
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:HABP2-LAPTM4B (FusionGDB2 ID:35545) |
Fusion Gene Summary for HABP2-LAPTM4B |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: HABP2-LAPTM4B | Fusion gene ID: 35545 | Hgene | Tgene | Gene symbol | HABP2 | LAPTM4B | Gene ID | 3026 | 55353 |
| Gene name | hyaluronan binding protein 2 | lysosomal protein transmembrane 4 beta | |
| Synonyms | FSAP|HABP|HGFAL|NMTC5|PHBP | LAPTM4beta|LC27 | |
| Cytomap | 10q25.3 | 8q22.1 | |
| Type of gene | protein-coding | protein-coding | |
| Description | hyaluronan-binding protein 2factor VII activating proteinfactor VII-activating proteasefactor seven-activating proteasehepatocyte growth factor activator-like proteinhyaluronic acid binding protein 2plasma hyaluronan binding protein | lysosomal-associated transmembrane protein 4Blysosomal associated protein transmembrane 4 betalysosome-associated transmembrane protein 4-beta | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | Q14520 | Q86VI4 | |
| Ensembl transtripts involved in fusion gene | ENST00000537906, ENST00000351270, ENST00000541666, ENST00000460714, ENST00000542051, | ENST00000445593, ENST00000521545, | |
| Fusion gene scores | * DoF score | 3 X 2 X 3=18 | 14 X 8 X 8=896 |
| # samples | 3 | 16 | |
| ** MAII score | log2(3/18*10)=0.736965594166206 effective Gene in Pan-Cancer Fusion Genes (eGinPCFGs). DoF>8 and MAII>0 | log2(16/896*10)=-2.48542682717024 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: HABP2 [Title/Abstract] AND LAPTM4B [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | HABP2(115312949)-LAPTM4B(98817581), # samples:3 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | HABP2 | GO:0006508 | proteolysis | 11217080 |
| Tgene | LAPTM4B | GO:0032509 | endosome transport via multivesicular body sorting pathway | 25588945 |
| Tgene | LAPTM4B | GO:0032911 | negative regulation of transforming growth factor beta1 production | 26126825 |
 Fusion gene breakpoints across HABP2 (5'-gene) Fusion gene breakpoints across HABP2 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
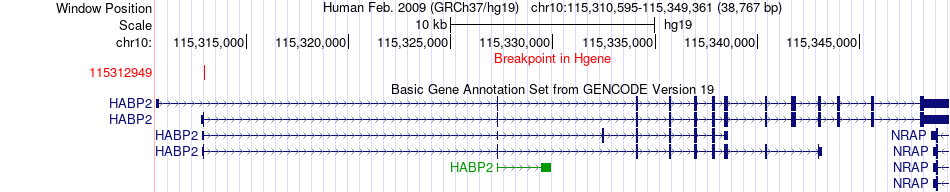 |
 Fusion gene breakpoints across LAPTM4B (3'-gene) Fusion gene breakpoints across LAPTM4B (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
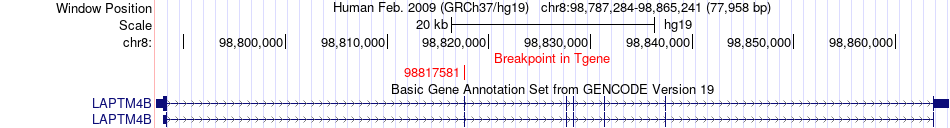 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | LIHC | TCGA-RC-A6M4-01A | HABP2 | chr10 | 115312949 | - | LAPTM4B | chr8 | 98817581 | + |
| ChimerDB4 | LIHC | TCGA-RC-A6M4-01A | HABP2 | chr10 | 115312949 | + | LAPTM4B | chr8 | 98817581 | + |
Top |
Fusion Gene ORF analysis for HABP2-LAPTM4B |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5UTR-3CDS | ENST00000537906 | ENST00000445593 | HABP2 | chr10 | 115312949 | + | LAPTM4B | chr8 | 98817581 | + |
| 5UTR-3CDS | ENST00000537906 | ENST00000521545 | HABP2 | chr10 | 115312949 | + | LAPTM4B | chr8 | 98817581 | + |
| In-frame | ENST00000351270 | ENST00000445593 | HABP2 | chr10 | 115312949 | + | LAPTM4B | chr8 | 98817581 | + |
| In-frame | ENST00000351270 | ENST00000521545 | HABP2 | chr10 | 115312949 | + | LAPTM4B | chr8 | 98817581 | + |
| In-frame | ENST00000541666 | ENST00000445593 | HABP2 | chr10 | 115312949 | + | LAPTM4B | chr8 | 98817581 | + |
| In-frame | ENST00000541666 | ENST00000521545 | HABP2 | chr10 | 115312949 | + | LAPTM4B | chr8 | 98817581 | + |
| intron-3CDS | ENST00000460714 | ENST00000445593 | HABP2 | chr10 | 115312949 | + | LAPTM4B | chr8 | 98817581 | + |
| intron-3CDS | ENST00000460714 | ENST00000521545 | HABP2 | chr10 | 115312949 | + | LAPTM4B | chr8 | 98817581 | + |
| intron-3CDS | ENST00000542051 | ENST00000445593 | HABP2 | chr10 | 115312949 | + | LAPTM4B | chr8 | 98817581 | + |
| intron-3CDS | ENST00000542051 | ENST00000521545 | HABP2 | chr10 | 115312949 | + | LAPTM4B | chr8 | 98817581 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000351270 | HABP2 | chr10 | 115312949 | + | ENST00000445593 | LAPTM4B | chr8 | 98817581 | + | 2286 | 165 | 96 | 746 | 216 |
| ENST00000351270 | HABP2 | chr10 | 115312949 | + | ENST00000521545 | LAPTM4B | chr8 | 98817581 | + | 747 | 165 | 96 | 746 | 216 |
| ENST00000541666 | HABP2 | chr10 | 115312949 | + | ENST00000445593 | LAPTM4B | chr8 | 98817581 | + | 2240 | 119 | 50 | 700 | 216 |
| ENST00000541666 | HABP2 | chr10 | 115312949 | + | ENST00000521545 | LAPTM4B | chr8 | 98817581 | + | 701 | 119 | 50 | 700 | 217 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000351270 | ENST00000445593 | HABP2 | chr10 | 115312949 | + | LAPTM4B | chr8 | 98817581 | + | 0.001224481 | 0.99877554 |
| ENST00000351270 | ENST00000521545 | HABP2 | chr10 | 115312949 | + | LAPTM4B | chr8 | 98817581 | + | 0.000638845 | 0.9993612 |
| ENST00000541666 | ENST00000445593 | HABP2 | chr10 | 115312949 | + | LAPTM4B | chr8 | 98817581 | + | 0.000954869 | 0.9990451 |
| ENST00000541666 | ENST00000521545 | HABP2 | chr10 | 115312949 | + | LAPTM4B | chr8 | 98817581 | + | 0.000589841 | 0.99941015 |
Top |
Fusion Genomic Features for HABP2-LAPTM4B |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| HABP2 | chr10 | 115312949 | + | LAPTM4B | chr8 | 98817580 | + | 6.83E-07 | 0.9999993 |
| HABP2 | chr10 | 115312949 | + | LAPTM4B | chr8 | 98817580 | + | 6.83E-07 | 0.9999993 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
 |
Top |
Fusion Protein Features for HABP2-LAPTM4B |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr10:115312949/chr8:98817581) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| HABP2 | LAPTM4B |
| FUNCTION: Cleaves the alpha-chain at multiple sites and the beta-chain between 'Lys-53' and 'Lys-54' but not the gamma-chain of fibrinogen and therefore does not initiate the formation of the fibrin clot and does not cause the fibrinolysis directly. It does not cleave (activate) prothrombin and plasminogen but converts the inactive single chain urinary plasminogen activator (pro-urokinase) to the active two chain form. Activates coagulation factor VII (PubMed:8827452, PubMed:10754382, PubMed:11217080). May function as a tumor suppressor negatively regulating cell proliferation and cell migration (PubMed:26222560). {ECO:0000269|PubMed:10754382, ECO:0000269|PubMed:11217080, ECO:0000269|PubMed:26222560, ECO:0000269|PubMed:8827452}. | FUNCTION: Required for optimal lysosomal function (PubMed:21224396). Blocks EGF-stimulated EGFR intraluminal sorting and degradation. Conversely by binding with the phosphatidylinositol 4,5-bisphosphate, regulates its PIP5K1C interaction, inhibits HGS ubiquitination and relieves LAPTM4B inhibition of EGFR degradation (PubMed:25588945). Recruits SLC3A2 and SLC7A5 (the Leu transporter) to the lysosome, promoting entry of leucine and other essential amino acid (EAA) into the lysosome, stimulating activation of proton-transporting vacuolar (V)-ATPase protein pump (V-ATPase) and hence mTORC1 activation (PubMed:25998567). Plays a role as negative regulator of TGFB1 production in regulatory T cells (PubMed:26126825). Binds ceramide and facilitates its exit from late endosome in order to control cell death pathways (PubMed:26280656). {ECO:0000269|PubMed:21224396, ECO:0000269|PubMed:25588945, ECO:0000269|PubMed:25998567, ECO:0000269|PubMed:26126825, ECO:0000269|PubMed:26280656}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | LAPTM4B | chr10:115312949 | chr8:98817581 | ENST00000445593 | 0 | 7 | 163_183 | 124 | 318.0 | Transmembrane | Helical | |
| Tgene | LAPTM4B | chr10:115312949 | chr8:98817581 | ENST00000445593 | 0 | 7 | 191_211 | 124 | 318.0 | Transmembrane | Helical | |
| Tgene | LAPTM4B | chr10:115312949 | chr8:98817581 | ENST00000445593 | 0 | 7 | 244_264 | 124 | 318.0 | Transmembrane | Helical | |
| Tgene | LAPTM4B | chr10:115312949 | chr8:98817581 | ENST00000521545 | 0 | 7 | 117_137 | 33 | 227.0 | Transmembrane | Helical | |
| Tgene | LAPTM4B | chr10:115312949 | chr8:98817581 | ENST00000521545 | 0 | 7 | 163_183 | 33 | 227.0 | Transmembrane | Helical | |
| Tgene | LAPTM4B | chr10:115312949 | chr8:98817581 | ENST00000521545 | 0 | 7 | 191_211 | 33 | 227.0 | Transmembrane | Helical | |
| Tgene | LAPTM4B | chr10:115312949 | chr8:98817581 | ENST00000521545 | 0 | 7 | 244_264 | 33 | 227.0 | Transmembrane | Helical |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | HABP2 | chr10:115312949 | chr8:98817581 | ENST00000351270 | + | 1 | 13 | 111_148 | 23 | 561.0 | Domain | EGF-like 2 |
| Hgene | HABP2 | chr10:115312949 | chr8:98817581 | ENST00000351270 | + | 1 | 13 | 150_188 | 23 | 561.0 | Domain | EGF-like 3 |
| Hgene | HABP2 | chr10:115312949 | chr8:98817581 | ENST00000351270 | + | 1 | 13 | 193_276 | 23 | 561.0 | Domain | Kringle |
| Hgene | HABP2 | chr10:115312949 | chr8:98817581 | ENST00000351270 | + | 1 | 13 | 314_555 | 23 | 561.0 | Domain | Peptidase S1 |
| Hgene | HABP2 | chr10:115312949 | chr8:98817581 | ENST00000351270 | + | 1 | 13 | 73_109 | 23 | 561.0 | Domain | EGF-like 1 |
| Hgene | HABP2 | chr10:115312949 | chr8:98817581 | ENST00000542051 | + | 1 | 13 | 111_148 | 0 | 535.0 | Domain | EGF-like 2 |
| Hgene | HABP2 | chr10:115312949 | chr8:98817581 | ENST00000542051 | + | 1 | 13 | 150_188 | 0 | 535.0 | Domain | EGF-like 3 |
| Hgene | HABP2 | chr10:115312949 | chr8:98817581 | ENST00000542051 | + | 1 | 13 | 193_276 | 0 | 535.0 | Domain | Kringle |
| Hgene | HABP2 | chr10:115312949 | chr8:98817581 | ENST00000542051 | + | 1 | 13 | 314_555 | 0 | 535.0 | Domain | Peptidase S1 |
| Hgene | HABP2 | chr10:115312949 | chr8:98817581 | ENST00000542051 | + | 1 | 13 | 73_109 | 0 | 535.0 | Domain | EGF-like 1 |
| Tgene | LAPTM4B | chr10:115312949 | chr8:98817581 | ENST00000445593 | 0 | 7 | 117_137 | 124 | 318.0 | Transmembrane | Helical |
Top |
Fusion Gene Sequence for HABP2-LAPTM4B |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >35545_35545_1_HABP2-LAPTM4B_HABP2_chr10_115312949_ENST00000351270_LAPTM4B_chr8_98817581_ENST00000445593_length(transcript)=2286nt_BP=165nt CCTGAATCCTTGGAGACTGACATTTTTCCCCCCTAAAGGCATAGACAACAAAAGAAATTTTATTGAGAGGAAAACACAAGTCCTTAAACT GCAAAGATGTTTGCCAGGATGTCTGATCTCCATGTTCTGCTGTTAATGGCTCTGGTGGGAAAGACAGCCTGTGGGATCATCAATGCTGTG GTACTGTTGATTTTATTGAGTGCCCTGGCTGATCCGGATCAGTATAACTTTTCAAGTTCTGAACTGGGAGGTGACTTTGAGTTCATGGAT GATGCCAACATGTGCATTGCCATTGCGATTTCTCTTCTCATGATCCTGATATGTGCTATGGCTACTTACGGAGCGTACAAGCAACGCGCA GCCTGGATCATCCCATTCTTCTGTTACCAGATCTTTGACTTTGCCCTGAACATGTTGGTTGCAATCACTGTGCTTATTTATCCAAACTCC ATTCAGGAATACATACGGCAACTGCCTCCTAATTTTCCCTACAGAGATGATGTCATGTCAGTGAATCCTACCTGTTTGGTCCTTATTATT CTTCTGTTTATTAGCATTATCTTGACTTTTAAGGGTTACTTGATTAGCTGTGTTTGGAACTGCTACCGATACATCAATGGTAGGAACTCC TCTGATGTCCTGGTTTATGTTACCAGCAATGACACTACGGTGCTGCTACCCCCGTATGATGATGCCACTGTGAATGGTGCTGCCAAGGAG CCACCGCCACCTTACGTGTCTGCCTAAGCCTTCAAGTGGGCGGAGCTGAGGGCAGCAGCTTGACTTTGCAGACATCTGAGCAATAGTTCT GTTATTTCACTTTTGCCATGAGCCTCTCTGAGCTTGTTTGTTGCTGAAATGCTACTTTTTAAAATTTAGATGTTAGATTGAAAACTGTAG TTTTCAACATATGCTTTGCTGGAACACTGTGATAGATTAACTGTAGAATTCTTCCTGTACGATTGGGGATATAATGGGCTTCACTAACCT TCCCTAGGCATTGAAACTTCCCCCAAATCTGATGGACCTAGAAGTCTGCTTTTGTACCTGCTGGGCCCCAAAGTTGGGCATTTTTCTCTC TGTTCCCTCTCTTTTGAAAATGTAAAATAAAACCAAAAATAGACAACTTTTTCTTCAGCCATTCCAGCATAGAGAACAAAACCTTATGGA AACAGGAATGTCAATTGTGTAATCATTGTTCTAATTAGGTAAATAGAAGTCCTTATGTATGTGTTACAAGAATTTCCCCCACAACATCCT TTATGACTGAAGTTCAATGACAGTTTGTGTTTGGTGGTAAAGGATTTTCTCCATGGCCTGAATTAAGACCATTAGAAAGCACCAGGCCGT GGGAGCAGTGACCATCTGCTGACTGTTCTTGTGGATCTTGTGTCCAGGGACATGGGGTGACATGCCTCGTATGTGTTAGAGGGTGGAATG GATGTGTTTGGCGCTGCATGGGATCTGGTGCCCCTCTTCTCCTGGATTCACATCCCCACCCAGGGCCCGCTTTTACTAAGTGTTCTGCCC TAGATTGGTTCAAGGAGGTCATCCAACTGACTTTATCAAGTGGAATTGGGATATATTTGATATACTTCTGCCTAACAACATGGAAAAGGG TTTTCTTTTCCCTGCAAGCTACATCCTACTGCTTTGAACTTCCAAGTATGTCTAGTCACCTTTTAAAATGTAAACATTTTCAGAAAAATG AGGATTGCCTTCCTTGTATGCGCTTTTTACCTTGACTACCTGAATTGCAAGGGATTTTTATATATTCATATGTTACAAAGTCAGCAACTC TCCTGTTGGTTCATTATTGAATGTGCTGTAAATTAAGTTGTTTGCAATTAAAACAAGGTTTGCCCACATCCAAGATGACCTTGTGATTTT GTGCTGATTGTGTCTGAGGACCTTTCCCTCCACATATGGTCTGGCAGATGCACCCAGTTCAGCCTAAGGAGTAGGCTTTTTTTTGGGGGG GGGAGGTCGGGTGGGGGGGATTTTTAATCTTTTAATTTTCAAGATGGTTAAAATATTGAAATGTTTAAGTGGATAACTATATTATTCATA AAATGACTGAGTGAAAACTAATACACTATGAGATGAAAAGTACTTGTCAGGGATTTTCAAGCTTTTGCTCAAGTAATTACTATGAAATAA CAATTTCTAGAAATGAAGAACAATCCGTGGAGAATAAATTACCATTGGTGTGGGGGAAAAAAGCCAAACAGAAGTAGAAAAAGGTGTAGC >35545_35545_1_HABP2-LAPTM4B_HABP2_chr10_115312949_ENST00000351270_LAPTM4B_chr8_98817581_ENST00000445593_length(amino acids)=216AA_BP=10 MFARMSDLHVLLLMALVGKTACGIINAVVLLILLSALADPDQYNFSSSELGGDFEFMDDANMCIAIAISLLMILICAMATYGAYKQRAAW IIPFFCYQIFDFALNMLVAITVLIYPNSIQEYIRQLPPNFPYRDDVMSVNPTCLVLIILLFISIILTFKGYLISCVWNCYRYINGRNSSD -------------------------------------------------------------- >35545_35545_2_HABP2-LAPTM4B_HABP2_chr10_115312949_ENST00000351270_LAPTM4B_chr8_98817581_ENST00000521545_length(transcript)=747nt_BP=165nt CCTGAATCCTTGGAGACTGACATTTTTCCCCCCTAAAGGCATAGACAACAAAAGAAATTTTATTGAGAGGAAAACACAAGTCCTTAAACT GCAAAGATGTTTGCCAGGATGTCTGATCTCCATGTTCTGCTGTTAATGGCTCTGGTGGGAAAGACAGCCTGTGGGATCATCAATGCTGTG GTACTGTTGATTTTATTGAGTGCCCTGGCTGATCCGGATCAGTATAACTTTTCAAGTTCTGAACTGGGAGGTGACTTTGAGTTCATGGAT GATGCCAACATGTGCATTGCCATTGCGATTTCTCTTCTCATGATCCTGATATGTGCTATGGCTACTTACGGAGCGTACAAGCAACGCGCA GCCTGGATCATCCCATTCTTCTGTTACCAGATCTTTGACTTTGCCCTGAACATGTTGGTTGCAATCACTGTGCTTATTTATCCAAACTCC ATTCAGGAATACATACGGCAACTGCCTCCTAATTTTCCCTACAGAGATGATGTCATGTCAGTGAATCCTACCTGTTTGGTCCTTATTATT CTTCTGTTTATTAGCATTATCTTGACTTTTAAGGGTTACTTGATTAGCTGTGTTTGGAACTGCTACCGATACATCAATGGTAGGAACTCC TCTGATGTCCTGGTTTATGTTACCAGCAATGACACTACGGTGCTGCTACCCCCGTATGATGATGCCACTGTGAATGGTGCTGCCAAGGAG >35545_35545_2_HABP2-LAPTM4B_HABP2_chr10_115312949_ENST00000351270_LAPTM4B_chr8_98817581_ENST00000521545_length(amino acids)=216AA_BP=10 MFARMSDLHVLLLMALVGKTACGIINAVVLLILLSALADPDQYNFSSSELGGDFEFMDDANMCIAIAISLLMILICAMATYGAYKQRAAW IIPFFCYQIFDFALNMLVAITVLIYPNSIQEYIRQLPPNFPYRDDVMSVNPTCLVLIILLFISIILTFKGYLISCVWNCYRYINGRNSSD -------------------------------------------------------------- >35545_35545_3_HABP2-LAPTM4B_HABP2_chr10_115312949_ENST00000541666_LAPTM4B_chr8_98817581_ENST00000445593_length(transcript)=2240nt_BP=119nt AACAAAAGAAATTTTATTGAGAGGAAAACACAAGTCCTTAAACTGCAAAGATGTTTGCCAGGATGTCTGATCTCCATGTTCTGCTGTTAA TGGCTCTGGTGGGAAAGACAGCCTGTGGGATCATCAATGCTGTGGTACTGTTGATTTTATTGAGTGCCCTGGCTGATCCGGATCAGTATA ACTTTTCAAGTTCTGAACTGGGAGGTGACTTTGAGTTCATGGATGATGCCAACATGTGCATTGCCATTGCGATTTCTCTTCTCATGATCC TGATATGTGCTATGGCTACTTACGGAGCGTACAAGCAACGCGCAGCCTGGATCATCCCATTCTTCTGTTACCAGATCTTTGACTTTGCCC TGAACATGTTGGTTGCAATCACTGTGCTTATTTATCCAAACTCCATTCAGGAATACATACGGCAACTGCCTCCTAATTTTCCCTACAGAG ATGATGTCATGTCAGTGAATCCTACCTGTTTGGTCCTTATTATTCTTCTGTTTATTAGCATTATCTTGACTTTTAAGGGTTACTTGATTA GCTGTGTTTGGAACTGCTACCGATACATCAATGGTAGGAACTCCTCTGATGTCCTGGTTTATGTTACCAGCAATGACACTACGGTGCTGC TACCCCCGTATGATGATGCCACTGTGAATGGTGCTGCCAAGGAGCCACCGCCACCTTACGTGTCTGCCTAAGCCTTCAAGTGGGCGGAGC TGAGGGCAGCAGCTTGACTTTGCAGACATCTGAGCAATAGTTCTGTTATTTCACTTTTGCCATGAGCCTCTCTGAGCTTGTTTGTTGCTG AAATGCTACTTTTTAAAATTTAGATGTTAGATTGAAAACTGTAGTTTTCAACATATGCTTTGCTGGAACACTGTGATAGATTAACTGTAG AATTCTTCCTGTACGATTGGGGATATAATGGGCTTCACTAACCTTCCCTAGGCATTGAAACTTCCCCCAAATCTGATGGACCTAGAAGTC TGCTTTTGTACCTGCTGGGCCCCAAAGTTGGGCATTTTTCTCTCTGTTCCCTCTCTTTTGAAAATGTAAAATAAAACCAAAAATAGACAA CTTTTTCTTCAGCCATTCCAGCATAGAGAACAAAACCTTATGGAAACAGGAATGTCAATTGTGTAATCATTGTTCTAATTAGGTAAATAG AAGTCCTTATGTATGTGTTACAAGAATTTCCCCCACAACATCCTTTATGACTGAAGTTCAATGACAGTTTGTGTTTGGTGGTAAAGGATT TTCTCCATGGCCTGAATTAAGACCATTAGAAAGCACCAGGCCGTGGGAGCAGTGACCATCTGCTGACTGTTCTTGTGGATCTTGTGTCCA GGGACATGGGGTGACATGCCTCGTATGTGTTAGAGGGTGGAATGGATGTGTTTGGCGCTGCATGGGATCTGGTGCCCCTCTTCTCCTGGA TTCACATCCCCACCCAGGGCCCGCTTTTACTAAGTGTTCTGCCCTAGATTGGTTCAAGGAGGTCATCCAACTGACTTTATCAAGTGGAAT TGGGATATATTTGATATACTTCTGCCTAACAACATGGAAAAGGGTTTTCTTTTCCCTGCAAGCTACATCCTACTGCTTTGAACTTCCAAG TATGTCTAGTCACCTTTTAAAATGTAAACATTTTCAGAAAAATGAGGATTGCCTTCCTTGTATGCGCTTTTTACCTTGACTACCTGAATT GCAAGGGATTTTTATATATTCATATGTTACAAAGTCAGCAACTCTCCTGTTGGTTCATTATTGAATGTGCTGTAAATTAAGTTGTTTGCA ATTAAAACAAGGTTTGCCCACATCCAAGATGACCTTGTGATTTTGTGCTGATTGTGTCTGAGGACCTTTCCCTCCACATATGGTCTGGCA GATGCACCCAGTTCAGCCTAAGGAGTAGGCTTTTTTTTGGGGGGGGGAGGTCGGGTGGGGGGGATTTTTAATCTTTTAATTTTCAAGATG GTTAAAATATTGAAATGTTTAAGTGGATAACTATATTATTCATAAAATGACTGAGTGAAAACTAATACACTATGAGATGAAAAGTACTTG TCAGGGATTTTCAAGCTTTTGCTCAAGTAATTACTATGAAATAACAATTTCTAGAAATGAAGAACAATCCGTGGAGAATAAATTACCATT >35545_35545_3_HABP2-LAPTM4B_HABP2_chr10_115312949_ENST00000541666_LAPTM4B_chr8_98817581_ENST00000445593_length(amino acids)=216AA_BP=10 MFARMSDLHVLLLMALVGKTACGIINAVVLLILLSALADPDQYNFSSSELGGDFEFMDDANMCIAIAISLLMILICAMATYGAYKQRAAW IIPFFCYQIFDFALNMLVAITVLIYPNSIQEYIRQLPPNFPYRDDVMSVNPTCLVLIILLFISIILTFKGYLISCVWNCYRYINGRNSSD -------------------------------------------------------------- >35545_35545_4_HABP2-LAPTM4B_HABP2_chr10_115312949_ENST00000541666_LAPTM4B_chr8_98817581_ENST00000521545_length(transcript)=701nt_BP=119nt AACAAAAGAAATTTTATTGAGAGGAAAACACAAGTCCTTAAACTGCAAAGATGTTTGCCAGGATGTCTGATCTCCATGTTCTGCTGTTAA TGGCTCTGGTGGGAAAGACAGCCTGTGGGATCATCAATGCTGTGGTACTGTTGATTTTATTGAGTGCCCTGGCTGATCCGGATCAGTATA ACTTTTCAAGTTCTGAACTGGGAGGTGACTTTGAGTTCATGGATGATGCCAACATGTGCATTGCCATTGCGATTTCTCTTCTCATGATCC TGATATGTGCTATGGCTACTTACGGAGCGTACAAGCAACGCGCAGCCTGGATCATCCCATTCTTCTGTTACCAGATCTTTGACTTTGCCC TGAACATGTTGGTTGCAATCACTGTGCTTATTTATCCAAACTCCATTCAGGAATACATACGGCAACTGCCTCCTAATTTTCCCTACAGAG ATGATGTCATGTCAGTGAATCCTACCTGTTTGGTCCTTATTATTCTTCTGTTTATTAGCATTATCTTGACTTTTAAGGGTTACTTGATTA GCTGTGTTTGGAACTGCTACCGATACATCAATGGTAGGAACTCCTCTGATGTCCTGGTTTATGTTACCAGCAATGACACTACGGTGCTGC >35545_35545_4_HABP2-LAPTM4B_HABP2_chr10_115312949_ENST00000541666_LAPTM4B_chr8_98817581_ENST00000521545_length(amino acids)=217AA_BP=10 MFARMSDLHVLLLMALVGKTACGIINAVVLLILLSALADPDQYNFSSSELGGDFEFMDDANMCIAIAISLLMILICAMATYGAYKQRAAW IIPFFCYQIFDFALNMLVAITVLIYPNSIQEYIRQLPPNFPYRDDVMSVNPTCLVLIILLFISIILTFKGYLISCVWNCYRYINGRNSSD -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for HABP2-LAPTM4B |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for HABP2-LAPTM4B |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for HABP2-LAPTM4B |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
