
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:HADH-HPGD (FusionGDB2 ID:35582) |
Fusion Gene Summary for HADH-HPGD |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: HADH-HPGD | Fusion gene ID: 35582 | Hgene | Tgene | Gene symbol | HADH | HPGD | Gene ID | 3033 | 3248 |
| Gene name | hydroxyacyl-CoA dehydrogenase | 15-hydroxyprostaglandin dehydrogenase | |
| Synonyms | HAD|HADH1|HADHSC|HCDH|HHF4|MSCHAD|SCHAD | 15-PGDH|PGDH|PGDH1|PHOAR1|SDR36C1 | |
| Cytomap | 4q25 | 4q34.1 | |
| Type of gene | protein-coding | protein-coding | |
| Description | hydroxyacyl-coenzyme A dehydrogenase, mitochondrialL-3-hydroxyacyl-Coenzyme A dehydrogenase, short chainmedium and short-chain L-3-hydroxyacyl-coenzyme A dehydrogenaseshort-chain 3-hydroxyacyl-CoA dehydrogenasetestis secretory sperm-binding protein Li | 15-hydroxyprostaglandin dehydrogenase [NAD(+)]15-hydroxyprostaglandin dehydrogenase (NAD(+))NAD+-dependent 15-hydroxyprostaglandin dehydrogenasehydroxyprostaglandin dehydrogenase 15-(NAD)prostaglandin dehydrogenase 1short chain dehydrogenase/reductas | |
| Modification date | 20200320 | 20200313 | |
| UniProtAcc | Q16836 | O60760 | |
| Ensembl transtripts involved in fusion gene | ENST00000309522, ENST00000403312, ENST00000603302, ENST00000505878, ENST00000454409, ENST00000510728, | ENST00000504433, ENST00000510901, ENST00000541923, ENST00000542498, ENST00000422112, ENST00000296521, ENST00000296522, | |
| Fusion gene scores | * DoF score | 7 X 6 X 3=126 | 2 X 2 X 2=8 |
| # samples | 7 | 2 | |
| ** MAII score | log2(7/126*10)=-0.84799690655495 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(2/8*10)=1.32192809488736 | |
| Context | PubMed: HADH [Title/Abstract] AND HPGD [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | HADH(108911220)-HPGD(175429943), # samples:4 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Tgene | HPGD | GO:0006693 | prostaglandin metabolic process | 16828555|21072165 |
| Tgene | HPGD | GO:0007179 | transforming growth factor beta receptor signaling pathway | 15574495 |
| Tgene | HPGD | GO:0007565 | female pregnancy | 15531523 |
| Tgene | HPGD | GO:0007567 | parturition | 15531523 |
| Tgene | HPGD | GO:0045786 | negative regulation of cell cycle | 15574495 |
 Fusion gene breakpoints across HADH (5'-gene) Fusion gene breakpoints across HADH (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
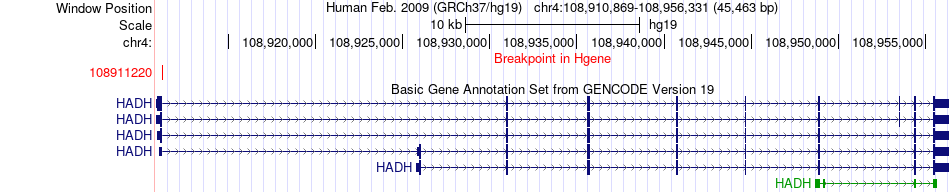 |
 Fusion gene breakpoints across HPGD (3'-gene) Fusion gene breakpoints across HPGD (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
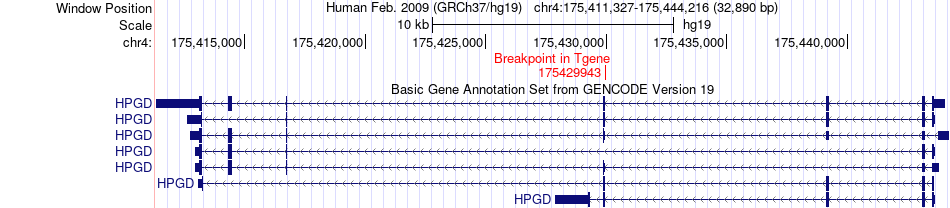 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | UCEC | TCGA-AJ-A3NF-01A | HADH | chr4 | 108911220 | - | HPGD | chr4 | 175429943 | - |
| ChimerDB4 | UCEC | TCGA-AJ-A3NF-01A | HADH | chr4 | 108911220 | + | HPGD | chr4 | 175429943 | - |
| ChimerDB4 | UCEC | TCGA-AJ-A3NF | HADH | chr4 | 108911220 | + | HPGD | chr4 | 175429943 | - |
Top |
Fusion Gene ORF analysis for HADH-HPGD |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-5UTR | ENST00000309522 | ENST00000504433 | HADH | chr4 | 108911220 | + | HPGD | chr4 | 175429943 | - |
| 5CDS-5UTR | ENST00000309522 | ENST00000510901 | HADH | chr4 | 108911220 | + | HPGD | chr4 | 175429943 | - |
| 5CDS-5UTR | ENST00000309522 | ENST00000541923 | HADH | chr4 | 108911220 | + | HPGD | chr4 | 175429943 | - |
| 5CDS-5UTR | ENST00000309522 | ENST00000542498 | HADH | chr4 | 108911220 | + | HPGD | chr4 | 175429943 | - |
| 5CDS-5UTR | ENST00000403312 | ENST00000504433 | HADH | chr4 | 108911220 | + | HPGD | chr4 | 175429943 | - |
| 5CDS-5UTR | ENST00000403312 | ENST00000510901 | HADH | chr4 | 108911220 | + | HPGD | chr4 | 175429943 | - |
| 5CDS-5UTR | ENST00000403312 | ENST00000541923 | HADH | chr4 | 108911220 | + | HPGD | chr4 | 175429943 | - |
| 5CDS-5UTR | ENST00000403312 | ENST00000542498 | HADH | chr4 | 108911220 | + | HPGD | chr4 | 175429943 | - |
| 5CDS-5UTR | ENST00000603302 | ENST00000504433 | HADH | chr4 | 108911220 | + | HPGD | chr4 | 175429943 | - |
| 5CDS-5UTR | ENST00000603302 | ENST00000510901 | HADH | chr4 | 108911220 | + | HPGD | chr4 | 175429943 | - |
| 5CDS-5UTR | ENST00000603302 | ENST00000541923 | HADH | chr4 | 108911220 | + | HPGD | chr4 | 175429943 | - |
| 5CDS-5UTR | ENST00000603302 | ENST00000542498 | HADH | chr4 | 108911220 | + | HPGD | chr4 | 175429943 | - |
| 5CDS-intron | ENST00000309522 | ENST00000422112 | HADH | chr4 | 108911220 | + | HPGD | chr4 | 175429943 | - |
| 5CDS-intron | ENST00000403312 | ENST00000422112 | HADH | chr4 | 108911220 | + | HPGD | chr4 | 175429943 | - |
| 5CDS-intron | ENST00000603302 | ENST00000422112 | HADH | chr4 | 108911220 | + | HPGD | chr4 | 175429943 | - |
| 5UTR-3CDS | ENST00000505878 | ENST00000296521 | HADH | chr4 | 108911220 | + | HPGD | chr4 | 175429943 | - |
| 5UTR-3CDS | ENST00000505878 | ENST00000296522 | HADH | chr4 | 108911220 | + | HPGD | chr4 | 175429943 | - |
| 5UTR-5UTR | ENST00000505878 | ENST00000504433 | HADH | chr4 | 108911220 | + | HPGD | chr4 | 175429943 | - |
| 5UTR-5UTR | ENST00000505878 | ENST00000510901 | HADH | chr4 | 108911220 | + | HPGD | chr4 | 175429943 | - |
| 5UTR-5UTR | ENST00000505878 | ENST00000541923 | HADH | chr4 | 108911220 | + | HPGD | chr4 | 175429943 | - |
| 5UTR-5UTR | ENST00000505878 | ENST00000542498 | HADH | chr4 | 108911220 | + | HPGD | chr4 | 175429943 | - |
| 5UTR-intron | ENST00000505878 | ENST00000422112 | HADH | chr4 | 108911220 | + | HPGD | chr4 | 175429943 | - |
| In-frame | ENST00000309522 | ENST00000296521 | HADH | chr4 | 108911220 | + | HPGD | chr4 | 175429943 | - |
| In-frame | ENST00000309522 | ENST00000296522 | HADH | chr4 | 108911220 | + | HPGD | chr4 | 175429943 | - |
| In-frame | ENST00000403312 | ENST00000296521 | HADH | chr4 | 108911220 | + | HPGD | chr4 | 175429943 | - |
| In-frame | ENST00000403312 | ENST00000296522 | HADH | chr4 | 108911220 | + | HPGD | chr4 | 175429943 | - |
| In-frame | ENST00000603302 | ENST00000296521 | HADH | chr4 | 108911220 | + | HPGD | chr4 | 175429943 | - |
| In-frame | ENST00000603302 | ENST00000296522 | HADH | chr4 | 108911220 | + | HPGD | chr4 | 175429943 | - |
| intron-3CDS | ENST00000454409 | ENST00000296521 | HADH | chr4 | 108911220 | + | HPGD | chr4 | 175429943 | - |
| intron-3CDS | ENST00000454409 | ENST00000296522 | HADH | chr4 | 108911220 | + | HPGD | chr4 | 175429943 | - |
| intron-3CDS | ENST00000510728 | ENST00000296521 | HADH | chr4 | 108911220 | + | HPGD | chr4 | 175429943 | - |
| intron-3CDS | ENST00000510728 | ENST00000296522 | HADH | chr4 | 108911220 | + | HPGD | chr4 | 175429943 | - |
| intron-5UTR | ENST00000454409 | ENST00000504433 | HADH | chr4 | 108911220 | + | HPGD | chr4 | 175429943 | - |
| intron-5UTR | ENST00000454409 | ENST00000510901 | HADH | chr4 | 108911220 | + | HPGD | chr4 | 175429943 | - |
| intron-5UTR | ENST00000454409 | ENST00000541923 | HADH | chr4 | 108911220 | + | HPGD | chr4 | 175429943 | - |
| intron-5UTR | ENST00000454409 | ENST00000542498 | HADH | chr4 | 108911220 | + | HPGD | chr4 | 175429943 | - |
| intron-5UTR | ENST00000510728 | ENST00000504433 | HADH | chr4 | 108911220 | + | HPGD | chr4 | 175429943 | - |
| intron-5UTR | ENST00000510728 | ENST00000510901 | HADH | chr4 | 108911220 | + | HPGD | chr4 | 175429943 | - |
| intron-5UTR | ENST00000510728 | ENST00000541923 | HADH | chr4 | 108911220 | + | HPGD | chr4 | 175429943 | - |
| intron-5UTR | ENST00000510728 | ENST00000542498 | HADH | chr4 | 108911220 | + | HPGD | chr4 | 175429943 | - |
| intron-intron | ENST00000454409 | ENST00000422112 | HADH | chr4 | 108911220 | + | HPGD | chr4 | 175429943 | - |
| intron-intron | ENST00000510728 | ENST00000422112 | HADH | chr4 | 108911220 | + | HPGD | chr4 | 175429943 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000603302 | HADH | chr4 | 108911220 | + | ENST00000296522 | HPGD | chr4 | 175429943 | - | 2607 | 351 | 42 | 827 | 261 |
| ENST00000603302 | HADH | chr4 | 108911220 | + | ENST00000296521 | HPGD | chr4 | 175429943 | - | 1162 | 351 | 42 | 563 | 173 |
| ENST00000403312 | HADH | chr4 | 108911220 | + | ENST00000296522 | HPGD | chr4 | 175429943 | - | 2607 | 351 | 42 | 827 | 261 |
| ENST00000403312 | HADH | chr4 | 108911220 | + | ENST00000296521 | HPGD | chr4 | 175429943 | - | 1162 | 351 | 42 | 563 | 173 |
| ENST00000309522 | HADH | chr4 | 108911220 | + | ENST00000296522 | HPGD | chr4 | 175429943 | - | 2537 | 281 | 41 | 757 | 238 |
| ENST00000309522 | HADH | chr4 | 108911220 | + | ENST00000296521 | HPGD | chr4 | 175429943 | - | 1092 | 281 | 41 | 493 | 150 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000603302 | ENST00000296522 | HADH | chr4 | 108911220 | + | HPGD | chr4 | 175429943 | - | 0.000706739 | 0.9992932 |
| ENST00000603302 | ENST00000296521 | HADH | chr4 | 108911220 | + | HPGD | chr4 | 175429943 | - | 0.040315192 | 0.95968485 |
| ENST00000403312 | ENST00000296522 | HADH | chr4 | 108911220 | + | HPGD | chr4 | 175429943 | - | 0.000706739 | 0.9992932 |
| ENST00000403312 | ENST00000296521 | HADH | chr4 | 108911220 | + | HPGD | chr4 | 175429943 | - | 0.040315192 | 0.95968485 |
| ENST00000309522 | ENST00000296522 | HADH | chr4 | 108911220 | + | HPGD | chr4 | 175429943 | - | 0.000345029 | 0.999655 |
| ENST00000309522 | ENST00000296521 | HADH | chr4 | 108911220 | + | HPGD | chr4 | 175429943 | - | 0.006506751 | 0.99349326 |
Top |
Fusion Genomic Features for HADH-HPGD |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
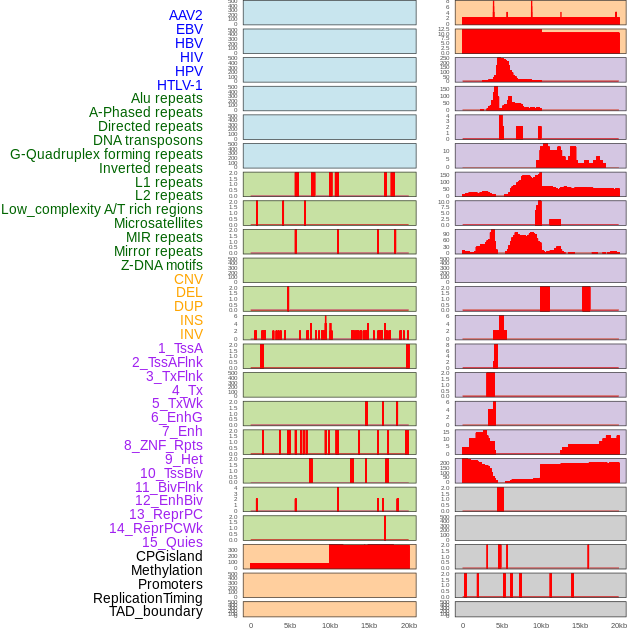 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for HADH-HPGD |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr4:108911220/chr4:175429943) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| HADH | HPGD |
| FUNCTION: Mitochondrial fatty acid beta-oxidation enzyme that catalyzes the third step of the beta-oxidation cycle for medium and short-chain 3-hydroxy fatty acyl-CoAs (C4 to C10) (PubMed:10231530, PubMed:11489939, PubMed:16725361). Plays a role in the control of insulin secretion by inhibiting the activation of glutamate dehydrogenase 1 (GLUD1), an enzyme that has an important role in regulating amino acid-induced insulin secretion (By similarity). {ECO:0000250|UniProtKB:Q61425, ECO:0000269|PubMed:10231530, ECO:0000269|PubMed:11489939, ECO:0000269|PubMed:16725361}. | FUNCTION: Bifunctional enzyme which catalyzes both the conversion of PGH2 to PGD2, a prostaglandin involved in smooth muscle contraction/relaxation and a potent inhibitor of platelet aggregation, and the conjugation of glutathione with a wide range of aryl halides and organic isothiocyanates. Also exhibits low glutathione-peroxidase activity towards cumene hydroperoxide. {ECO:0000269|PubMed:10824118, ECO:0000269|PubMed:11672424, ECO:0000269|PubMed:12244105, ECO:0000269|PubMed:12627223, ECO:0000269|PubMed:15113825, ECO:0000269|PubMed:16547010, ECO:0000269|PubMed:19939518, ECO:0000269|PubMed:9353279, ECO:0000269|PubMed:9425264}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | HADH | chr4:108911220 | chr4:175429943 | ENST00000309522 | + | 1 | 8 | 34_39 | 44 | 315.0 | Nucleotide binding | NAD |
| Hgene | HADH | chr4:108911220 | chr4:175429943 | ENST00000603302 | + | 1 | 9 | 34_39 | 44 | 332.0 | Nucleotide binding | NAD |
| Tgene | HPGD | chr4:108911220 | chr4:175429943 | ENST00000296521 | 2 | 6 | 151_155 | 108 | 179.0 | Nucleotide binding | NAD | |
| Tgene | HPGD | chr4:108911220 | chr4:175429943 | ENST00000296522 | 2 | 7 | 151_155 | 108 | 267.0 | Nucleotide binding | NAD | |
| Tgene | HPGD | chr4:108911220 | chr4:175429943 | ENST00000422112 | 0 | 5 | 12_20 | 0 | 199.0 | Nucleotide binding | NAD | |
| Tgene | HPGD | chr4:108911220 | chr4:175429943 | ENST00000422112 | 0 | 5 | 151_155 | 0 | 199.0 | Nucleotide binding | NAD | |
| Tgene | HPGD | chr4:108911220 | chr4:175429943 | ENST00000422112 | 0 | 5 | 36_37 | 0 | 199.0 | Nucleotide binding | NAD | |
| Tgene | HPGD | chr4:108911220 | chr4:175429943 | ENST00000422112 | 0 | 5 | 63_65 | 0 | 199.0 | Nucleotide binding | NAD | |
| Tgene | HPGD | chr4:108911220 | chr4:175429943 | ENST00000510901 | 2 | 7 | 12_20 | 0 | 146.0 | Nucleotide binding | NAD | |
| Tgene | HPGD | chr4:108911220 | chr4:175429943 | ENST00000510901 | 2 | 7 | 151_155 | 0 | 146.0 | Nucleotide binding | NAD | |
| Tgene | HPGD | chr4:108911220 | chr4:175429943 | ENST00000510901 | 2 | 7 | 36_37 | 0 | 146.0 | Nucleotide binding | NAD | |
| Tgene | HPGD | chr4:108911220 | chr4:175429943 | ENST00000510901 | 2 | 7 | 63_65 | 0 | 146.0 | Nucleotide binding | NAD | |
| Tgene | HPGD | chr4:108911220 | chr4:175429943 | ENST00000541923 | 1 | 6 | 12_20 | 0 | 146.0 | Nucleotide binding | NAD | |
| Tgene | HPGD | chr4:108911220 | chr4:175429943 | ENST00000541923 | 1 | 6 | 151_155 | 0 | 146.0 | Nucleotide binding | NAD | |
| Tgene | HPGD | chr4:108911220 | chr4:175429943 | ENST00000541923 | 1 | 6 | 36_37 | 0 | 146.0 | Nucleotide binding | NAD | |
| Tgene | HPGD | chr4:108911220 | chr4:175429943 | ENST00000541923 | 1 | 6 | 63_65 | 0 | 146.0 | Nucleotide binding | NAD | |
| Tgene | HPGD | chr4:108911220 | chr4:175429943 | ENST00000542498 | 2 | 5 | 151_155 | 108 | 144.0 | Nucleotide binding | NAD |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | HPGD | chr4:108911220 | chr4:175429943 | ENST00000296521 | 2 | 6 | 12_20 | 108 | 179.0 | Nucleotide binding | NAD | |
| Tgene | HPGD | chr4:108911220 | chr4:175429943 | ENST00000296521 | 2 | 6 | 36_37 | 108 | 179.0 | Nucleotide binding | NAD | |
| Tgene | HPGD | chr4:108911220 | chr4:175429943 | ENST00000296521 | 2 | 6 | 63_65 | 108 | 179.0 | Nucleotide binding | NAD | |
| Tgene | HPGD | chr4:108911220 | chr4:175429943 | ENST00000296522 | 2 | 7 | 12_20 | 108 | 267.0 | Nucleotide binding | NAD | |
| Tgene | HPGD | chr4:108911220 | chr4:175429943 | ENST00000296522 | 2 | 7 | 36_37 | 108 | 267.0 | Nucleotide binding | NAD | |
| Tgene | HPGD | chr4:108911220 | chr4:175429943 | ENST00000296522 | 2 | 7 | 63_65 | 108 | 267.0 | Nucleotide binding | NAD | |
| Tgene | HPGD | chr4:108911220 | chr4:175429943 | ENST00000542498 | 2 | 5 | 12_20 | 108 | 144.0 | Nucleotide binding | NAD | |
| Tgene | HPGD | chr4:108911220 | chr4:175429943 | ENST00000542498 | 2 | 5 | 36_37 | 108 | 144.0 | Nucleotide binding | NAD | |
| Tgene | HPGD | chr4:108911220 | chr4:175429943 | ENST00000542498 | 2 | 5 | 63_65 | 108 | 144.0 | Nucleotide binding | NAD |
Top |
Fusion Gene Sequence for HADH-HPGD |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >35582_35582_1_HADH-HPGD_HADH_chr4_108911220_ENST00000309522_HPGD_chr4_175429943_ENST00000296521_length(transcript)=1092nt_BP=281nt CCCCGCCTTGTGGCGTCACGGGGACGCCGGGGGCGCGCGGGCTGCAGGGCCGCGTAGGTCCCCGCCCCCAGAGTCTGGCTTTCCGCGGCT GCCCGCCTCGCGCGTCTTCCCTGCCCGGGTCTCCTCGCTGTCGCCGCCGCTGCCACACCATGGCCTTCGTCACCAGGCAGTTCATGCGTT CCGTGTCCTCCTCGTCCACCGCCTCGGCCTCGGCCAAGAAGATAATCGTCAAGCACGTGACGGTCATCGGCGGCGGGCTGATGGGCGCCG GCATTGCCCAGGTTTCTGTTATCAGTGGAACCTATCTTGGTTTGGATTACATGAGTAAGCAAAATGGAGGTGAAGGCGGCATCATTATCA ATATGTCATCTTTAGCAGGACTCATGCCCGTTGCACAGCAGCCGGTTTATTGTGCTTCAAAGCATGGCATAGTTGGATTCACACGCTCAG CAGCGCCCACCATTGATTGCCAATGGATTGATAACACTCATTGAAGATGATGCTTTAAATGGTGCTATTATGAAGATCACAACTTCTAAG GGAATTCATTTTCAAGACTATGATACAACTCCATTTCAAGCAAAAACCCAATGAACAGCTTATGTGTTAGCCATAGCTGAAAATAAGCAC AAATAGCTTATATTCAGATCCTATCTTCATTTGAATATAGCTTTTAAATGAAATGTTACAGTTTGAAGTTTTCCTTCATGCACTTGGTGA TAAACGTTTTCTAAATTTTTAGTTAAGTATATGGATAAAAAGTTATGAACTATTAAAAATGTGATGTGGACCAAAGGCTAGGTTGTAATC TTGATAGTCTAAAAAATGATCAAAACAAATGATTTTCAAGGAATATTCAATATTCTGCCTTTCAGAAAGTGTATTTATATCTGTGCTTCA TAAATATTAATGTTCTTCAGAACATCATTTTAAAGGAGATACTTGAATTGTTATTTAAATCAAACCAGATGTAAAACACTCACATACAAG TTCATACTTTAAAAGAGGAAAGCTACTTAACAATGACAAATATTTCACAATAATAATTTTTACTTATATACCATCTTTCAACTGAACATT >35582_35582_1_HADH-HPGD_HADH_chr4_108911220_ENST00000309522_HPGD_chr4_175429943_ENST00000296521_length(amino acids)=150AA_BP=80 MQGRVGPRPQSLAFRGCPPRASSLPGSPRCRRRCHTMAFVTRQFMRSVSSSSTASASAKKIIVKHVTVIGGGLMGAGIAQVSVISGTYLG -------------------------------------------------------------- >35582_35582_2_HADH-HPGD_HADH_chr4_108911220_ENST00000309522_HPGD_chr4_175429943_ENST00000296522_length(transcript)=2537nt_BP=281nt CCCCGCCTTGTGGCGTCACGGGGACGCCGGGGGCGCGCGGGCTGCAGGGCCGCGTAGGTCCCCGCCCCCAGAGTCTGGCTTTCCGCGGCT GCCCGCCTCGCGCGTCTTCCCTGCCCGGGTCTCCTCGCTGTCGCCGCCGCTGCCACACCATGGCCTTCGTCACCAGGCAGTTCATGCGTT CCGTGTCCTCCTCGTCCACCGCCTCGGCCTCGGCCAAGAAGATAATCGTCAAGCACGTGACGGTCATCGGCGGCGGGCTGATGGGCGCCG GCATTGCCCAGGTTTCTGTTATCAGTGGAACCTATCTTGGTTTGGATTACATGAGTAAGCAAAATGGAGGTGAAGGCGGCATCATTATCA ATATGTCATCTTTAGCAGGACTCATGCCCGTTGCACAGCAGCCGGTTTATTGTGCTTCAAAGCATGGCATAGTTGGATTCACACGCTCAG CAGCGTTGGCTGCTAATCTTATGAACAGTGGTGTGAGACTGAATGCCATTTGTCCAGGCTTTGTTAACACAGCCATCCTTGAATCAATTG AAAAAGAAGAAAACATGGGACAATATATAGAATATAAGGATCATATCAAGGATATGATTAAATACTATGGAATTTTGGACCCACCATTGA TTGCCAATGGATTGATAACACTCATTGAAGATGATGCTTTAAATGGTGCTATTATGAAGATCACAACTTCTAAGGGAATTCATTTTCAAG ACTATGATACAACTCCATTTCAAGCAAAAACCCAATGAACAGCTTATGTGTTAGCCATAGCTGAAAATAAGCACAAATAGCTTATATTCA GATCCTATCTTCATTTGAATATAGCTTTTAAATGAAATGTTACAGTTTGAAGTTTTCCTTCATGCACTTGGTGATAAACGTTTTCTAAAT TTTTAGTTAAGTATATGGATAAAAAGTTATGAACTATTAAAAATGTGATGTGGACCAAAGGCTAGGTTGTAATCTTGATAGTCTAAAAAA TGATCAAAACAAATGATTTTCAAGGAATATTCAATATTCTGCCTTTCAGAAAGTGTATTTATATCTGTGCTTCATAAATATTAATGTTCT TCAGAACATCATTTTAAAGGAGATACTTGAATTGTTATTTAAATCAAACCAGATGTAAAACACTCACATACAAGTTCATACTTTAAAAGA GGAAAGCTACTTAACAATGACAAATATTTCACAATAATAATTTTTACTTATATACCATCTTTCAACTGAACATTTCAGTTCTTCCAAGAG CTTCTTAGAGTAGTATATTTTGGGGGCAGTCAAGGAATAAACTACAGTGTAAACATATCCCAGATGAAAACTGCTGTATGGAAAAATGAC AGAAAGTAACTGATTGACACTGTTGATTCACAGTTCAGCCTCCTATCTGGGAAAGACATTTCTTTCCTCTGCTCACTTTAAGAACTTTTA CCGACTCCAAAAATCTCAGGAATTAAACTTTTAACAGTTACAGCAATAAAGAATAGTTAGTACTCCAAAAATATTATATTTAAGATGCTC AACAAGAAAAAAATGCAAATGTAATATTTTTTTCAAATTACTTCTTTATTGACTTGTCCAAATTTCAAAAGTGCCTACCCTTCAATAAAA CTTTTTTATTCTGATCTCCATAAATTACTTAGTCTTCTATGTATAGCTATCAAGGAAATAAAACCAATTTTGCCACAGCCACAACTGTAA ATGTTTTTGTACCCATGCTGAAACTCATAACAACACAGACATAAAAATAGCTGTGAGGTTTTGCTTTTTTTGTTGTCAGCTATCTTAAGA ATCATTAAATACACCTGCTTTGGGTAAAACTCTTTGCAAGCAGTAATTAACACTAGTAACAGTGAAAGCACAAGATTTCCAAATCAGTCG TTTTCTCAAAAAAATATCGTATAAGTGACTCATCCTGTCTGCTAACTCCAGACCTCCCAGCTTGAAGCCAAATCTTTCCATGTGAGATTG ATATGGATTTCCTAGAAGTACTGGAATGTTGTCATATCTTGCCCTATTTTAATTCTGCTATAGAAAACAATTGCCTTCACTTTTAAGGAG TAATTTGAATATTAATAACTCTGGTCTAGATTTTCATATAATGTATTAAAGACAAAGTAGTGAACATCAATGAACATCTGATAGAGATAA ACTGTAATCAGGCATAAGCTTGTTTGTATGTTCTGGCAGTGACTAATCAGTAAATGATGTCGGTTTGCCCAGTATCACTTATCTTCTGTA TTTTTCCTCTGTCGTGTAAATAGTATAACCTTTTCATTTATGGACAATTTTTTGGACTAGTAGCCTTCAATATACATTCTGCTTTGAATT AATTTTTTCAAATCAATAAATTATGTAGACATTTAAAATCAAATATCAAGTAGAATTGAAAAATGTGAGTTACATAAGTTAAAAACTTAC TTTAAATCTTACCTTCTATAGGTAGCTCTAAATAAATTCATATGGTTATATGGCATCTCTGGTGTATACTGATTGAGAAAATAATTAAAC >35582_35582_2_HADH-HPGD_HADH_chr4_108911220_ENST00000309522_HPGD_chr4_175429943_ENST00000296522_length(amino acids)=238AA_BP=80 MQGRVGPRPQSLAFRGCPPRASSLPGSPRCRRRCHTMAFVTRQFMRSVSSSSTASASAKKIIVKHVTVIGGGLMGAGIAQVSVISGTYLG LDYMSKQNGGEGGIIINMSSLAGLMPVAQQPVYCASKHGIVGFTRSAALAANLMNSGVRLNAICPGFVNTAILESIEKEENMGQYIEYKD -------------------------------------------------------------- >35582_35582_3_HADH-HPGD_HADH_chr4_108911220_ENST00000403312_HPGD_chr4_175429943_ENST00000296521_length(transcript)=1162nt_BP=351nt CGTGTATACCCGCTCAACGCTGGGACGTTACAGCCAGGGCCAATGGGCAGAGCGGGACTCGAGGCCCCGCCCCCGCCTTGTGGCGTCACG GGGACGCCGGGGGCGCGCGGGCTGCAGGGCCGCGTAGGTCCCCGCCCCCAGAGTCTGGCTTTCCGCGGCTGCCCGCCTCGCGCGTCTTCC CTGCCCGGGTCTCCTCGCTGTCGCCGCCGCTGCCACACCATGGCCTTCGTCACCAGGCAGTTCATGCGTTCCGTGTCCTCCTCGTCCACC GCCTCGGCCTCGGCCAAGAAGATAATCGTCAAGCACGTGACGGTCATCGGCGGCGGGCTGATGGGCGCCGGCATTGCCCAGGTTTCTGTT ATCAGTGGAACCTATCTTGGTTTGGATTACATGAGTAAGCAAAATGGAGGTGAAGGCGGCATCATTATCAATATGTCATCTTTAGCAGGA CTCATGCCCGTTGCACAGCAGCCGGTTTATTGTGCTTCAAAGCATGGCATAGTTGGATTCACACGCTCAGCAGCGCCCACCATTGATTGC CAATGGATTGATAACACTCATTGAAGATGATGCTTTAAATGGTGCTATTATGAAGATCACAACTTCTAAGGGAATTCATTTTCAAGACTA TGATACAACTCCATTTCAAGCAAAAACCCAATGAACAGCTTATGTGTTAGCCATAGCTGAAAATAAGCACAAATAGCTTATATTCAGATC CTATCTTCATTTGAATATAGCTTTTAAATGAAATGTTACAGTTTGAAGTTTTCCTTCATGCACTTGGTGATAAACGTTTTCTAAATTTTT AGTTAAGTATATGGATAAAAAGTTATGAACTATTAAAAATGTGATGTGGACCAAAGGCTAGGTTGTAATCTTGATAGTCTAAAAAATGAT CAAAACAAATGATTTTCAAGGAATATTCAATATTCTGCCTTTCAGAAAGTGTATTTATATCTGTGCTTCATAAATATTAATGTTCTTCAG AACATCATTTTAAAGGAGATACTTGAATTGTTATTTAAATCAAACCAGATGTAAAACACTCACATACAAGTTCATACTTTAAAAGAGGAA >35582_35582_3_HADH-HPGD_HADH_chr4_108911220_ENST00000403312_HPGD_chr4_175429943_ENST00000296521_length(amino acids)=173AA_BP=103 MGRAGLEAPPPPCGVTGTPGARGLQGRVGPRPQSLAFRGCPPRASSLPGSPRCRRRCHTMAFVTRQFMRSVSSSSTASASAKKIIVKHVT -------------------------------------------------------------- >35582_35582_4_HADH-HPGD_HADH_chr4_108911220_ENST00000403312_HPGD_chr4_175429943_ENST00000296522_length(transcript)=2607nt_BP=351nt CGTGTATACCCGCTCAACGCTGGGACGTTACAGCCAGGGCCAATGGGCAGAGCGGGACTCGAGGCCCCGCCCCCGCCTTGTGGCGTCACG GGGACGCCGGGGGCGCGCGGGCTGCAGGGCCGCGTAGGTCCCCGCCCCCAGAGTCTGGCTTTCCGCGGCTGCCCGCCTCGCGCGTCTTCC CTGCCCGGGTCTCCTCGCTGTCGCCGCCGCTGCCACACCATGGCCTTCGTCACCAGGCAGTTCATGCGTTCCGTGTCCTCCTCGTCCACC GCCTCGGCCTCGGCCAAGAAGATAATCGTCAAGCACGTGACGGTCATCGGCGGCGGGCTGATGGGCGCCGGCATTGCCCAGGTTTCTGTT ATCAGTGGAACCTATCTTGGTTTGGATTACATGAGTAAGCAAAATGGAGGTGAAGGCGGCATCATTATCAATATGTCATCTTTAGCAGGA CTCATGCCCGTTGCACAGCAGCCGGTTTATTGTGCTTCAAAGCATGGCATAGTTGGATTCACACGCTCAGCAGCGTTGGCTGCTAATCTT ATGAACAGTGGTGTGAGACTGAATGCCATTTGTCCAGGCTTTGTTAACACAGCCATCCTTGAATCAATTGAAAAAGAAGAAAACATGGGA CAATATATAGAATATAAGGATCATATCAAGGATATGATTAAATACTATGGAATTTTGGACCCACCATTGATTGCCAATGGATTGATAACA CTCATTGAAGATGATGCTTTAAATGGTGCTATTATGAAGATCACAACTTCTAAGGGAATTCATTTTCAAGACTATGATACAACTCCATTT CAAGCAAAAACCCAATGAACAGCTTATGTGTTAGCCATAGCTGAAAATAAGCACAAATAGCTTATATTCAGATCCTATCTTCATTTGAAT ATAGCTTTTAAATGAAATGTTACAGTTTGAAGTTTTCCTTCATGCACTTGGTGATAAACGTTTTCTAAATTTTTAGTTAAGTATATGGAT AAAAAGTTATGAACTATTAAAAATGTGATGTGGACCAAAGGCTAGGTTGTAATCTTGATAGTCTAAAAAATGATCAAAACAAATGATTTT CAAGGAATATTCAATATTCTGCCTTTCAGAAAGTGTATTTATATCTGTGCTTCATAAATATTAATGTTCTTCAGAACATCATTTTAAAGG AGATACTTGAATTGTTATTTAAATCAAACCAGATGTAAAACACTCACATACAAGTTCATACTTTAAAAGAGGAAAGCTACTTAACAATGA CAAATATTTCACAATAATAATTTTTACTTATATACCATCTTTCAACTGAACATTTCAGTTCTTCCAAGAGCTTCTTAGAGTAGTATATTT TGGGGGCAGTCAAGGAATAAACTACAGTGTAAACATATCCCAGATGAAAACTGCTGTATGGAAAAATGACAGAAAGTAACTGATTGACAC TGTTGATTCACAGTTCAGCCTCCTATCTGGGAAAGACATTTCTTTCCTCTGCTCACTTTAAGAACTTTTACCGACTCCAAAAATCTCAGG AATTAAACTTTTAACAGTTACAGCAATAAAGAATAGTTAGTACTCCAAAAATATTATATTTAAGATGCTCAACAAGAAAAAAATGCAAAT GTAATATTTTTTTCAAATTACTTCTTTATTGACTTGTCCAAATTTCAAAAGTGCCTACCCTTCAATAAAACTTTTTTATTCTGATCTCCA TAAATTACTTAGTCTTCTATGTATAGCTATCAAGGAAATAAAACCAATTTTGCCACAGCCACAACTGTAAATGTTTTTGTACCCATGCTG AAACTCATAACAACACAGACATAAAAATAGCTGTGAGGTTTTGCTTTTTTTGTTGTCAGCTATCTTAAGAATCATTAAATACACCTGCTT TGGGTAAAACTCTTTGCAAGCAGTAATTAACACTAGTAACAGTGAAAGCACAAGATTTCCAAATCAGTCGTTTTCTCAAAAAAATATCGT ATAAGTGACTCATCCTGTCTGCTAACTCCAGACCTCCCAGCTTGAAGCCAAATCTTTCCATGTGAGATTGATATGGATTTCCTAGAAGTA CTGGAATGTTGTCATATCTTGCCCTATTTTAATTCTGCTATAGAAAACAATTGCCTTCACTTTTAAGGAGTAATTTGAATATTAATAACT CTGGTCTAGATTTTCATATAATGTATTAAAGACAAAGTAGTGAACATCAATGAACATCTGATAGAGATAAACTGTAATCAGGCATAAGCT TGTTTGTATGTTCTGGCAGTGACTAATCAGTAAATGATGTCGGTTTGCCCAGTATCACTTATCTTCTGTATTTTTCCTCTGTCGTGTAAA TAGTATAACCTTTTCATTTATGGACAATTTTTTGGACTAGTAGCCTTCAATATACATTCTGCTTTGAATTAATTTTTTCAAATCAATAAA TTATGTAGACATTTAAAATCAAATATCAAGTAGAATTGAAAAATGTGAGTTACATAAGTTAAAAACTTACTTTAAATCTTACCTTCTATA >35582_35582_4_HADH-HPGD_HADH_chr4_108911220_ENST00000403312_HPGD_chr4_175429943_ENST00000296522_length(amino acids)=261AA_BP=103 MGRAGLEAPPPPCGVTGTPGARGLQGRVGPRPQSLAFRGCPPRASSLPGSPRCRRRCHTMAFVTRQFMRSVSSSSTASASAKKIIVKHVT VIGGGLMGAGIAQVSVISGTYLGLDYMSKQNGGEGGIIINMSSLAGLMPVAQQPVYCASKHGIVGFTRSAALAANLMNSGVRLNAICPGF -------------------------------------------------------------- >35582_35582_5_HADH-HPGD_HADH_chr4_108911220_ENST00000603302_HPGD_chr4_175429943_ENST00000296521_length(transcript)=1162nt_BP=351nt CGTGTATACCCGCTCAACGCTGGGACGTTACAGCCAGGGCCAATGGGCAGAGCGGGACTCGAGGCCCCGCCCCCGCCTTGTGGCGTCACG GGGACGCCGGGGGCGCGCGGGCTGCAGGGCCGCGTAGGTCCCCGCCCCCAGAGTCTGGCTTTCCGCGGCTGCCCGCCTCGCGCGTCTTCC CTGCCCGGGTCTCCTCGCTGTCGCCGCCGCTGCCACACCATGGCCTTCGTCACCAGGCAGTTCATGCGTTCCGTGTCCTCCTCGTCCACC GCCTCGGCCTCGGCCAAGAAGATAATCGTCAAGCACGTGACGGTCATCGGCGGCGGGCTGATGGGCGCCGGCATTGCCCAGGTTTCTGTT ATCAGTGGAACCTATCTTGGTTTGGATTACATGAGTAAGCAAAATGGAGGTGAAGGCGGCATCATTATCAATATGTCATCTTTAGCAGGA CTCATGCCCGTTGCACAGCAGCCGGTTTATTGTGCTTCAAAGCATGGCATAGTTGGATTCACACGCTCAGCAGCGCCCACCATTGATTGC CAATGGATTGATAACACTCATTGAAGATGATGCTTTAAATGGTGCTATTATGAAGATCACAACTTCTAAGGGAATTCATTTTCAAGACTA TGATACAACTCCATTTCAAGCAAAAACCCAATGAACAGCTTATGTGTTAGCCATAGCTGAAAATAAGCACAAATAGCTTATATTCAGATC CTATCTTCATTTGAATATAGCTTTTAAATGAAATGTTACAGTTTGAAGTTTTCCTTCATGCACTTGGTGATAAACGTTTTCTAAATTTTT AGTTAAGTATATGGATAAAAAGTTATGAACTATTAAAAATGTGATGTGGACCAAAGGCTAGGTTGTAATCTTGATAGTCTAAAAAATGAT CAAAACAAATGATTTTCAAGGAATATTCAATATTCTGCCTTTCAGAAAGTGTATTTATATCTGTGCTTCATAAATATTAATGTTCTTCAG AACATCATTTTAAAGGAGATACTTGAATTGTTATTTAAATCAAACCAGATGTAAAACACTCACATACAAGTTCATACTTTAAAAGAGGAA >35582_35582_5_HADH-HPGD_HADH_chr4_108911220_ENST00000603302_HPGD_chr4_175429943_ENST00000296521_length(amino acids)=173AA_BP=103 MGRAGLEAPPPPCGVTGTPGARGLQGRVGPRPQSLAFRGCPPRASSLPGSPRCRRRCHTMAFVTRQFMRSVSSSSTASASAKKIIVKHVT -------------------------------------------------------------- >35582_35582_6_HADH-HPGD_HADH_chr4_108911220_ENST00000603302_HPGD_chr4_175429943_ENST00000296522_length(transcript)=2607nt_BP=351nt CGTGTATACCCGCTCAACGCTGGGACGTTACAGCCAGGGCCAATGGGCAGAGCGGGACTCGAGGCCCCGCCCCCGCCTTGTGGCGTCACG GGGACGCCGGGGGCGCGCGGGCTGCAGGGCCGCGTAGGTCCCCGCCCCCAGAGTCTGGCTTTCCGCGGCTGCCCGCCTCGCGCGTCTTCC CTGCCCGGGTCTCCTCGCTGTCGCCGCCGCTGCCACACCATGGCCTTCGTCACCAGGCAGTTCATGCGTTCCGTGTCCTCCTCGTCCACC GCCTCGGCCTCGGCCAAGAAGATAATCGTCAAGCACGTGACGGTCATCGGCGGCGGGCTGATGGGCGCCGGCATTGCCCAGGTTTCTGTT ATCAGTGGAACCTATCTTGGTTTGGATTACATGAGTAAGCAAAATGGAGGTGAAGGCGGCATCATTATCAATATGTCATCTTTAGCAGGA CTCATGCCCGTTGCACAGCAGCCGGTTTATTGTGCTTCAAAGCATGGCATAGTTGGATTCACACGCTCAGCAGCGTTGGCTGCTAATCTT ATGAACAGTGGTGTGAGACTGAATGCCATTTGTCCAGGCTTTGTTAACACAGCCATCCTTGAATCAATTGAAAAAGAAGAAAACATGGGA CAATATATAGAATATAAGGATCATATCAAGGATATGATTAAATACTATGGAATTTTGGACCCACCATTGATTGCCAATGGATTGATAACA CTCATTGAAGATGATGCTTTAAATGGTGCTATTATGAAGATCACAACTTCTAAGGGAATTCATTTTCAAGACTATGATACAACTCCATTT CAAGCAAAAACCCAATGAACAGCTTATGTGTTAGCCATAGCTGAAAATAAGCACAAATAGCTTATATTCAGATCCTATCTTCATTTGAAT ATAGCTTTTAAATGAAATGTTACAGTTTGAAGTTTTCCTTCATGCACTTGGTGATAAACGTTTTCTAAATTTTTAGTTAAGTATATGGAT AAAAAGTTATGAACTATTAAAAATGTGATGTGGACCAAAGGCTAGGTTGTAATCTTGATAGTCTAAAAAATGATCAAAACAAATGATTTT CAAGGAATATTCAATATTCTGCCTTTCAGAAAGTGTATTTATATCTGTGCTTCATAAATATTAATGTTCTTCAGAACATCATTTTAAAGG AGATACTTGAATTGTTATTTAAATCAAACCAGATGTAAAACACTCACATACAAGTTCATACTTTAAAAGAGGAAAGCTACTTAACAATGA CAAATATTTCACAATAATAATTTTTACTTATATACCATCTTTCAACTGAACATTTCAGTTCTTCCAAGAGCTTCTTAGAGTAGTATATTT TGGGGGCAGTCAAGGAATAAACTACAGTGTAAACATATCCCAGATGAAAACTGCTGTATGGAAAAATGACAGAAAGTAACTGATTGACAC TGTTGATTCACAGTTCAGCCTCCTATCTGGGAAAGACATTTCTTTCCTCTGCTCACTTTAAGAACTTTTACCGACTCCAAAAATCTCAGG AATTAAACTTTTAACAGTTACAGCAATAAAGAATAGTTAGTACTCCAAAAATATTATATTTAAGATGCTCAACAAGAAAAAAATGCAAAT GTAATATTTTTTTCAAATTACTTCTTTATTGACTTGTCCAAATTTCAAAAGTGCCTACCCTTCAATAAAACTTTTTTATTCTGATCTCCA TAAATTACTTAGTCTTCTATGTATAGCTATCAAGGAAATAAAACCAATTTTGCCACAGCCACAACTGTAAATGTTTTTGTACCCATGCTG AAACTCATAACAACACAGACATAAAAATAGCTGTGAGGTTTTGCTTTTTTTGTTGTCAGCTATCTTAAGAATCATTAAATACACCTGCTT TGGGTAAAACTCTTTGCAAGCAGTAATTAACACTAGTAACAGTGAAAGCACAAGATTTCCAAATCAGTCGTTTTCTCAAAAAAATATCGT ATAAGTGACTCATCCTGTCTGCTAACTCCAGACCTCCCAGCTTGAAGCCAAATCTTTCCATGTGAGATTGATATGGATTTCCTAGAAGTA CTGGAATGTTGTCATATCTTGCCCTATTTTAATTCTGCTATAGAAAACAATTGCCTTCACTTTTAAGGAGTAATTTGAATATTAATAACT CTGGTCTAGATTTTCATATAATGTATTAAAGACAAAGTAGTGAACATCAATGAACATCTGATAGAGATAAACTGTAATCAGGCATAAGCT TGTTTGTATGTTCTGGCAGTGACTAATCAGTAAATGATGTCGGTTTGCCCAGTATCACTTATCTTCTGTATTTTTCCTCTGTCGTGTAAA TAGTATAACCTTTTCATTTATGGACAATTTTTTGGACTAGTAGCCTTCAATATACATTCTGCTTTGAATTAATTTTTTCAAATCAATAAA TTATGTAGACATTTAAAATCAAATATCAAGTAGAATTGAAAAATGTGAGTTACATAAGTTAAAAACTTACTTTAAATCTTACCTTCTATA >35582_35582_6_HADH-HPGD_HADH_chr4_108911220_ENST00000603302_HPGD_chr4_175429943_ENST00000296522_length(amino acids)=261AA_BP=103 MGRAGLEAPPPPCGVTGTPGARGLQGRVGPRPQSLAFRGCPPRASSLPGSPRCRRRCHTMAFVTRQFMRSVSSSSTASASAKKIIVKHVT VIGGGLMGAGIAQVSVISGTYLGLDYMSKQNGGEGGIIINMSSLAGLMPVAQQPVYCASKHGIVGFTRSAALAANLMNSGVRLNAICPGF -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for HADH-HPGD |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for HADH-HPGD |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for HADH-HPGD |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
