
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:HAMP-GPI (FusionGDB2 ID:35593) |
Fusion Gene Summary for HAMP-GPI |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: HAMP-GPI | Fusion gene ID: 35593 | Hgene | Tgene | Gene symbol | HAMP | GPI | Gene ID | 57817 | 10007 |
| Gene name | hepcidin antimicrobial peptide | glucosamine-6-phosphate deaminase 1 | |
| Synonyms | HEPC|HFE2B|LEAP1|PLTR | GNP1|GNPDA|GNPI|GPI|HLN | |
| Cytomap | 19q13.12 | 5q31.3 | |
| Type of gene | protein-coding | protein-coding | |
| Description | hepcidinhepcidin preproproteinliver-expressed antimicrobial peptide 1putative liver tumor regressor | glucosamine-6-phosphate isomerase 1GNPDA 1glcN6P deaminase 1oscillin | |
| Modification date | 20200329 | 20200313 | |
| UniProtAcc | P81172 | Q8IV16 | |
| Ensembl transtripts involved in fusion gene | ENST00000222304, ENST00000598398, | ENST00000356487, ENST00000415930, ENST00000586425, | |
| Fusion gene scores | * DoF score | 1 X 1 X 1=1 | 19 X 17 X 10=3230 |
| # samples | 1 | 23 | |
| ** MAII score | log2(1/1*10)=3.32192809488736 | log2(23/3230*10)=-3.81182839863691 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: HAMP [Title/Abstract] AND GPI [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | HAMP(35773570)-GPI(34884153), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | HAMP | GO:0006879 | cellular iron ion homeostasis | 15514116 |
 Fusion gene breakpoints across HAMP (5'-gene) Fusion gene breakpoints across HAMP (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
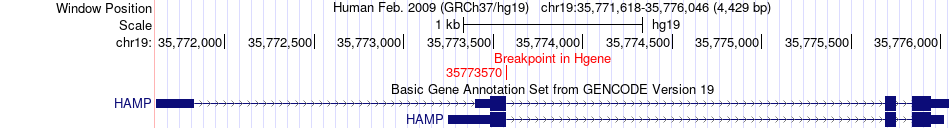 |
 Fusion gene breakpoints across GPI (3'-gene) Fusion gene breakpoints across GPI (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
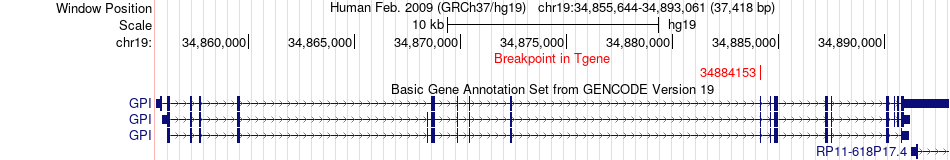 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | LIHC | TCGA-DD-AAEG-01A | HAMP | chr19 | 35773570 | + | GPI | chr19 | 34884153 | + |
Top |
Fusion Gene ORF analysis for HAMP-GPI |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| In-frame | ENST00000222304 | ENST00000356487 | HAMP | chr19 | 35773570 | + | GPI | chr19 | 34884153 | + |
| In-frame | ENST00000222304 | ENST00000415930 | HAMP | chr19 | 35773570 | + | GPI | chr19 | 34884153 | + |
| In-frame | ENST00000222304 | ENST00000586425 | HAMP | chr19 | 35773570 | + | GPI | chr19 | 34884153 | + |
| In-frame | ENST00000598398 | ENST00000356487 | HAMP | chr19 | 35773570 | + | GPI | chr19 | 34884153 | + |
| In-frame | ENST00000598398 | ENST00000415930 | HAMP | chr19 | 35773570 | + | GPI | chr19 | 34884153 | + |
| In-frame | ENST00000598398 | ENST00000586425 | HAMP | chr19 | 35773570 | + | GPI | chr19 | 34884153 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000598398 | HAMP | chr19 | 35773570 | + | ENST00000415930 | GPI | chr19 | 34884153 | + | 3379 | 386 | 227 | 1258 | 343 |
| ENST00000598398 | HAMP | chr19 | 35773570 | + | ENST00000356487 | GPI | chr19 | 34884153 | + | 1551 | 386 | 1276 | 230 | 348 |
| ENST00000598398 | HAMP | chr19 | 35773570 | + | ENST00000586425 | GPI | chr19 | 34884153 | + | 1356 | 386 | 1033 | 230 | 267 |
| ENST00000222304 | HAMP | chr19 | 35773570 | + | ENST00000415930 | GPI | chr19 | 34884153 | + | 3315 | 322 | 163 | 1194 | 343 |
| ENST00000222304 | HAMP | chr19 | 35773570 | + | ENST00000356487 | GPI | chr19 | 34884153 | + | 1487 | 322 | 1212 | 166 | 348 |
| ENST00000222304 | HAMP | chr19 | 35773570 | + | ENST00000586425 | GPI | chr19 | 34884153 | + | 1292 | 322 | 969 | 166 | 267 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000598398 | ENST00000415930 | HAMP | chr19 | 35773570 | + | GPI | chr19 | 34884153 | + | 0.005355045 | 0.994645 |
| ENST00000598398 | ENST00000356487 | HAMP | chr19 | 35773570 | + | GPI | chr19 | 34884153 | + | 0.011113044 | 0.9888869 |
| ENST00000598398 | ENST00000586425 | HAMP | chr19 | 35773570 | + | GPI | chr19 | 34884153 | + | 0.050407995 | 0.94959193 |
| ENST00000222304 | ENST00000415930 | HAMP | chr19 | 35773570 | + | GPI | chr19 | 34884153 | + | 0.005606839 | 0.9943931 |
| ENST00000222304 | ENST00000356487 | HAMP | chr19 | 35773570 | + | GPI | chr19 | 34884153 | + | 0.014048969 | 0.985951 |
| ENST00000222304 | ENST00000586425 | HAMP | chr19 | 35773570 | + | GPI | chr19 | 34884153 | + | 0.06762425 | 0.9323757 |
Top |
Fusion Genomic Features for HAMP-GPI |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| HAMP | chr19 | 35773570 | + | GPI | chr19 | 34884152 | + | 0.004218205 | 0.9957818 |
| HAMP | chr19 | 35773570 | + | GPI | chr19 | 34884152 | + | 0.004218205 | 0.9957818 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
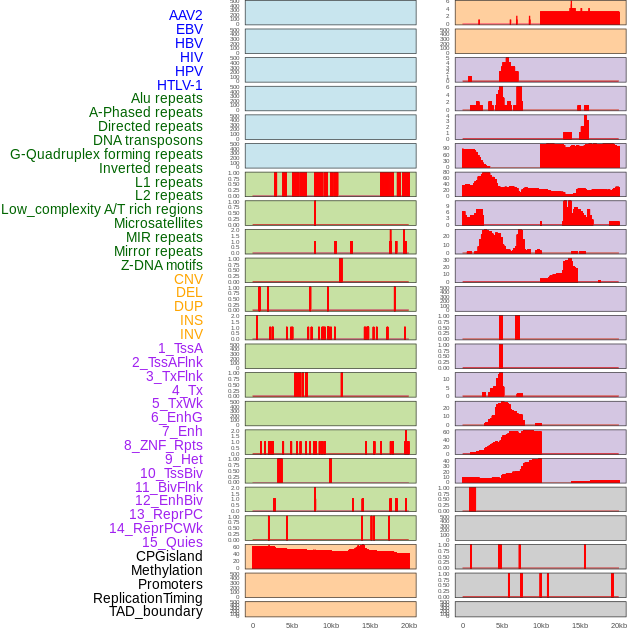 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
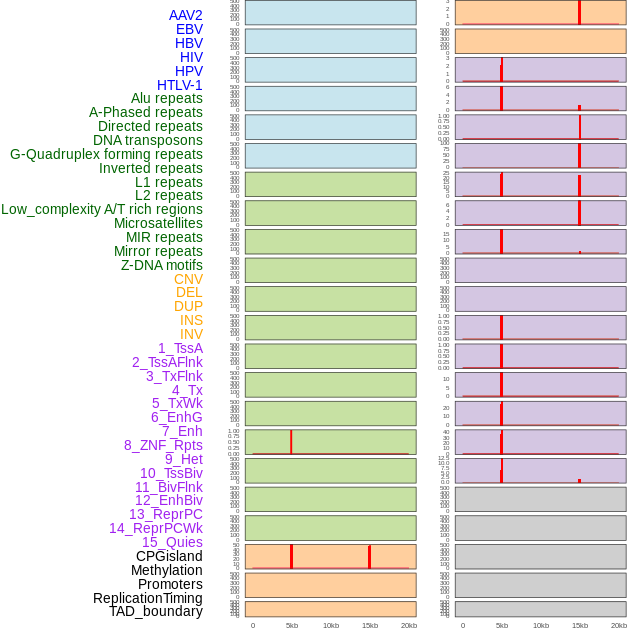 |
Top |
Fusion Protein Features for HAMP-GPI |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr19:35773570/chr19:34884153) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| HAMP | GPI |
| FUNCTION: Liver-produced hormone that constitutes the main circulating regulator of iron absorption and distribution across tissues. Acts by promoting endocytosis and degradation of ferroportin, leading to the retention of iron in iron-exporting cells and decreased flow of iron into plasma. Controls the major flows of iron into plasma: absorption of dietary iron in the intestine, recycling of iron by macrophages, which phagocytose old erythrocytes and other cells, and mobilization of stored iron from hepatocytes (PubMed:22306005). {ECO:0000269|PubMed:22306005}.; FUNCTION: Has strong antimicrobial activity against E.coli ML35P N.cinerea and weaker against S.epidermidis, S.aureus and group b streptococcus bacteria. Active against the fungus C.albicans. No activity against P.aeruginosa (PubMed:11113131, PubMed:11034317). {ECO:0000269|PubMed:11034317, ECO:0000269|PubMed:11113131}. | FUNCTION: Mediates the transport of lipoprotein lipase LPL from the basolateral to the apical surface of endothelial cells in capillaries (By similarity). Anchors LPL on the surface of endothelial cells in the lumen of blood capillaries (By similarity). Protects LPL against loss of activity, and against ANGPTL4-mediated unfolding (PubMed:27929370, PubMed:29899144). Thereby, plays an important role in lipolytic processing of chylomicrons by LPL, triglyceride metabolism and lipid homeostasis (PubMed:19304573, PubMed:21314738). Binds chylomicrons and phospholipid particles that contain APOA5 (PubMed:17997385, PubMed:19304573). Binds high-density lipoprotein (HDL) and plays a role in the uptake of lipids from HDL (By similarity). {ECO:0000250|UniProtKB:Q9D1N2, ECO:0000269|PubMed:17997385, ECO:0000269|PubMed:19304573, ECO:0000269|PubMed:21314738, ECO:0000269|PubMed:27929370, ECO:0000269|PubMed:29899144}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | GPI | chr19:35773570 | chr19:34884153 | ENST00000356487 | 8 | 18 | 159_160 | 268 | 559.0 | Region | Glucose-6-phosphate binding | |
| Tgene | GPI | chr19:35773570 | chr19:34884153 | ENST00000356487 | 8 | 18 | 210_215 | 268 | 559.0 | Region | Glucose-6-phosphate binding | |
| Tgene | GPI | chr19:35773570 | chr19:34884153 | ENST00000415930 | 8 | 18 | 159_160 | 279 | 570.0 | Region | Glucose-6-phosphate binding | |
| Tgene | GPI | chr19:35773570 | chr19:34884153 | ENST00000415930 | 8 | 18 | 210_215 | 279 | 570.0 | Region | Glucose-6-phosphate binding |
Top |
Fusion Gene Sequence for HAMP-GPI |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >35593_35593_1_HAMP-GPI_HAMP_chr19_35773570_ENST00000222304_GPI_chr19_34884153_ENST00000356487_length(transcript)=1487nt_BP=322nt AACACACCTCTGCCGGCTGAGGGTGACACAACCCTGTTCCCTGTCGCTCTGTTCCCGCTTATCTCTCCCGCCTTTTCGGCGCCACCACCT TCTTGGAAATGAGACAGAGCAAAGGGGAGGGGGCTCAGACCACCGCCTCCCCTGGCAGGCCCCATAAAAGCGACTGTCACTCGGTCCCAG ACACCAGAGCAAGCTCAAGACCCAGCAGTGGGACAGCCAGACAGACGGCACGATGGCACTGAGCTCCCAGATCTGGGCCGCTTGCCTCCT GCTCCTCCTCCTCCTCGCCAGCCTGACCAGTGGCTCTGTTTTCCCACAACAGTGGGTGGGAGGACGCTACTCGCTGTGGTCGGCCATCGG ACTCTCCATTGCCCTGCACGTGGGTTTTGACAACTTCGAGCAGCTGCTCTCGGGGGCTCACTGGATGGACCAGCACTTCCGCACGACGCC CCTGGAGAAGAACGCCCCCGTCTTGCTGGCCCTGCTGGGTATCTGGTACATCAACTGCTTTGGGTGTGAGACACACGCCATGCTGCCCTA TGACCAGTACCTGCACCGCTTTGCTGCGTACTTCCAGCAGGGCGACATGGAGTCCAATGGGAAATACATCACCAAATCTGGAACCCGTGT GGACCACCAGACAGGCCCCATTGTGTGGGGGGAGCCAGGGACCAATGGCCAGCATGCTTTTTACCAGCTCATCCACCAAGGCACCAAGAT GATACCCTGTGACTTCCTCATCCCGGTCCAGACCCAGCACCCCATACGGAAGGGTCTGCATCACAAGATCCTCCTGGCCAACTTCTTGGC CCAGACAGAGGCCCTGATGAGGGGAAAATCGACGGAGGAGGCCCGAAAGGAGCTCCAGGCTGCGGGCAAGAGTCCAGAGGACCTTGAGAG GCTGCTGCCACATAAGGTCTTTGAAGGAAATCGCCCAACCAACTCTATTGTGTTCACCAAGCTCACACCATTCATGCTTGGAGCCTTGGT CGCCATGTATGAGCACAAGATCTTCGTTCAGGGCATCATCTGGGACATCAACAGCTTTGACCAGTGGGGAGTGGAGCTGGGAAAGCAGCT GGCTAAGAAAATAGAGCCTGAGCTTGATGGCAGTGCTCAAGTGACCTCTCACGACGCTTCTACCAATGGGCTCATCAACTTCATCAAGCA GCAGCGCGAGGCCAGAGTCCAATAAACTCGTGCTCATCTGCAGCCTCCTCTGTGACTCCCCTTTCTCTTCTCGTCCCTCCTCCCCGGAGC CGGCACTGCATGTTCCTGGACACCACCCAGAGCACCCTCTGGTTGTGGGCTTGGACCACGAGCCCTTAGCAGGGAAGGCTGGTCTCCCCC AGCCTAACCCCCAGCCCCTCCATGTCTATGCTCCCTCTGTGTTAGAATTGGCTGAAGTGTTTTTGTGCAGCTGACTTTTCTGACCCATGT >35593_35593_1_HAMP-GPI_HAMP_chr19_35773570_ENST00000222304_GPI_chr19_34884153_ENST00000356487_length(amino acids)=348AA_BP=1 MQMSTSLLDSGLALLLDEVDEPIGRSVVRGHLSTAIKLRLYFLSQLLSQLHSPLVKAVDVPDDALNEDLVLIHGDQGSKHEWCELGEHNR VGWAISFKDLMWQQPLKVLWTLARSLELLSGLLRRFSPHQGLCLGQEVGQEDLVMQTLPYGVLGLDRDEEVTGYHLGALVDELVKSMLAI GPWLPPHNGACLVVHTGSRFGDVFPIGLHVALLEVRSKAVQVLVIGQHGVCLTPKAVDVPDTQQGQQDGGVLLQGRRAEVLVHPVSPREQ -------------------------------------------------------------- >35593_35593_2_HAMP-GPI_HAMP_chr19_35773570_ENST00000222304_GPI_chr19_34884153_ENST00000415930_length(transcript)=3315nt_BP=322nt AACACACCTCTGCCGGCTGAGGGTGACACAACCCTGTTCCCTGTCGCTCTGTTCCCGCTTATCTCTCCCGCCTTTTCGGCGCCACCACCT TCTTGGAAATGAGACAGAGCAAAGGGGAGGGGGCTCAGACCACCGCCTCCCCTGGCAGGCCCCATAAAAGCGACTGTCACTCGGTCCCAG ACACCAGAGCAAGCTCAAGACCCAGCAGTGGGACAGCCAGACAGACGGCACGATGGCACTGAGCTCCCAGATCTGGGCCGCTTGCCTCCT GCTCCTCCTCCTCCTCGCCAGCCTGACCAGTGGCTCTGTTTTCCCACAACAGTGGGTGGGAGGACGCTACTCGCTGTGGTCGGCCATCGG ACTCTCCATTGCCCTGCACGTGGGTTTTGACAACTTCGAGCAGCTGCTCTCGGGGGCTCACTGGATGGACCAGCACTTCCGCACGACGCC CCTGGAGAAGAACGCCCCCGTCTTGCTGGCCCTGCTGGGTATCTGGTACATCAACTGCTTTGGGTGTGAGACACACGCCATGCTGCCCTA TGACCAGTACCTGCACCGCTTTGCTGCGTACTTCCAGCAGGGCGACATGGAGTCCAATGGGAAATACATCACCAAATCTGGAACCCGTGT GGACCACCAGACAGGCCCCATTGTGTGGGGGGAGCCAGGGACCAATGGCCAGCATGCTTTTTACCAGCTCATCCACCAAGGCACCAAGAT GATACCCTGTGACTTCCTCATCCCGGTCCAGACCCAGCACCCCATACGGAAGGGTCTGCATCACAAGATCCTCCTGGCCAACTTCTTGGC CCAGACAGAGGCCCTGATGAGGGGAAAATCGACGGAGGAGGCCCGAAAGGAGCTCCAGGCTGCGGGCAAGAGTCCAGAGGACCTTGAGAG GCTGCTGCCACATAAGGTCTTTGAAGGAAATCGCCCAACCAACTCTATTGTGTTCACCAAGCTCACACCATTCATGCTTGGAGCCTTGGT CGCCATGTATGAGCACAAGATCTTCGTTCAGGGCATCATCTGGGACATCAACAGCTTTGACCAGTGGGGAGTGGAGCTGGGAAAGCAGCT GGCTAAGAAAATAGAGCCTGAGCTTGATGGCAGTGCTCAAGTGACCTCTCACGACGCTTCTACCAATGGGCTCATCAACTTCATCAAGCA GCAGCGCGAGGCCAGAGTCCAATAAACTCGTGCTCATCTGCAGCCTCCTCTGTGACTCCCCTTTCTCTTCTCGTCCCTCCTCCCCGGAGC CGGCACTGCATGTTCCTGGACACCACCCAGAGCACCCTCTGGTTGTGGGCTTGGACCACGAGCCCTTAGCAGGGAAGGCTGGTCTCCCCC AGCCTAACCCCCAGCCCCTCCATGTCTATGCTCCCTCTGTGTTAGAATTGGCTGAAGTGTTTTTGTGCAGCTGACTTTTCTGACCCATGT TCACGTTGTTCACATCCCATGTAGAAAAATAAAGATGCCACGGAGGAGGTTGTAGGCTCAGCCTCTGATTTTTTTTTTCCTGTGATGGTG CTTTATGTAGCAGAGGGCAGGAGCGCTCAGCAGGACGCAGGCTGTGCCTCTGCGGACACTTAACACTAAGTGGTGAGCGGGTCTAGAGTG GAGCAAGGTGCCCTGAGAAGACAATAGTGGGGTGGGGGCACAATCAGTCAGGACGGCAACTTGGCCTGTGTCACCAAATCCCAAGACTGT TTTCCACTCCTCACCTCTGTGACTGCAGAAATTGGATACTCTGTTCACTCGATGGTTCTAAAAACTGCATTGAGATTATGTTTGTTTCGG GTGAATTCCTGGACAAGACCGAGGATGACTGCCATCTCCTGGCAAGACGCTCAGGTAGTTCTTTTGCTTTAAAAGGCAGATATTGAAAAC TGGAATTTTTTTTTTTTGAGTCTCGCTCTGTCACCCAGACTGGAGTGCAGTGGTGCAATCTCGGCTCACTGCAACCTCCGCCTCCCGGGT TCAAGCTATTCTCCTGCCTCAGCCTCCCGAGTAGCTGGGATTACACGGCGCACACCACCATACCCAGCTAATTTTTGTATTTTTAGTAGT GAAGGGGTTTTACCATGTTGGGCAGGCTGGTCTTGAACTCCTGACCTCAGGTGATCTGCCCGCCTCAGCCTCCCACAGTGCTGGGATTAC AGGTATGAGCCACCACGCCCGGCCCATTTTTTTTTTTTTTTTGACAACTTTTTTTTTTTTTTGAGACAGGGTCTTGTTCCATTGCCCAGA CTGGAGTGCAGTGGCATGATCACAGCTCACTGCAGCCAGTAATCCTCTTGCCTCAGCCTCCCAAGTAGTTGAGACTACAGGTTGTACCAC TATGCCCTGCTAGTTTTTTCATTTTTTGTAGAGAGACGGGTCTTTTTTTTTTTTGAGACGGAGTCTCGCTCTGTCGCCCAAGCTGGAGTG CAGTAGCACGGTCTCAGCTCATTGCAAGCTCCGCCTCCCAGGTTCACGCCATTCTCCTGCCTCAGACTCCTGTGTAGCTGGGAGTACAGG CACCTGCCACCATGCCCGGCTAATTTTTTATATATTTTTTTAGCAGAGACAGTGTCTCACTGTGTTAGTCAGGATGGTCTCGATCTCCTG ACCTCGTGATCCGCCCGCCTCAGCCTCCCAAAGTGCTGGGATTACAGGCGTGAGCCACCGCGCCCAGCAAGGCGGGTCTTGCTGTGTTTC CCAGACTAGACTGGTCTTGAATTCCAGGGCTCAAGAGATCTCCCACCTCAGCCTCCCACAGTGCTGGGATTACAGGCGTGAGCCGCCACA CCCAGCCTATTCAAAATTTTTTTTTCTTAGAGACAGGGTCTTTGTTGCCCAGGCTGGACTGCAGTGATACAATCATAGCTGACTGAAGCC TCAAATTCCCAGGCTAAGGTGATCTTCTCACCTCAGCCTTCCAAGTAGCTGGGTCCGCAGATGCATGCCAGTACACCCAGCTCATTTAAA AAAAAATTTTTTTCTTTTTTGAGAGTCTTGCTTTGTTGCCCAGGCTGGAGTGCAGTGGTGTGATCTCGGCTCACTGCAAGCTCCACCTCC CGGCTTCACGCCATTCTCCTGCCTCAGCCTCCCGAGTAGCTGGGACTACAGGTGCCCGCCACCACACCCGGCTAATTTTTTGTATTTTTA GTAGAGACGGGGTTTCACCGTGTTAGCCAGGATGGTCTTGATCTCCTGACCTCATGATCCGCCTGCTTTGGCCTCCCAAAGTGCTGGGAT >35593_35593_2_HAMP-GPI_HAMP_chr19_35773570_ENST00000222304_GPI_chr19_34884153_ENST00000415930_length(amino acids)=343AA_BP=31 MSLGPRHQSKLKTQQWDSQTDGTMALSSQIWAACLLLLLLLASLTSGSVFPQQWVGGRYSLWSAIGLSIALHVGFDNFEQLLSGAHWMDQ HFRTTPLEKNAPVLLALLGIWYINCFGCETHAMLPYDQYLHRFAAYFQQGDMESNGKYITKSGTRVDHQTGPIVWGEPGTNGQHAFYQLI HQGTKMIPCDFLIPVQTQHPIRKGLHHKILLANFLAQTEALMRGKSTEEARKELQAAGKSPEDLERLLPHKVFEGNRPTNSIVFTKLTPF -------------------------------------------------------------- >35593_35593_3_HAMP-GPI_HAMP_chr19_35773570_ENST00000222304_GPI_chr19_34884153_ENST00000586425_length(transcript)=1292nt_BP=322nt AACACACCTCTGCCGGCTGAGGGTGACACAACCCTGTTCCCTGTCGCTCTGTTCCCGCTTATCTCTCCCGCCTTTTCGGCGCCACCACCT TCTTGGAAATGAGACAGAGCAAAGGGGAGGGGGCTCAGACCACCGCCTCCCCTGGCAGGCCCCATAAAAGCGACTGTCACTCGGTCCCAG ACACCAGAGCAAGCTCAAGACCCAGCAGTGGGACAGCCAGACAGACGGCACGATGGCACTGAGCTCCCAGATCTGGGCCGCTTGCCTCCT GCTCCTCCTCCTCCTCGCCAGCCTGACCAGTGGCTCTGTTTTCCCACAACAGTGGGTGGGAGGACGCTACTCGCTGTGGTCGGCCATCGG ACTCTCCATTGCCCTGCACGTGGGTTTTGACAACTTCGAGCAGCTGCTCTCGGGGGCTCACTGGATGGACCAGCACTTCCGCACGACGCC CCTGGAGAAGAACGCCCCCGTCTTGCTGGCCCTGCTGGGTATCTGGTACATCAACTGCTTTGGGTGTGAGACACACGCCATGCTGCCCTA TGACCAGTACCTGCACCGCTTTGCTGCGTACTTCCAGCAGGGCGACATGGAGTCCAATGGGAAATACATCACCAAATCTGGAACCCGTGT GGACCACCAGACAGGCCCCATTGTGTGGGGGGAGCCAGGGACCAATGGCCAGCATGCTTTTTACCAGCTCATCCACCAAGGCACCAAGAT GATACCCTGTGACTTCCTCATCCCGGTCCAGACCCAGCACCCCATACGGAAGGGTCTGCATCACAAGATCCTCCTGGCCAACTTCTTGGC CCAGACAGAGGCCCTGATGAGGGGAAAATCGACGGAGGAGGCCCGAAAGGAGCTCCAGGCTGCGGGCAAGAGTCCAGAGGACCTTGAGAG GCTGCTGCCACATAAGAGTGGAGCTGGGAAAGCAGCTGGCTAAGAAAATAGAGCCTGAGCTTGATGGCAGTGCTCAAGTGACCTCTCACG ACGCTTCTACCAATGGGCTCATCAACTTCATCAAGCAGCAGCGCGAGGCCAGAGTCCAATAAACTCGTGCTCATCTGCAGCCTCCTCTGT GACTCCCCTTTCTCTTCTCGTCCCTCCTCCCCGGAGCCGGCACTGCATGTTCCTGGACACCACCCAGAGCACCCTCTGGTTGTGGGCTTG GACCACGAGCCCTTAGCAGGGAAGGCTGGTCTCCCCCAGCCTAACCCCCAGCCCCTCCATGTCTATGCTCCCTCTGTGTTAGAATTGGCT >35593_35593_3_HAMP-GPI_HAMP_chr19_35773570_ENST00000222304_GPI_chr19_34884153_ENST00000586425_length(amino acids)=267AA_BP=1 MPSSSGSIFLASCFPSSTLMWQQPLKVLWTLARSLELLSGLLRRFSPHQGLCLGQEVGQEDLVMQTLPYGVLGLDRDEEVTGYHLGALVD ELVKSMLAIGPWLPPHNGACLVVHTGSRFGDVFPIGLHVALLEVRSKAVQVLVIGQHGVCLTPKAVDVPDTQQGQQDGGVLLQGRRAEVL -------------------------------------------------------------- >35593_35593_4_HAMP-GPI_HAMP_chr19_35773570_ENST00000598398_GPI_chr19_34884153_ENST00000356487_length(transcript)=1551nt_BP=386nt GAGCCTCCCACGTGGTGTGGATGAGGAGGCAGATGGCAGGGCCTGTGCATTTCTGTGCTTGAGTGGGCCTTGAAAGTGGTTCAGCAACCA GGAAGAAGTGTTCATTCCTCGACAACAACATCCCCGGGCTCTGGTGACTTGGCTGACACTGGATGGCCCTGGAATGAAAAAGGCAAAGAG GCAAAATGTGCAAGGGCCCATCTGGAACCAAGGCCCCATAAAAGCGACTGTCACTCGGTCCCAGACACCAGAGCAAGCTCAAGACCCAGC AGTGGGACAGCCAGACAGACGGCACGATGGCACTGAGCTCCCAGATCTGGGCCGCTTGCCTCCTGCTCCTCCTCCTCCTCGCCAGCCTGA CCAGTGGCTCTGTTTTCCCACAACAGTGGGTGGGAGGACGCTACTCGCTGTGGTCGGCCATCGGACTCTCCATTGCCCTGCACGTGGGTT TTGACAACTTCGAGCAGCTGCTCTCGGGGGCTCACTGGATGGACCAGCACTTCCGCACGACGCCCCTGGAGAAGAACGCCCCCGTCTTGC TGGCCCTGCTGGGTATCTGGTACATCAACTGCTTTGGGTGTGAGACACACGCCATGCTGCCCTATGACCAGTACCTGCACCGCTTTGCTG CGTACTTCCAGCAGGGCGACATGGAGTCCAATGGGAAATACATCACCAAATCTGGAACCCGTGTGGACCACCAGACAGGCCCCATTGTGT GGGGGGAGCCAGGGACCAATGGCCAGCATGCTTTTTACCAGCTCATCCACCAAGGCACCAAGATGATACCCTGTGACTTCCTCATCCCGG TCCAGACCCAGCACCCCATACGGAAGGGTCTGCATCACAAGATCCTCCTGGCCAACTTCTTGGCCCAGACAGAGGCCCTGATGAGGGGAA AATCGACGGAGGAGGCCCGAAAGGAGCTCCAGGCTGCGGGCAAGAGTCCAGAGGACCTTGAGAGGCTGCTGCCACATAAGGTCTTTGAAG GAAATCGCCCAACCAACTCTATTGTGTTCACCAAGCTCACACCATTCATGCTTGGAGCCTTGGTCGCCATGTATGAGCACAAGATCTTCG TTCAGGGCATCATCTGGGACATCAACAGCTTTGACCAGTGGGGAGTGGAGCTGGGAAAGCAGCTGGCTAAGAAAATAGAGCCTGAGCTTG ATGGCAGTGCTCAAGTGACCTCTCACGACGCTTCTACCAATGGGCTCATCAACTTCATCAAGCAGCAGCGCGAGGCCAGAGTCCAATAAA CTCGTGCTCATCTGCAGCCTCCTCTGTGACTCCCCTTTCTCTTCTCGTCCCTCCTCCCCGGAGCCGGCACTGCATGTTCCTGGACACCAC CCAGAGCACCCTCTGGTTGTGGGCTTGGACCACGAGCCCTTAGCAGGGAAGGCTGGTCTCCCCCAGCCTAACCCCCAGCCCCTCCATGTC TATGCTCCCTCTGTGTTAGAATTGGCTGAAGTGTTTTTGTGCAGCTGACTTTTCTGACCCATGTTCACGTTGTTCACATCCCATGTAGAA >35593_35593_4_HAMP-GPI_HAMP_chr19_35773570_ENST00000598398_GPI_chr19_34884153_ENST00000356487_length(amino acids)=348AA_BP=1 MQMSTSLLDSGLALLLDEVDEPIGRSVVRGHLSTAIKLRLYFLSQLLSQLHSPLVKAVDVPDDALNEDLVLIHGDQGSKHEWCELGEHNR VGWAISFKDLMWQQPLKVLWTLARSLELLSGLLRRFSPHQGLCLGQEVGQEDLVMQTLPYGVLGLDRDEEVTGYHLGALVDELVKSMLAI GPWLPPHNGACLVVHTGSRFGDVFPIGLHVALLEVRSKAVQVLVIGQHGVCLTPKAVDVPDTQQGQQDGGVLLQGRRAEVLVHPVSPREQ -------------------------------------------------------------- >35593_35593_5_HAMP-GPI_HAMP_chr19_35773570_ENST00000598398_GPI_chr19_34884153_ENST00000415930_length(transcript)=3379nt_BP=386nt GAGCCTCCCACGTGGTGTGGATGAGGAGGCAGATGGCAGGGCCTGTGCATTTCTGTGCTTGAGTGGGCCTTGAAAGTGGTTCAGCAACCA GGAAGAAGTGTTCATTCCTCGACAACAACATCCCCGGGCTCTGGTGACTTGGCTGACACTGGATGGCCCTGGAATGAAAAAGGCAAAGAG GCAAAATGTGCAAGGGCCCATCTGGAACCAAGGCCCCATAAAAGCGACTGTCACTCGGTCCCAGACACCAGAGCAAGCTCAAGACCCAGC AGTGGGACAGCCAGACAGACGGCACGATGGCACTGAGCTCCCAGATCTGGGCCGCTTGCCTCCTGCTCCTCCTCCTCCTCGCCAGCCTGA CCAGTGGCTCTGTTTTCCCACAACAGTGGGTGGGAGGACGCTACTCGCTGTGGTCGGCCATCGGACTCTCCATTGCCCTGCACGTGGGTT TTGACAACTTCGAGCAGCTGCTCTCGGGGGCTCACTGGATGGACCAGCACTTCCGCACGACGCCCCTGGAGAAGAACGCCCCCGTCTTGC TGGCCCTGCTGGGTATCTGGTACATCAACTGCTTTGGGTGTGAGACACACGCCATGCTGCCCTATGACCAGTACCTGCACCGCTTTGCTG CGTACTTCCAGCAGGGCGACATGGAGTCCAATGGGAAATACATCACCAAATCTGGAACCCGTGTGGACCACCAGACAGGCCCCATTGTGT GGGGGGAGCCAGGGACCAATGGCCAGCATGCTTTTTACCAGCTCATCCACCAAGGCACCAAGATGATACCCTGTGACTTCCTCATCCCGG TCCAGACCCAGCACCCCATACGGAAGGGTCTGCATCACAAGATCCTCCTGGCCAACTTCTTGGCCCAGACAGAGGCCCTGATGAGGGGAA AATCGACGGAGGAGGCCCGAAAGGAGCTCCAGGCTGCGGGCAAGAGTCCAGAGGACCTTGAGAGGCTGCTGCCACATAAGGTCTTTGAAG GAAATCGCCCAACCAACTCTATTGTGTTCACCAAGCTCACACCATTCATGCTTGGAGCCTTGGTCGCCATGTATGAGCACAAGATCTTCG TTCAGGGCATCATCTGGGACATCAACAGCTTTGACCAGTGGGGAGTGGAGCTGGGAAAGCAGCTGGCTAAGAAAATAGAGCCTGAGCTTG ATGGCAGTGCTCAAGTGACCTCTCACGACGCTTCTACCAATGGGCTCATCAACTTCATCAAGCAGCAGCGCGAGGCCAGAGTCCAATAAA CTCGTGCTCATCTGCAGCCTCCTCTGTGACTCCCCTTTCTCTTCTCGTCCCTCCTCCCCGGAGCCGGCACTGCATGTTCCTGGACACCAC CCAGAGCACCCTCTGGTTGTGGGCTTGGACCACGAGCCCTTAGCAGGGAAGGCTGGTCTCCCCCAGCCTAACCCCCAGCCCCTCCATGTC TATGCTCCCTCTGTGTTAGAATTGGCTGAAGTGTTTTTGTGCAGCTGACTTTTCTGACCCATGTTCACGTTGTTCACATCCCATGTAGAA AAATAAAGATGCCACGGAGGAGGTTGTAGGCTCAGCCTCTGATTTTTTTTTTCCTGTGATGGTGCTTTATGTAGCAGAGGGCAGGAGCGC TCAGCAGGACGCAGGCTGTGCCTCTGCGGACACTTAACACTAAGTGGTGAGCGGGTCTAGAGTGGAGCAAGGTGCCCTGAGAAGACAATA GTGGGGTGGGGGCACAATCAGTCAGGACGGCAACTTGGCCTGTGTCACCAAATCCCAAGACTGTTTTCCACTCCTCACCTCTGTGACTGC AGAAATTGGATACTCTGTTCACTCGATGGTTCTAAAAACTGCATTGAGATTATGTTTGTTTCGGGTGAATTCCTGGACAAGACCGAGGAT GACTGCCATCTCCTGGCAAGACGCTCAGGTAGTTCTTTTGCTTTAAAAGGCAGATATTGAAAACTGGAATTTTTTTTTTTTGAGTCTCGC TCTGTCACCCAGACTGGAGTGCAGTGGTGCAATCTCGGCTCACTGCAACCTCCGCCTCCCGGGTTCAAGCTATTCTCCTGCCTCAGCCTC CCGAGTAGCTGGGATTACACGGCGCACACCACCATACCCAGCTAATTTTTGTATTTTTAGTAGTGAAGGGGTTTTACCATGTTGGGCAGG CTGGTCTTGAACTCCTGACCTCAGGTGATCTGCCCGCCTCAGCCTCCCACAGTGCTGGGATTACAGGTATGAGCCACCACGCCCGGCCCA TTTTTTTTTTTTTTTTGACAACTTTTTTTTTTTTTTGAGACAGGGTCTTGTTCCATTGCCCAGACTGGAGTGCAGTGGCATGATCACAGC TCACTGCAGCCAGTAATCCTCTTGCCTCAGCCTCCCAAGTAGTTGAGACTACAGGTTGTACCACTATGCCCTGCTAGTTTTTTCATTTTT TGTAGAGAGACGGGTCTTTTTTTTTTTTGAGACGGAGTCTCGCTCTGTCGCCCAAGCTGGAGTGCAGTAGCACGGTCTCAGCTCATTGCA AGCTCCGCCTCCCAGGTTCACGCCATTCTCCTGCCTCAGACTCCTGTGTAGCTGGGAGTACAGGCACCTGCCACCATGCCCGGCTAATTT TTTATATATTTTTTTAGCAGAGACAGTGTCTCACTGTGTTAGTCAGGATGGTCTCGATCTCCTGACCTCGTGATCCGCCCGCCTCAGCCT CCCAAAGTGCTGGGATTACAGGCGTGAGCCACCGCGCCCAGCAAGGCGGGTCTTGCTGTGTTTCCCAGACTAGACTGGTCTTGAATTCCA GGGCTCAAGAGATCTCCCACCTCAGCCTCCCACAGTGCTGGGATTACAGGCGTGAGCCGCCACACCCAGCCTATTCAAAATTTTTTTTTC TTAGAGACAGGGTCTTTGTTGCCCAGGCTGGACTGCAGTGATACAATCATAGCTGACTGAAGCCTCAAATTCCCAGGCTAAGGTGATCTT CTCACCTCAGCCTTCCAAGTAGCTGGGTCCGCAGATGCATGCCAGTACACCCAGCTCATTTAAAAAAAAATTTTTTTCTTTTTTGAGAGT CTTGCTTTGTTGCCCAGGCTGGAGTGCAGTGGTGTGATCTCGGCTCACTGCAAGCTCCACCTCCCGGCTTCACGCCATTCTCCTGCCTCA GCCTCCCGAGTAGCTGGGACTACAGGTGCCCGCCACCACACCCGGCTAATTTTTTGTATTTTTAGTAGAGACGGGGTTTCACCGTGTTAG CCAGGATGGTCTTGATCTCCTGACCTCATGATCCGCCTGCTTTGGCCTCCCAAAGTGCTGGGATTACAGGCGTGAGTCACCGCGCCCGGC >35593_35593_5_HAMP-GPI_HAMP_chr19_35773570_ENST00000598398_GPI_chr19_34884153_ENST00000415930_length(amino acids)=343AA_BP=31 MSLGPRHQSKLKTQQWDSQTDGTMALSSQIWAACLLLLLLLASLTSGSVFPQQWVGGRYSLWSAIGLSIALHVGFDNFEQLLSGAHWMDQ HFRTTPLEKNAPVLLALLGIWYINCFGCETHAMLPYDQYLHRFAAYFQQGDMESNGKYITKSGTRVDHQTGPIVWGEPGTNGQHAFYQLI HQGTKMIPCDFLIPVQTQHPIRKGLHHKILLANFLAQTEALMRGKSTEEARKELQAAGKSPEDLERLLPHKVFEGNRPTNSIVFTKLTPF -------------------------------------------------------------- >35593_35593_6_HAMP-GPI_HAMP_chr19_35773570_ENST00000598398_GPI_chr19_34884153_ENST00000586425_length(transcript)=1356nt_BP=386nt GAGCCTCCCACGTGGTGTGGATGAGGAGGCAGATGGCAGGGCCTGTGCATTTCTGTGCTTGAGTGGGCCTTGAAAGTGGTTCAGCAACCA GGAAGAAGTGTTCATTCCTCGACAACAACATCCCCGGGCTCTGGTGACTTGGCTGACACTGGATGGCCCTGGAATGAAAAAGGCAAAGAG GCAAAATGTGCAAGGGCCCATCTGGAACCAAGGCCCCATAAAAGCGACTGTCACTCGGTCCCAGACACCAGAGCAAGCTCAAGACCCAGC AGTGGGACAGCCAGACAGACGGCACGATGGCACTGAGCTCCCAGATCTGGGCCGCTTGCCTCCTGCTCCTCCTCCTCCTCGCCAGCCTGA CCAGTGGCTCTGTTTTCCCACAACAGTGGGTGGGAGGACGCTACTCGCTGTGGTCGGCCATCGGACTCTCCATTGCCCTGCACGTGGGTT TTGACAACTTCGAGCAGCTGCTCTCGGGGGCTCACTGGATGGACCAGCACTTCCGCACGACGCCCCTGGAGAAGAACGCCCCCGTCTTGC TGGCCCTGCTGGGTATCTGGTACATCAACTGCTTTGGGTGTGAGACACACGCCATGCTGCCCTATGACCAGTACCTGCACCGCTTTGCTG CGTACTTCCAGCAGGGCGACATGGAGTCCAATGGGAAATACATCACCAAATCTGGAACCCGTGTGGACCACCAGACAGGCCCCATTGTGT GGGGGGAGCCAGGGACCAATGGCCAGCATGCTTTTTACCAGCTCATCCACCAAGGCACCAAGATGATACCCTGTGACTTCCTCATCCCGG TCCAGACCCAGCACCCCATACGGAAGGGTCTGCATCACAAGATCCTCCTGGCCAACTTCTTGGCCCAGACAGAGGCCCTGATGAGGGGAA AATCGACGGAGGAGGCCCGAAAGGAGCTCCAGGCTGCGGGCAAGAGTCCAGAGGACCTTGAGAGGCTGCTGCCACATAAGAGTGGAGCTG GGAAAGCAGCTGGCTAAGAAAATAGAGCCTGAGCTTGATGGCAGTGCTCAAGTGACCTCTCACGACGCTTCTACCAATGGGCTCATCAAC TTCATCAAGCAGCAGCGCGAGGCCAGAGTCCAATAAACTCGTGCTCATCTGCAGCCTCCTCTGTGACTCCCCTTTCTCTTCTCGTCCCTC CTCCCCGGAGCCGGCACTGCATGTTCCTGGACACCACCCAGAGCACCCTCTGGTTGTGGGCTTGGACCACGAGCCCTTAGCAGGGAAGGC TGGTCTCCCCCAGCCTAACCCCCAGCCCCTCCATGTCTATGCTCCCTCTGTGTTAGAATTGGCTGAAGTGTTTTTGTGCAGCTGACTTTT >35593_35593_6_HAMP-GPI_HAMP_chr19_35773570_ENST00000598398_GPI_chr19_34884153_ENST00000586425_length(amino acids)=267AA_BP=1 MPSSSGSIFLASCFPSSTLMWQQPLKVLWTLARSLELLSGLLRRFSPHQGLCLGQEVGQEDLVMQTLPYGVLGLDRDEEVTGYHLGALVD ELVKSMLAIGPWLPPHNGACLVVHTGSRFGDVFPIGLHVALLEVRSKAVQVLVIGQHGVCLTPKAVDVPDTQQGQQDGGVLLQGRRAEVL -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for HAMP-GPI |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for HAMP-GPI |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for HAMP-GPI |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
