTATCACATTTAGACATTATTACTCAAAATATTCCAGAAAAGAAACTAATTCTGAAGCAAGCCCTTATAGTTGTGCT GCAGTGGTGTTTCAATCACAATTTTAGTGTTCGACTGTATGCTTTAGTTGCTCTTAAGAAACTCTGGACTGTGTGTAAAGTGTTAAGTGT TGAAGAATTTGATGCCCTGACTCCTGTGATTGAATCCAGCCTCCATCAAGTGGAAAGCATGCACGGAGCAGGGAATGCCAAGAAGAATTG GCAACGCATTCAGGAGCATTTCTTTTTTGCAACATTTCACCCACTCAAGGATTATTGTCTAGAGACCATATTTTACATCCTTCCACGCCT TTCAGGCCTTATTGAAGATGAATGGATCACCATTGATAAATTTACCAGATTCACTGATGTTCCTTTAGCTGCGGGATTTCAGTGGTACCT TTCTCAAACTCAACTTAGTAAACTAAAACCAGGTGACTGGTCTCAGCAAGACATAGGTACTAATTTGGTCGAAGCAGATAACCAAGCGGA GTGGACCGACGTTCAGAAGAAGATTATCCCGTGGAACAGTCGTGTTTCCGACTTAGACCTGGAGCTCCTGTTTCAGGATCGTGCTGCCAG ACTTGGAAAGTCAATTAGTAGACTCATCGTTGTGGCCTCGCTCATCGACAAACCGACCAATTTAGGAGGACTGTGCAGGACCTGTGAGGT ATTTGGGGCTTCAGTGCTCGTTGTTGGCAGCCTTCAGTGTATCAGCGACAAACAGTTTCAGCACCTCAGTGTCTCTGCAGAACAGTGGCT TCCTCTAGTGGAGGTAAAACCACCTCAGCTAATTGATTATCTGCAGCAGAAGAAAACAGAAGGTTATACCATCATTGGAGTGGAACAAAC TGCCAAAAGTTTAGACCTAACCCAATATTGCTTTCCTGAGAAATCTCTGCTCTTGTTGGGAAATGAACGTGAGGGAATTCCAGCAAATCT GATCCAACAGTTGGACGTTTGTGTGGAAATTCCTCAACAGGGCATTATCCGCTCCCTGAATGTCCATGTGAGTGGAGCCCTGCTGATCTG GGAGTACACCAGGCAGCAGCTGCTCTCGCACGGAGATACCAAGCCATGATGTGCCTTCCTTAGTGAACTGCTGCTGCTGTTCAGACTTTT TTAAAAAAAACTATTTGGACTAAAGAAACAGATTCTGAAATTTATTGTGATAATTTGTATTTCTTTTTTCTTGCAATTTAATGCCAAAAG TTTGCCATGTGCCTTAAACATATTACTATATATTTTCCCCTTTAATAAACACTTTTTGTTAAATTGTATTCTTCCTTTAATAAAATATTT >35368_35368_1_GUK1-TARBP1_GUK1_chr1_228328064_ENST00000366723_TARBP1_chr1_234603396_ENST00000040877_length(amino acids)=1275AA_BP=20 MLRRPLAGLAAAALGRAPPDGCWLFHPSWHMCIYKRMFESENKILSKEGVIHFLELYETKILPFSPEFSEFIIGPLMDALSESSLYSRSP GQPIGSCSPLGLKLQKFLVTYISLLPEEIKSSFLLKFIRKMTSRHWCAVPILFLSKALANVPRHKALGIDGLLALRDVIHCTMITHQILL RGAAQCYLLQTAMNLLDVEKVSLSDVSTFLMSLRQEESLGRGTSLWTELCDWLRVNESYFKPSPTCSSIGLHKTSLNAYVKSIVQEYVKS SAWETGENCFMPDWFEAKLVSLMVLLAVDVEGMKTQYSGKQRTENVLRIFLDPLLDVLMKFSTNAYMPLLKTDRCLQLLLKLLNTCRLKG SSAQDDEVSTVLQNFFMSTTESISEFILRRLTMNELNSVSDLDRCHLYLMVLTELINLHLKVGWKRGNPIWRVISLLKNASIQHLQEMDS GQEPTVGSQIQRVVSMAALAMVCEAIDQKPELQLDSLHAGPLESFLSSLQLNQTLQKPHAEEQSSYAHPLECSSVLEESSSSQGWGKIVA QYIHDQWVCLSFLLKKYHTLIPTTGSEILEPFLPAVQMPIRTLQSALEALTVLSSDQVLPVFHCLKVLVPKLLTSSESLCIESFDMAWKI ISSLSNTQLIFWANLKAFVQFVFDNKVLTIAAKIKGQAYFKIKEIMYKIIEMSAIKTGVFNTLISYCCQSWIVSASNVSQGSLSSAKNYS ELILEACIFGTVFRRDQRLVQDVQTFIENLGHDCAANIVMENTKREDHYVRICAVKFLCLLDGSNMSHKLFIEDLAIKLLDKDELVSKSK KRYYVNSLQHRVKNRVWQTLLVLFPRLDQNFLNGIIDRIFQAGFTNNQASIKYFIEWIIILILHKFPQFLPKFWDCFSYGEENLKTSICT FLAVLSHLDIITQNIPEKKLILKQALIVVLQWCFNHNFSVRLYALVALKKLWTVCKVLSVEEFDALTPVIESSLHQVESMHGAGNAKKNW QRIQEHFFFATFHPLKDYCLETIFYILPRLSGLIEDEWITIDKFTRFTDVPLAAGFQWYLSQTQLSKLKPGDWSQQDIGTNLVEADNQAE WTDVQKKIIPWNSRVSDLDLELLFQDRAARLGKSISRLIVVASLIDKPTNLGGLCRTCEVFGASVLVVGSLQCISDKQFQHLSVSAEQWL PLVEVKPPQLIDYLQQKKTEGYTIIGVEQTAKSLDLTQYCFPEKSLLLLGNEREGIPANLIQQLDVCVEIPQQGIIRSLNVHVSGALLIW -------------------------------------------------------------- >35368_35368_2_GUK1-TARBP1_GUK1_chr1_228328064_ENST00000366728_TARBP1_chr1_234603396_ENST00000040877_length(transcript)=4105nt_BP=74nt GAGGTGGCCCCGGATGCTGCGGCGCCCGCTGGCCGGGCTGGCTGCGGCCGCCCTGGGCCGGGCCCCACCGGACGGATGTTGGCTCTTTCA CCCATCCTGGCATATGTGTATTTATAAAAGAATGTTTGAAAGTGAAAACAAAATCCTGTCCAAAGAAGGTGTTATCCATTTTTTGGAGCT GTATGAAACAAAGATTCTTCCATTTTCACCAGAATTTTCTGAGTTTATTATTGGACCATTAATGGATGCGCTTTCAGAGAGCTCTCTGTA TAGCAGGTCCCCAGGCCAGCCAATAGGAAGCTGTTCTCCATTGGGACTGAAATTACAGAAGTTTTTAGTCACTTATATTTCTCTTCTTCC AGAAGAAATAAAGAGTAGCTTCCTATTGAAGTTTATTCGGAAGATGACAAGTAGGCATTGGTGTGCTGTTCCCATTTTGTTTCTATCTAA GGCTTTGGCAAATGTCCCAAGACATAAGGCCCTGGGTATAGATGGGCTTCTTGCTCTCAGGGATGTTATTCATTGCACTATGATCACACA TCAGATTCTCCTGAGAGGGGCAGCCCAATGCTACCTTCTTCAAACAGCTATGAATTTGCTAGATGTGGAGAAAGTGTCACTTTCTGATGT CTCAACTTTTCTCATGTCTCTGAGACAAGAGGAATCCTTAGGACGAGGAACTTCATTGTGGACAGAGCTGTGTGACTGGCTACGTGTTAA TGAAAGCTATTTTAAGCCATCCCCTACGTGTAGCTCCATTGGACTTCACAAGACATCTTTAAATGCTTATGTAAAGAGCATTGTTCAAGA GTATGTTAAGTCATCTGCTTGGGAAACAGGAGAAAACTGCTTTATGCCTGATTGGTTTGAAGCCAAGCTTGTTTCTCTGATGGTCTTGCT GGCTGTGGATGTGGAAGGAATGAAGACTCAGTATAGCGGAAAGCAGAGAACAGAGAATGTATTGCGGATATTCTTAGACCCTCTTCTGGA TGTGCTTATGAAGTTTAGTACCAATGCCTACATGCCCTTGCTGAAGACTGACAGATGCCTCCAGCTGCTGTTGAAGCTGTTGAACACATG CAGGTTGAAAGGTTCCAGTGCCCAAGATGATGAGGTGTCTACTGTTCTTCAGAACTTTTTCATGTCTACTACAGAGAGCATTTCTGAATT TATTCTCAGAAGACTTACTATGAATGAGCTAAATAGTGTTTCAGATCTGGATCGTTGCCATTTATACCTGATGGTGTTAACTGAGCTTAT AAATCTGCATTTGAAGGTTGGGTGGAAAAGGGGTAACCCTATCTGGAGAGTTATTTCTCTTTTGAAAAATGCATCCATTCAGCATCTTCA AGAGATGGACAGTGGACAGGAGCCAACAGTTGGAAGTCAGATTCAGAGAGTAGTGAGCATGGCTGCCTTGGCCATGGTGTGTGAGGCCAT AGACCAGAAGCCTGAGCTGCAGCTGGACTCTCTCCATGCTGGGCCCCTGGAAAGCTTCCTTTCCTCTCTTCAGCTCAATCAGACGCTGCA GAAGCCCCACGCAGAGGAGCAGAGCAGTTATGCTCACCCCTTGGAGTGCAGCAGTGTTTTGGAAGAATCGTCATCTTCCCAAGGATGGGG AAAAATAGTTGCACAATATATTCATGATCAATGGGTGTGCCTCTCTTTCCTGTTGAAAAAATATCACACCCTTATACCAACCACAGGGAG TGAAATTCTGGAACCGTTTCTACCTGCCGTTCAGATGCCAATAAGGACTTTGCAGTCTGCACTAGAAGCCCTCACAGTTCTTTCTTCTGA TCAAGTTTTACCAGTGTTCCATTGCTTGAAAGTGTTGGTTCCCAAGCTTCTGACTTCCTCTGAATCACTCTGCATAGAGTCTTTTGACAT GGCGTGGAAAATTATATCTTCTTTAAGCAACACTCAGCTGATATTCTGGGCTAATTTAAAAGCTTTTGTTCAGTTTGTTTTTGATAACAA AGTTCTTACCATTGCTGCCAAAATCAAGGGCCAGGCATATTTCAAAATAAAAGAGATTATGTACAAGATAATTGAAATGTCTGCTATAAA GACTGGAGTCTTCAATACACTGATAAGTTACTGCTGTCAGTCTTGGATAGTGTCTGCTTCAAATGTGTCCCAAGGATCTTTATCAAGTGC TAAAAATTATAGCGAACTTATCCTTGAGGCTTGTATATTTGGAACTGTGTTTAGGCGTGATCAAAGACTTGTTCAGGATGTACAGACCTT CATAGAAAACCTTGGACATGACTGTGCGGCAAATATTGTTATGGAAAATACTAAGAGAGAAGACCATTATGTGAGAATTTGTGCTGTCAA ATTCCTGTGTTTATTAGATGGCTCCAATATGTCCCACAAGTTGTTTATTGAGGATCTTGCAATCAAGCTATTAGATAAAGATGAATTAGT GTCCAAGTCCAAAAAACGCTACTATGTGAATTCTCTACAGCACAGAGTGAAAAACCGAGTGTGGCAGACTCTGCTGGTACTTTTCCCTAG ACTTGACCAGAATTTCTTGAATGGAATTATTGACAGGATTTTCCAGGCTGGTTTCACCAACAATCAAGCATCCATAAAATATTTTATAGA ATGGATTATTATATTGATTCTTCATAAATTCCCTCAATTTCTTCCAAAGTTCTGGGATTGTTTTTCTTATGGTGAAGAAAATCTTAAAAC AAGCATTTGTACGTTTTTAGCAGTTTTATCACATTTAGACATTATTACTCAAAATATTCCAGAAAAGAAACTAATTCTGAAGCAAGCCCT TATAGTTGTGCTGCAGTGGTGTTTCAATCACAATTTTAGTGTTCGACTGTATGCTTTAGTTGCTCTTAAGAAACTCTGGACTGTGTGTAA AGTGTTAAGTGTTGAAGAATTTGATGCCCTGACTCCTGTGATTGAATCCAGCCTCCATCAAGTGGAAAGCATGCACGGAGCAGGGAATGC CAAGAAGAATTGGCAACGCATTCAGGAGCATTTCTTTTTTGCAACATTTCACCCACTCAAGGATTATTGTCTAGAGACCATATTTTACAT CCTTCCACGCCTTTCAGGCCTTATTGAAGATGAATGGATCACCATTGATAAATTTACCAGATTCACTGATGTTCCTTTAGCTGCGGGATT TCAGTGGTACCTTTCTCAAACTCAACTTAGTAAACTAAAACCAGGTGACTGGTCTCAGCAAGACATAGGTACTAATTTGGTCGAAGCAGA TAACCAAGCGGAGTGGACCGACGTTCAGAAGAAGATTATCCCGTGGAACAGTCGTGTTTCCGACTTAGACCTGGAGCTCCTGTTTCAGGA TCGTGCTGCCAGACTTGGAAAGTCAATTAGTAGACTCATCGTTGTGGCCTCGCTCATCGACAAACCGACCAATTTAGGAGGACTGTGCAG GACCTGTGAGGTATTTGGGGCTTCAGTGCTCGTTGTTGGCAGCCTTCAGTGTATCAGCGACAAACAGTTTCAGCACCTCAGTGTCTCTGC AGAACAGTGGCTTCCTCTAGTGGAGGTAAAACCACCTCAGCTAATTGATTATCTGCAGCAGAAGAAAACAGAAGGTTATACCATCATTGG AGTGGAACAAACTGCCAAAAGTTTAGACCTAACCCAATATTGCTTTCCTGAGAAATCTCTGCTCTTGTTGGGAAATGAACGTGAGGGAAT TCCAGCAAATCTGATCCAACAGTTGGACGTTTGTGTGGAAATTCCTCAACAGGGCATTATCCGCTCCCTGAATGTCCATGTGAGTGGAGC CCTGCTGATCTGGGAGTACACCAGGCAGCAGCTGCTCTCGCACGGAGATACCAAGCCATGATGTGCCTTCCTTAGTGAACTGCTGCTGCT GTTCAGACTTTTTTAAAAAAAACTATTTGGACTAAAGAAACAGATTCTGAAATTTATTGTGATAATTTGTATTTCTTTTTTCTTGCAATT TAATGCCAAAAGTTTGCCATGTGCCTTAAACATATTACTATATATTTTCCCCTTTAATAAACACTTTTTGTTAAATTGTATTCTTCCTTT >35368_35368_2_GUK1-TARBP1_GUK1_chr1_228328064_ENST00000366728_TARBP1_chr1_234603396_ENST00000040877_length(amino acids)=1275AA_BP=20 MLRRPLAGLAAAALGRAPPDGCWLFHPSWHMCIYKRMFESENKILSKEGVIHFLELYETKILPFSPEFSEFIIGPLMDALSESSLYSRSP GQPIGSCSPLGLKLQKFLVTYISLLPEEIKSSFLLKFIRKMTSRHWCAVPILFLSKALANVPRHKALGIDGLLALRDVIHCTMITHQILL RGAAQCYLLQTAMNLLDVEKVSLSDVSTFLMSLRQEESLGRGTSLWTELCDWLRVNESYFKPSPTCSSIGLHKTSLNAYVKSIVQEYVKS SAWETGENCFMPDWFEAKLVSLMVLLAVDVEGMKTQYSGKQRTENVLRIFLDPLLDVLMKFSTNAYMPLLKTDRCLQLLLKLLNTCRLKG SSAQDDEVSTVLQNFFMSTTESISEFILRRLTMNELNSVSDLDRCHLYLMVLTELINLHLKVGWKRGNPIWRVISLLKNASIQHLQEMDS GQEPTVGSQIQRVVSMAALAMVCEAIDQKPELQLDSLHAGPLESFLSSLQLNQTLQKPHAEEQSSYAHPLECSSVLEESSSSQGWGKIVA QYIHDQWVCLSFLLKKYHTLIPTTGSEILEPFLPAVQMPIRTLQSALEALTVLSSDQVLPVFHCLKVLVPKLLTSSESLCIESFDMAWKI ISSLSNTQLIFWANLKAFVQFVFDNKVLTIAAKIKGQAYFKIKEIMYKIIEMSAIKTGVFNTLISYCCQSWIVSASNVSQGSLSSAKNYS ELILEACIFGTVFRRDQRLVQDVQTFIENLGHDCAANIVMENTKREDHYVRICAVKFLCLLDGSNMSHKLFIEDLAIKLLDKDELVSKSK KRYYVNSLQHRVKNRVWQTLLVLFPRLDQNFLNGIIDRIFQAGFTNNQASIKYFIEWIIILILHKFPQFLPKFWDCFSYGEENLKTSICT FLAVLSHLDIITQNIPEKKLILKQALIVVLQWCFNHNFSVRLYALVALKKLWTVCKVLSVEEFDALTPVIESSLHQVESMHGAGNAKKNW QRIQEHFFFATFHPLKDYCLETIFYILPRLSGLIEDEWITIDKFTRFTDVPLAAGFQWYLSQTQLSKLKPGDWSQQDIGTNLVEADNQAE WTDVQKKIIPWNSRVSDLDLELLFQDRAARLGKSISRLIVVASLIDKPTNLGGLCRTCEVFGASVLVVGSLQCISDKQFQHLSVSAEQWL PLVEVKPPQLIDYLQQKKTEGYTIIGVEQTAKSLDLTQYCFPEKSLLLLGNEREGIPANLIQQLDVCVEIPQQGIIRSLNVHVSGALLIW -------------------------------------------------------------- >35368_35368_3_GUK1-TARBP1_GUK1_chr1_228328064_ENST00000391865_TARBP1_chr1_234603396_ENST00000040877_length(transcript)=4167nt_BP=136nt AGTCTCGCGCGCGGGCGGAGGTGGGCCGGTGCGGCGCTGTCACGTAGGTTCAGTGGGCGGAAGAGGTGGCCCCGGATGCTGCGGCGCCCG CTGGCCGGGCTGGCTGCGGCCGCCCTGGGCCGGGCCCCACCGGACGGATGTTGGCTCTTTCACCCATCCTGGCATATGTGTATTTATAAA AGAATGTTTGAAAGTGAAAACAAAATCCTGTCCAAAGAAGGTGTTATCCATTTTTTGGAGCTGTATGAAACAAAGATTCTTCCATTTTCA CCAGAATTTTCTGAGTTTATTATTGGACCATTAATGGATGCGCTTTCAGAGAGCTCTCTGTATAGCAGGTCCCCAGGCCAGCCAATAGGA AGCTGTTCTCCATTGGGACTGAAATTACAGAAGTTTTTAGTCACTTATATTTCTCTTCTTCCAGAAGAAATAAAGAGTAGCTTCCTATTG AAGTTTATTCGGAAGATGACAAGTAGGCATTGGTGTGCTGTTCCCATTTTGTTTCTATCTAAGGCTTTGGCAAATGTCCCAAGACATAAG GCCCTGGGTATAGATGGGCTTCTTGCTCTCAGGGATGTTATTCATTGCACTATGATCACACATCAGATTCTCCTGAGAGGGGCAGCCCAA TGCTACCTTCTTCAAACAGCTATGAATTTGCTAGATGTGGAGAAAGTGTCACTTTCTGATGTCTCAACTTTTCTCATGTCTCTGAGACAA GAGGAATCCTTAGGACGAGGAACTTCATTGTGGACAGAGCTGTGTGACTGGCTACGTGTTAATGAAAGCTATTTTAAGCCATCCCCTACG TGTAGCTCCATTGGACTTCACAAGACATCTTTAAATGCTTATGTAAAGAGCATTGTTCAAGAGTATGTTAAGTCATCTGCTTGGGAAACA GGAGAAAACTGCTTTATGCCTGATTGGTTTGAAGCCAAGCTTGTTTCTCTGATGGTCTTGCTGGCTGTGGATGTGGAAGGAATGAAGACT CAGTATAGCGGAAAGCAGAGAACAGAGAATGTATTGCGGATATTCTTAGACCCTCTTCTGGATGTGCTTATGAAGTTTAGTACCAATGCC TACATGCCCTTGCTGAAGACTGACAGATGCCTCCAGCTGCTGTTGAAGCTGTTGAACACATGCAGGTTGAAAGGTTCCAGTGCCCAAGAT GATGAGGTGTCTACTGTTCTTCAGAACTTTTTCATGTCTACTACAGAGAGCATTTCTGAATTTATTCTCAGAAGACTTACTATGAATGAG CTAAATAGTGTTTCAGATCTGGATCGTTGCCATTTATACCTGATGGTGTTAACTGAGCTTATAAATCTGCATTTGAAGGTTGGGTGGAAA AGGGGTAACCCTATCTGGAGAGTTATTTCTCTTTTGAAAAATGCATCCATTCAGCATCTTCAAGAGATGGACAGTGGACAGGAGCCAACA GTTGGAAGTCAGATTCAGAGAGTAGTGAGCATGGCTGCCTTGGCCATGGTGTGTGAGGCCATAGACCAGAAGCCTGAGCTGCAGCTGGAC TCTCTCCATGCTGGGCCCCTGGAAAGCTTCCTTTCCTCTCTTCAGCTCAATCAGACGCTGCAGAAGCCCCACGCAGAGGAGCAGAGCAGT TATGCTCACCCCTTGGAGTGCAGCAGTGTTTTGGAAGAATCGTCATCTTCCCAAGGATGGGGAAAAATAGTTGCACAATATATTCATGAT CAATGGGTGTGCCTCTCTTTCCTGTTGAAAAAATATCACACCCTTATACCAACCACAGGGAGTGAAATTCTGGAACCGTTTCTACCTGCC GTTCAGATGCCAATAAGGACTTTGCAGTCTGCACTAGAAGCCCTCACAGTTCTTTCTTCTGATCAAGTTTTACCAGTGTTCCATTGCTTG AAAGTGTTGGTTCCCAAGCTTCTGACTTCCTCTGAATCACTCTGCATAGAGTCTTTTGACATGGCGTGGAAAATTATATCTTCTTTAAGC AACACTCAGCTGATATTCTGGGCTAATTTAAAAGCTTTTGTTCAGTTTGTTTTTGATAACAAAGTTCTTACCATTGCTGCCAAAATCAAG GGCCAGGCATATTTCAAAATAAAAGAGATTATGTACAAGATAATTGAAATGTCTGCTATAAAGACTGGAGTCTTCAATACACTGATAAGT TACTGCTGTCAGTCTTGGATAGTGTCTGCTTCAAATGTGTCCCAAGGATCTTTATCAAGTGCTAAAAATTATAGCGAACTTATCCTTGAG GCTTGTATATTTGGAACTGTGTTTAGGCGTGATCAAAGACTTGTTCAGGATGTACAGACCTTCATAGAAAACCTTGGACATGACTGTGCG GCAAATATTGTTATGGAAAATACTAAGAGAGAAGACCATTATGTGAGAATTTGTGCTGTCAAATTCCTGTGTTTATTAGATGGCTCCAAT ATGTCCCACAAGTTGTTTATTGAGGATCTTGCAATCAAGCTATTAGATAAAGATGAATTAGTGTCCAAGTCCAAAAAACGCTACTATGTG AATTCTCTACAGCACAGAGTGAAAAACCGAGTGTGGCAGACTCTGCTGGTACTTTTCCCTAGACTTGACCAGAATTTCTTGAATGGAATT ATTGACAGGATTTTCCAGGCTGGTTTCACCAACAATCAAGCATCCATAAAATATTTTATAGAATGGATTATTATATTGATTCTTCATAAA TTCCCTCAATTTCTTCCAAAGTTCTGGGATTGTTTTTCTTATGGTGAAGAAAATCTTAAAACAAGCATTTGTACGTTTTTAGCAGTTTTA TCACATTTAGACATTATTACTCAAAATATTCCAGAAAAGAAACTAATTCTGAAGCAAGCCCTTATAGTTGTGCTGCAGTGGTGTTTCAAT CACAATTTTAGTGTTCGACTGTATGCTTTAGTTGCTCTTAAGAAACTCTGGACTGTGTGTAAAGTGTTAAGTGTTGAAGAATTTGATGCC CTGACTCCTGTGATTGAATCCAGCCTCCATCAAGTGGAAAGCATGCACGGAGCAGGGAATGCCAAGAAGAATTGGCAACGCATTCAGGAG CATTTCTTTTTTGCAACATTTCACCCACTCAAGGATTATTGTCTAGAGACCATATTTTACATCCTTCCACGCCTTTCAGGCCTTATTGAA GATGAATGGATCACCATTGATAAATTTACCAGATTCACTGATGTTCCTTTAGCTGCGGGATTTCAGTGGTACCTTTCTCAAACTCAACTT AGTAAACTAAAACCAGGTGACTGGTCTCAGCAAGACATAGGTACTAATTTGGTCGAAGCAGATAACCAAGCGGAGTGGACCGACGTTCAG AAGAAGATTATCCCGTGGAACAGTCGTGTTTCCGACTTAGACCTGGAGCTCCTGTTTCAGGATCGTGCTGCCAGACTTGGAAAGTCAATT AGTAGACTCATCGTTGTGGCCTCGCTCATCGACAAACCGACCAATTTAGGAGGACTGTGCAGGACCTGTGAGGTATTTGGGGCTTCAGTG CTCGTTGTTGGCAGCCTTCAGTGTATCAGCGACAAACAGTTTCAGCACCTCAGTGTCTCTGCAGAACAGTGGCTTCCTCTAGTGGAGGTA AAACCACCTCAGCTAATTGATTATCTGCAGCAGAAGAAAACAGAAGGTTATACCATCATTGGAGTGGAACAAACTGCCAAAAGTTTAGAC CTAACCCAATATTGCTTTCCTGAGAAATCTCTGCTCTTGTTGGGAAATGAACGTGAGGGAATTCCAGCAAATCTGATCCAACAGTTGGAC GTTTGTGTGGAAATTCCTCAACAGGGCATTATCCGCTCCCTGAATGTCCATGTGAGTGGAGCCCTGCTGATCTGGGAGTACACCAGGCAG CAGCTGCTCTCGCACGGAGATACCAAGCCATGATGTGCCTTCCTTAGTGAACTGCTGCTGCTGTTCAGACTTTTTTAAAAAAAACTATTT GGACTAAAGAAACAGATTCTGAAATTTATTGTGATAATTTGTATTTCTTTTTTCTTGCAATTTAATGCCAAAAGTTTGCCATGTGCCTTA AACATATTACTATATATTTTCCCCTTTAATAAACACTTTTTGTTAAATTGTATTCTTCCTTTAATAAAATATTTTAAGCAATTGTGGAAA >35368_35368_3_GUK1-TARBP1_GUK1_chr1_228328064_ENST00000391865_TARBP1_chr1_234603396_ENST00000040877_length(amino acids)=1288AA_BP=33 MSRRFSGRKRWPRMLRRPLAGLAAAALGRAPPDGCWLFHPSWHMCIYKRMFESENKILSKEGVIHFLELYETKILPFSPEFSEFIIGPLM DALSESSLYSRSPGQPIGSCSPLGLKLQKFLVTYISLLPEEIKSSFLLKFIRKMTSRHWCAVPILFLSKALANVPRHKALGIDGLLALRD VIHCTMITHQILLRGAAQCYLLQTAMNLLDVEKVSLSDVSTFLMSLRQEESLGRGTSLWTELCDWLRVNESYFKPSPTCSSIGLHKTSLN AYVKSIVQEYVKSSAWETGENCFMPDWFEAKLVSLMVLLAVDVEGMKTQYSGKQRTENVLRIFLDPLLDVLMKFSTNAYMPLLKTDRCLQ LLLKLLNTCRLKGSSAQDDEVSTVLQNFFMSTTESISEFILRRLTMNELNSVSDLDRCHLYLMVLTELINLHLKVGWKRGNPIWRVISLL KNASIQHLQEMDSGQEPTVGSQIQRVVSMAALAMVCEAIDQKPELQLDSLHAGPLESFLSSLQLNQTLQKPHAEEQSSYAHPLECSSVLE ESSSSQGWGKIVAQYIHDQWVCLSFLLKKYHTLIPTTGSEILEPFLPAVQMPIRTLQSALEALTVLSSDQVLPVFHCLKVLVPKLLTSSE SLCIESFDMAWKIISSLSNTQLIFWANLKAFVQFVFDNKVLTIAAKIKGQAYFKIKEIMYKIIEMSAIKTGVFNTLISYCCQSWIVSASN VSQGSLSSAKNYSELILEACIFGTVFRRDQRLVQDVQTFIENLGHDCAANIVMENTKREDHYVRICAVKFLCLLDGSNMSHKLFIEDLAI KLLDKDELVSKSKKRYYVNSLQHRVKNRVWQTLLVLFPRLDQNFLNGIIDRIFQAGFTNNQASIKYFIEWIIILILHKFPQFLPKFWDCF SYGEENLKTSICTFLAVLSHLDIITQNIPEKKLILKQALIVVLQWCFNHNFSVRLYALVALKKLWTVCKVLSVEEFDALTPVIESSLHQV ESMHGAGNAKKNWQRIQEHFFFATFHPLKDYCLETIFYILPRLSGLIEDEWITIDKFTRFTDVPLAAGFQWYLSQTQLSKLKPGDWSQQD IGTNLVEADNQAEWTDVQKKIIPWNSRVSDLDLELLFQDRAARLGKSISRLIVVASLIDKPTNLGGLCRTCEVFGASVLVVGSLQCISDK QFQHLSVSAEQWLPLVEVKPPQLIDYLQQKKTEGYTIIGVEQTAKSLDLTQYCFPEKSLLLLGNEREGIPANLIQQLDVCVEIPQQGIIR -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for GUK1-TARBP1 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for GUK1-TARBP1 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for GUK1-TARBP1 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
