
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:HAT1-EXOC6B (FusionGDB2 ID:35628) |
Fusion Gene Summary for HAT1-EXOC6B |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: HAT1-EXOC6B | Fusion gene ID: 35628 | Hgene | Tgene | Gene symbol | HAT1 | EXOC6B | Gene ID | 8520 | 23233 |
| Gene name | histone acetyltransferase 1 | exocyst complex component 6B | |
| Synonyms | KAT1 | SEC15B|SEC15L2|SEMDJL3 | |
| Cytomap | 2q31.1 | 2p13.2 | |
| Type of gene | protein-coding | protein-coding | |
| Description | histone acetyltransferase type B catalytic subunit | exocyst complex component 6BSEC15 homolog BSEC15-like protein 2exocyst complex component Sec15B | |
| Modification date | 20200320 | 20200313 | |
| UniProtAcc | O14929 | Q9Y2D4 | |
| Ensembl transtripts involved in fusion gene | ENST00000264108, ENST00000392584, ENST00000460481, | ENST00000490919, ENST00000410104, ENST00000272427, | |
| Fusion gene scores | * DoF score | 6 X 6 X 4=144 | 10 X 8 X 6=480 |
| # samples | 6 | 11 | |
| ** MAII score | log2(6/144*10)=-1.26303440583379 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(11/480*10)=-2.12553088208386 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: HAT1 [Title/Abstract] AND EXOC6B [Title/Abstract] AND fusion [Title/Abstract] | ||
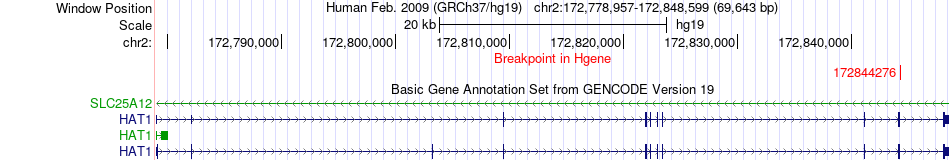
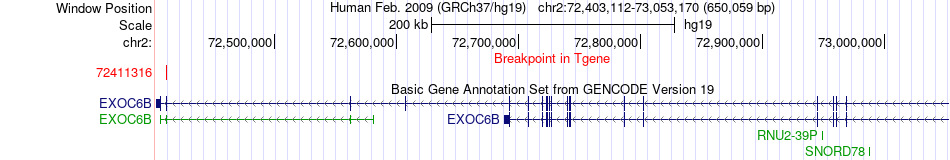
| Most frequent breakpoint | HAT1(172844276)-EXOC6B(72411316), # samples:4 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | HAT1 | GO:0006335 | DNA replication-dependent nucleosome assembly | 14718166 |
| Hgene | HAT1 | GO:0006336 | DNA replication-independent nucleosome assembly | 14718166 |
| Hgene | HAT1 | GO:0043967 | histone H4 acetylation | 22615379 |
 Fusion gene breakpoints across HAT1 (5'-gene) Fusion gene breakpoints across HAT1 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
 |
 Fusion gene breakpoints across EXOC6B (3'-gene) Fusion gene breakpoints across EXOC6B (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | LUAD | TCGA-05-4420-01A | HAT1 | chr2 | 172844276 | - | EXOC6B | chr2 | 72411316 | - |
| ChimerDB4 | LUAD | TCGA-05-4420-01A | HAT1 | chr2 | 172844276 | + | EXOC6B | chr2 | 72411316 | - |
| ChimerDB4 | LUAD | TCGA-05-4420 | HAT1 | chr2 | 172844276 | + | EXOC6B | chr2 | 72411316 | - |
Top |
Fusion Gene ORF analysis for HAT1-EXOC6B |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-5UTR | ENST00000264108 | ENST00000490919 | HAT1 | chr2 | 172844276 | + | EXOC6B | chr2 | 72411316 | - |
| 5CDS-5UTR | ENST00000392584 | ENST00000490919 | HAT1 | chr2 | 172844276 | + | EXOC6B | chr2 | 72411316 | - |
| 5CDS-intron | ENST00000264108 | ENST00000410104 | HAT1 | chr2 | 172844276 | + | EXOC6B | chr2 | 72411316 | - |
| 5CDS-intron | ENST00000392584 | ENST00000410104 | HAT1 | chr2 | 172844276 | + | EXOC6B | chr2 | 72411316 | - |
| In-frame | ENST00000264108 | ENST00000272427 | HAT1 | chr2 | 172844276 | + | EXOC6B | chr2 | 72411316 | - |
| In-frame | ENST00000392584 | ENST00000272427 | HAT1 | chr2 | 172844276 | + | EXOC6B | chr2 | 72411316 | - |
| intron-3CDS | ENST00000460481 | ENST00000272427 | HAT1 | chr2 | 172844276 | + | EXOC6B | chr2 | 72411316 | - |
| intron-5UTR | ENST00000460481 | ENST00000490919 | HAT1 | chr2 | 172844276 | + | EXOC6B | chr2 | 72411316 | - |
| intron-intron | ENST00000460481 | ENST00000410104 | HAT1 | chr2 | 172844276 | + | EXOC6B | chr2 | 72411316 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000392584 | HAT1 | chr2 | 172844276 | + | ENST00000272427 | EXOC6B | chr2 | 72411316 | - | 4660 | 1069 | 199 | 1308 | 369 |
| ENST00000264108 | HAT1 | chr2 | 172844276 | + | ENST00000272427 | EXOC6B | chr2 | 72411316 | - | 4719 | 1128 | 36 | 1367 | 443 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000392584 | ENST00000272427 | HAT1 | chr2 | 172844276 | + | EXOC6B | chr2 | 72411316 | - | 0.000834595 | 0.9991654 |
| ENST00000264108 | ENST00000272427 | HAT1 | chr2 | 172844276 | + | EXOC6B | chr2 | 72411316 | - | 0.001882516 | 0.99811757 |
Top |
Fusion Genomic Features for HAT1-EXOC6B |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
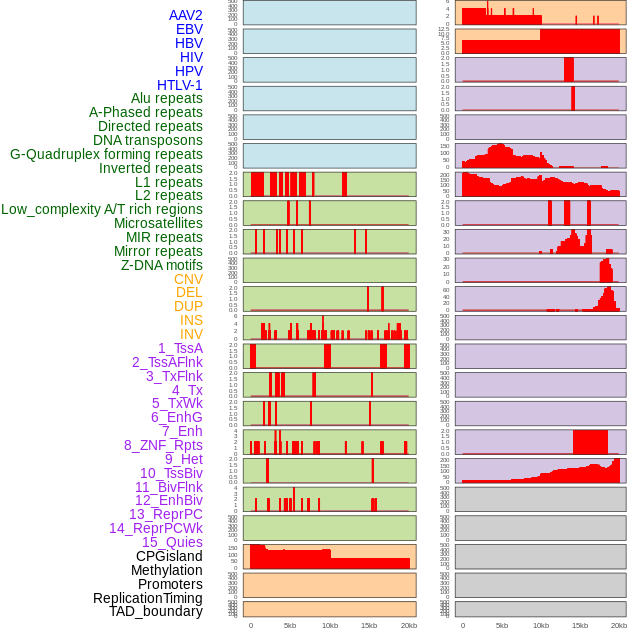
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for HAT1-EXOC6B |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr2:172844276/chr2:72411316) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| HAT1 | EXOC6B |
| FUNCTION: Histone acetyltransferase that plays a role in different biological processes including cell cycle progression, glucose metabolism, histone production or DNA damage repair (PubMed:31278053, PubMed:20953179, PubMed:23653357, PubMed:32081014). Coordinates histone production and acetylation via H4 promoter binding (PubMed:31278053). Acetylates histone H4 at 'Lys-5' (H4K5ac) and 'Lys-12' (H4K12ac) and, to a lesser extent, histone H2A at 'Lys-5' (H2AK5ac) (PubMed:22615379, PubMed:11585814). Drives H4 production by chromatin binding to support chromatin replication and acetylation. Since transcription of H4 genes is tightly coupled to S-phase, plays an important role in S-phase entry and progression (PubMed:31278053). Promotes homologous recombination in DNA repair by facilitating histone turnover and incorporation of acetylated H3.3 at sites of double-strand breaks (PubMed:23653357). In addition, acetylates other substrates such as chromatin-related proteins (PubMed:32081014). Acetylates also RSAD2 which mediates the interaction of ubiquitin ligase UBE4A with RSAD2 leading to RSAD2 ubiquitination and subsequent degradation (PubMed:31812350). {ECO:0000269|PubMed:11585814, ECO:0000269|PubMed:20953179, ECO:0000269|PubMed:22615379, ECO:0000269|PubMed:23653357, ECO:0000269|PubMed:31278053, ECO:0000269|PubMed:31812350, ECO:0000269|PubMed:32081014}.; FUNCTION: (Microbial infection) Contributes to hepatitis B virus (HBV) replication by acetylating histone H4 at the sites of 'Lys-5' and 'Lys-12' on the covalently closed circular DNA (cccDNA) minichromosome leading to its accumulation within the host cell. {ECO:0000269|PubMed:31695772}. | FUNCTION: Component of the exocyst complex involved in the docking of exocytic vesicles with fusion sites on the plasma membrane. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | HAT1 | chr2:172844276 | chr2:72411316 | ENST00000264108 | + | 10 | 11 | 241_243 | 364 | 420.0 | Region | Acetyl-CoA binding |
| Hgene | HAT1 | chr2:172844276 | chr2:72411316 | ENST00000264108 | + | 10 | 11 | 248_254 | 364 | 420.0 | Region | Acetyl-CoA binding |
| Hgene | HAT1 | chr2:172844276 | chr2:72411316 | ENST00000392584 | + | 9 | 10 | 241_243 | 279 | 335.0 | Region | Acetyl-CoA binding |
| Hgene | HAT1 | chr2:172844276 | chr2:72411316 | ENST00000392584 | + | 9 | 10 | 248_254 | 279 | 335.0 | Region | Acetyl-CoA binding |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | EXOC6B | chr2:172844276 | chr2:72411316 | ENST00000272427 | 19 | 22 | 50_119 | 732 | 812.0 | Coiled coil | Ontology_term=ECO:0000255 |
Top |
Fusion Gene Sequence for HAT1-EXOC6B |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >35628_35628_1_HAT1-EXOC6B_HAT1_chr2_172844276_ENST00000264108_EXOC6B_chr2_72411316_ENST00000272427_length(transcript)=4719nt_BP=1128nt CGTCCTTCCTCAGCCGCGGGTGATCGTAGCTCGGAAATGGCGGGATTTGGTGCTATGGAGAAATTTTTGGTAGAATATAAGAGTGCAGTG GAGAAGAAACTGGCAGAGTACAAATGTAACACCAACACAGCAATTGAACTAAAATTAGTTCGTTTTCCTGAAGATCTTGAAAATGACATT AGAACTTTCTTTCCTGAGTATACCCATCAACTCTTTGGGGATGATGAAACTGCTTTTGGTTACAAGGGTCTAAAGATCCTGTTATACTAT ATTGCTGGTAGCCTGTCAACAATGTTCCGTGTTGAATATGCATCTAAAGTTGATGAGAACTTTGACTGTGTAGAGGCAGATGATGTTGAG GGCAAAATTAGACAAATCATTCCACCTGGATTTTGCACAAACACGAATGATTTCCTTTCTTTACTGGAAAAGGAAGTTGATTTCAAGCCA TTCGGAACCTTACTTCATACCTACTCAGTTCTCAGTCCAACAGGAGGAGAAAACTTTACCTTTCAGATATATAAGGCTGACATGACATGT AGAGGCTTTCGAGAATATCATGAAAGGCTTCAGACCTTTTTGATGTGGTTTATTGAAACTGCTAGCTTTATTGACGTGGATGATGAAAGA TGGCACTACTTTCTAGTATTTGAGAAGTATAATAAGGATGGAGCTACGCTCTTTGCGACCGTAGGCTACATGACAGTCTATAATTACTAT GTGTACCCAGACAAAACCCGGCCACGTGTAAGTCAGATGCTGATTTTGACTCCATTTCAAGGTCAAGGCCATGGTGCTCAACTTCTTGAA ACAGTTCATAGATACTACACTGAATTTCCTACAGTTCTTGATATTACAGCGGAAGATCCATCCAAAAGCTATGTGAAATTACGAGACTTT GTGCTTGTGAAGCTTTGTCAAGATTTGCCCTGTTTTTCCCGGGAAAAATTAATGCAAGGATTCAATGAAGATATGGCGATAGAGGCACAA CAGAAGTTCAAAATAAATAAGCAACACGCTAGAAGGGTTTATGAAATTCTTCGACTACTGGTAACTGACATGAGTGATGCCGAACAATAC AGAAGCTACAGACTGGATATTAAAAGAAGACTAATTAGCCCATATAAGCTTTTGGATCTTTTCATCCAGTGGGATTGGTCAACCTACCTT GCTGACTATGGTCAGCCCAACTGCAAGTACCTCCGGGTAAACCCAGTGACTGCTCTGACCCTGCTTGAGAAGATGAAGGATACTAGCCGC AAGAACAACATGTTTGCACAGTTTCGGAAAAATGAGCGAGACAAGCAGAAACTCATTGACACCGTGGCCAAGCAGCTCCGAGGACTCATC AGCAGCCACCACTCATGAAGGGTGACCCCGGACCCACAGAGGCGACCTGCTGCAGCCTCTGTGTCAGTCTGGGGTCCAGCCCCAGCTGAC CCTGCATTTGTGCATTCTTCTTCAAAGAGAGCATATGTATTTTTTTATTAACCCCTGTGCAGTATTCACATAACATTTCCCTATAGTAAA AGAGGCACAAGAATAAGCAAGTACATGAGATGTTGCTAGGCTGGGACTCAGGGGAAGTTATTACTACACTATCTGTGTTATCTTAGAGAT GGTGGTATCATCAATTCCACAGTGATGAACCACCTTTCATCCATGGCTCTAAAGCTTAGGTCTGGGGCTTAGGAACCATAAAAAGTGGGT ATCTGGCGCTGGGAGGTAGAGTGAGTAAGCAAAAGGATACTAAAATATGTCCTGTTCACTCTTGGATGCCCATTCACCACATAGATGAGG CTGATTCTTCTCAATACAAGGAAAGGGCATGGCAAACATTACTATTTAGTACACTAAGGGCAGCTCTGGAGGGTCATCTCCTCAGTCTGC TGACTTTCACTGGCTTGTGAACAAGCATGGCATATGGCAAGATGACAGCTTCATTCCAATCCCCCAAGAATGGCGGTTTCTTTAGCCTGA TCTAACCCCAGGCTTTTATTGCAATTTTTTCTAGGGCAGGATACAGCTAAGTAGACAGCAGTAATAATTGGATCCCATGTTTAACTGCAT ATTGCCCCAGCTCTAGGAGTGTTGAGTCTCAACTCGAAGAGTTTTTCTGGATCTTGAAAGCCAGTTTTCATGGAATCAGAGACCCAATCC CTTCCCTGTTCATCTCATTTCCTTCACTTACAGTCAGAGCACTTGGTTCAGGACATAAATCAACAGTTACTTCCATAGGCCTTCACAGGG CGTTTCAGACTTAGCCTCCTCCAGTAATGCCATTGAAATAGTTCAGATTTCGGAAGCCTAAAAATGGCCTTTACCTTACCCTTAGGTGTA ACCTGAGGATTAGTTGGGATTGTTCCTTTATGTTTTAAAACCAGCTTTTTGGTTATCATCTTTAAGGAAATGTCACCAGCTCTGCACTGT GGGGGCAGCCATGCCTGGACTGGATGCCTCTAGGGATGGAGACATTGTTACCATAAAGTCGACAGAGCCAACCATGGCTTGTGGCTTGTT GCCTCTGGCCCAACTGTCCTTCACTAAATAATATCCTAGATATTATTTTGTTGCTCAAGCCCAAGGTTCCCACCCTCTTTTGAGATCCCC AAAGCCATCTTTGGCCTTGTCTAAGAAAGATTGCCTATAATCCTGTCCTCCAAACTCTAAAGCCTGGAGCAGCTTAGCGTGGAGGCTGCA CTCATTGACCCAGATGGACTCCTGAGACTTAACGCAAGGGCTTTGCTATGAGGTCAGTCGGGGGAAACTGTACTTTAAGACACTTCAGAA AGATGCTACTGGCTAGTAGCAGAAAGATGGCTAGTGGCAAGAGTATAATTGTGCATCACCAAACAATGACAAATAAGCCACACAGGCATT TCCAGCTTGGATTGATGGGAGGAGAGACTCATCAAACAGAATTCTTCACTACCTTTGTCAGCTACTGAGTTGCTTCTGGGGAGGGAAGTA CTTCCTTGCCCCTCCCCAACCCCCCTACCTCACCATATCCTATCATATCTTGATAGTCATGGGGAAGAGGATGTGCACACAGACATACAA ATTTCCTCAAAGCTGGAGAGACCAGGCTACATGTGAGCTCATAGATGCTGCTGAGGCTCATCCTGAGGGCTGGATGGTTGGCCAGGGTTT CAGAATGAGGGTAAGGGATGAGCACTGCCACCCAGAACTTGATAAGCCAAGAGAAGGCAGGAGCCAAAATGGCATCGCCAAGCTCTCATC TTTGTTGAAACAAGTGAGAAACGATTATATTAATCTGAAGTTCTATTGCTTGAGAAGAGTTCAAAACACTTCTCTTTGCTAGTACCCAGT TTGCAGCAAGATCTCTTCAGCTTTAAGTGCTGAGAGACTTTGAAGTGGAAATGGAGAGATGTGTAAGCATGGCACAGTGTTTGTCTTTTT AGAGGATCATCATTGCTGGTCTGGTCTCAAACCTCTGCCTTGGAAGAACTGTGCTATACAGGAAAAAGTGAACAAGGTGGAGGAATAGGG GTTATTCTGCACAGAAGTCCCATGAAAGGAAAAGCTTATTCAAGCTCAAGACATAAAGATGGACCCTTTGATATATCTTCTCTCCATTCT TAGTCTGCCTTCACCTTATCTAACAAAAAACAACATGCAGCTACTGAGTTGTATGTAGAGACATACTCTTGGTGGGCCAGGGAAGGTAAA AACAGGGACCAGCAATGTCAAGTTTTTGAACTAGAAAAGTGGGATGTTTTTAGTTTTGTCTATAACCTGTAGCTGGGCCAACCCTTGAGA GGACTTGTGCAAAGCATGTGTGCCCAGACCCATCAGACTCCTCAGATATGGCCTCAAATACCTAGAGATGTAGAAGTACCGTGGAGGGCC TCAATAAGAGGGCAAGCACACAGATGGCTAGAAGCCTGTATGGCTTCTGGTCAGGATGAACCCTTGGCCAAGTTCTTGAGGAAACCAGGT CTATATTCCTGAAGGGACAGTTCAGTTTGGGACTCTATTAGGTTTCAGTAGTAATATAGGCTGCTTGCATCATTTTAACATTCATAGGGA AAATCTCTTTGAGCAGGCTATAAATGAAGCTGTTGCAGGTAGGGAAGAACAGGACCTGAAAATGGGAACCAGGAAGTGAGATCCTGTTGG CAAAGGGAATGTAGAGCTTCCAGGGTGCATTGCCCACTGTTCTCATTGCATACCAGTCCTCCAAGTCAGTCTAGCCTTACTGTACACTTG ACAAGTGCTTACTCAGCAAGTCCCAGACCCACGGCCTTTTATCTCCCAAGACTGGCTTTGGCCATGACCTTCCTCATCAGGCCTCTGAGC ACACTCTCTAGGAACTGCTGCAGAAATTCCCCTACCTTAGCCTCCCTTCTCCCAGGACTCCTCTCTTGACTGCAATTGCCTCTTCTTCTT CTTCTTCTTCTTCCTTTTTGTTTTTTTTTTTTTTGCCTGCATTCACAATTACTTTTCACAAAGGTGTTGGAAATGGCATTTTTATTTTAC ACAGATAGTTGCACATATATAGAGAAAATTAGGAAAGTCCATTTTTAACTCTAAAGGGAACTTTTTTCTCTAAAACTTACTTGTAAAGCT ACTGATACAGATGGGCCGCATGGTTAGGGGAAAGCTCCTCTGTGAAAAGAGCATGTTTATTCCTCTTGTACATTTTCCTCTTTTAACTGC >35628_35628_1_HAT1-EXOC6B_HAT1_chr2_172844276_ENST00000264108_EXOC6B_chr2_72411316_ENST00000272427_length(amino acids)=443AA_BP=364 MAGFGAMEKFLVEYKSAVEKKLAEYKCNTNTAIELKLVRFPEDLENDIRTFFPEYTHQLFGDDETAFGYKGLKILLYYIAGSLSTMFRVE YASKVDENFDCVEADDVEGKIRQIIPPGFCTNTNDFLSLLEKEVDFKPFGTLLHTYSVLSPTGGENFTFQIYKADMTCRGFREYHERLQT FLMWFIETASFIDVDDERWHYFLVFEKYNKDGATLFATVGYMTVYNYYVYPDKTRPRVSQMLILTPFQGQGHGAQLLETVHRYYTEFPTV LDITAEDPSKSYVKLRDFVLVKLCQDLPCFSREKLMQGFNEDMAIEAQQKFKINKQHARRVYEILRLLVTDMSDAEQYRSYRLDIKRRLI -------------------------------------------------------------- >35628_35628_2_HAT1-EXOC6B_HAT1_chr2_172844276_ENST00000392584_EXOC6B_chr2_72411316_ENST00000272427_length(transcript)=4660nt_BP=1069nt GAGCGCGCGGGTTGATTCGTCCTTCCTCAGCCGCGGGTGATCGTAGCTCGGAAATGGCGGGATTTGGTGCTATGGAGAAATTTTTGGTAG AATATAAGAGTGCAGTGGAGAAGAAACTGGCAGAGTACAAATGTAACACCAACACAGCAATTGAACTAAAATTAGTGAAACTGCTTTTGG TTACAAGGGTCTAAAGATCCTGTTATACTATATTGCTGGTAGCCTGTCAACAATGTTCCGTGTTGAATATGCATCTAAAGTTGATGAGAA CTTTGACTGTGTAGAGGCAGATGATGTTGAGGGCAAAATTAGACAAATCATTCCACCTGGATTTTGCACAAACACGAATGATTTCCTTTC TTTACTGGAAAAGGAAGTTGATTTCAAGCCATTCGGAACCTTACTTCATACCTACTCAGTTCTCAGTCCAACAGGAGGAGAAAACTTTAC CTTTCAGATATATAAGGCTGACATGACATGTAGAGGCTTTCGAGAATATCATGAAAGGCTTCAGACCTTTTTGATGTGGTTTATTGAAAC TGCTAGCTTTATTGACGTGGATGATGAAAGATGGCACTACTTTCTAGTATTTGAGAAGTATAATAAGGATGGAGCTACGCTCTTTGCGAC CGTAGGCTACATGACAGTCTATAATTACTATGTGTACCCAGACAAAACCCGGCCACGTGTAAGTCAGATGCTGATTTTGACTCCATTTCA AGGTCAAGGCCATGGTGCTCAACTTCTTGAAACAGTTCATAGATACTACACTGAATTTCCTACAGTTCTTGATATTACAGCGGAAGATCC ATCCAAAAGCTATGTGAAATTACGAGACTTTGTGCTTGTGAAGCTTTGTCAAGATTTGCCCTGTTTTTCCCGGGAAAAATTAATGCAAGG ATTCAATGAAGATATGGCGATAGAGGCACAACAGAAGTTCAAAATAAATAAGCAACACGCTAGAAGGGTTTATGAAATTCTTCGACTACT GGTAACTGACATGAGTGATGCCGAACAATACAGAAGCTACAGACTGGATATTAAAAGAAGACTAATTAGCCCATATAAGCTTTTGGATCT TTTCATCCAGTGGGATTGGTCAACCTACCTTGCTGACTATGGTCAGCCCAACTGCAAGTACCTCCGGGTAAACCCAGTGACTGCTCTGAC CCTGCTTGAGAAGATGAAGGATACTAGCCGCAAGAACAACATGTTTGCACAGTTTCGGAAAAATGAGCGAGACAAGCAGAAACTCATTGA CACCGTGGCCAAGCAGCTCCGAGGACTCATCAGCAGCCACCACTCATGAAGGGTGACCCCGGACCCACAGAGGCGACCTGCTGCAGCCTC TGTGTCAGTCTGGGGTCCAGCCCCAGCTGACCCTGCATTTGTGCATTCTTCTTCAAAGAGAGCATATGTATTTTTTTATTAACCCCTGTG CAGTATTCACATAACATTTCCCTATAGTAAAAGAGGCACAAGAATAAGCAAGTACATGAGATGTTGCTAGGCTGGGACTCAGGGGAAGTT ATTACTACACTATCTGTGTTATCTTAGAGATGGTGGTATCATCAATTCCACAGTGATGAACCACCTTTCATCCATGGCTCTAAAGCTTAG GTCTGGGGCTTAGGAACCATAAAAAGTGGGTATCTGGCGCTGGGAGGTAGAGTGAGTAAGCAAAAGGATACTAAAATATGTCCTGTTCAC TCTTGGATGCCCATTCACCACATAGATGAGGCTGATTCTTCTCAATACAAGGAAAGGGCATGGCAAACATTACTATTTAGTACACTAAGG GCAGCTCTGGAGGGTCATCTCCTCAGTCTGCTGACTTTCACTGGCTTGTGAACAAGCATGGCATATGGCAAGATGACAGCTTCATTCCAA TCCCCCAAGAATGGCGGTTTCTTTAGCCTGATCTAACCCCAGGCTTTTATTGCAATTTTTTCTAGGGCAGGATACAGCTAAGTAGACAGC AGTAATAATTGGATCCCATGTTTAACTGCATATTGCCCCAGCTCTAGGAGTGTTGAGTCTCAACTCGAAGAGTTTTTCTGGATCTTGAAA GCCAGTTTTCATGGAATCAGAGACCCAATCCCTTCCCTGTTCATCTCATTTCCTTCACTTACAGTCAGAGCACTTGGTTCAGGACATAAA TCAACAGTTACTTCCATAGGCCTTCACAGGGCGTTTCAGACTTAGCCTCCTCCAGTAATGCCATTGAAATAGTTCAGATTTCGGAAGCCT AAAAATGGCCTTTACCTTACCCTTAGGTGTAACCTGAGGATTAGTTGGGATTGTTCCTTTATGTTTTAAAACCAGCTTTTTGGTTATCAT CTTTAAGGAAATGTCACCAGCTCTGCACTGTGGGGGCAGCCATGCCTGGACTGGATGCCTCTAGGGATGGAGACATTGTTACCATAAAGT CGACAGAGCCAACCATGGCTTGTGGCTTGTTGCCTCTGGCCCAACTGTCCTTCACTAAATAATATCCTAGATATTATTTTGTTGCTCAAG CCCAAGGTTCCCACCCTCTTTTGAGATCCCCAAAGCCATCTTTGGCCTTGTCTAAGAAAGATTGCCTATAATCCTGTCCTCCAAACTCTA AAGCCTGGAGCAGCTTAGCGTGGAGGCTGCACTCATTGACCCAGATGGACTCCTGAGACTTAACGCAAGGGCTTTGCTATGAGGTCAGTC GGGGGAAACTGTACTTTAAGACACTTCAGAAAGATGCTACTGGCTAGTAGCAGAAAGATGGCTAGTGGCAAGAGTATAATTGTGCATCAC CAAACAATGACAAATAAGCCACACAGGCATTTCCAGCTTGGATTGATGGGAGGAGAGACTCATCAAACAGAATTCTTCACTACCTTTGTC AGCTACTGAGTTGCTTCTGGGGAGGGAAGTACTTCCTTGCCCCTCCCCAACCCCCCTACCTCACCATATCCTATCATATCTTGATAGTCA TGGGGAAGAGGATGTGCACACAGACATACAAATTTCCTCAAAGCTGGAGAGACCAGGCTACATGTGAGCTCATAGATGCTGCTGAGGCTC ATCCTGAGGGCTGGATGGTTGGCCAGGGTTTCAGAATGAGGGTAAGGGATGAGCACTGCCACCCAGAACTTGATAAGCCAAGAGAAGGCA GGAGCCAAAATGGCATCGCCAAGCTCTCATCTTTGTTGAAACAAGTGAGAAACGATTATATTAATCTGAAGTTCTATTGCTTGAGAAGAG TTCAAAACACTTCTCTTTGCTAGTACCCAGTTTGCAGCAAGATCTCTTCAGCTTTAAGTGCTGAGAGACTTTGAAGTGGAAATGGAGAGA TGTGTAAGCATGGCACAGTGTTTGTCTTTTTAGAGGATCATCATTGCTGGTCTGGTCTCAAACCTCTGCCTTGGAAGAACTGTGCTATAC AGGAAAAAGTGAACAAGGTGGAGGAATAGGGGTTATTCTGCACAGAAGTCCCATGAAAGGAAAAGCTTATTCAAGCTCAAGACATAAAGA TGGACCCTTTGATATATCTTCTCTCCATTCTTAGTCTGCCTTCACCTTATCTAACAAAAAACAACATGCAGCTACTGAGTTGTATGTAGA GACATACTCTTGGTGGGCCAGGGAAGGTAAAAACAGGGACCAGCAATGTCAAGTTTTTGAACTAGAAAAGTGGGATGTTTTTAGTTTTGT CTATAACCTGTAGCTGGGCCAACCCTTGAGAGGACTTGTGCAAAGCATGTGTGCCCAGACCCATCAGACTCCTCAGATATGGCCTCAAAT ACCTAGAGATGTAGAAGTACCGTGGAGGGCCTCAATAAGAGGGCAAGCACACAGATGGCTAGAAGCCTGTATGGCTTCTGGTCAGGATGA ACCCTTGGCCAAGTTCTTGAGGAAACCAGGTCTATATTCCTGAAGGGACAGTTCAGTTTGGGACTCTATTAGGTTTCAGTAGTAATATAG GCTGCTTGCATCATTTTAACATTCATAGGGAAAATCTCTTTGAGCAGGCTATAAATGAAGCTGTTGCAGGTAGGGAAGAACAGGACCTGA AAATGGGAACCAGGAAGTGAGATCCTGTTGGCAAAGGGAATGTAGAGCTTCCAGGGTGCATTGCCCACTGTTCTCATTGCATACCAGTCC TCCAAGTCAGTCTAGCCTTACTGTACACTTGACAAGTGCTTACTCAGCAAGTCCCAGACCCACGGCCTTTTATCTCCCAAGACTGGCTTT GGCCATGACCTTCCTCATCAGGCCTCTGAGCACACTCTCTAGGAACTGCTGCAGAAATTCCCCTACCTTAGCCTCCCTTCTCCCAGGACT CCTCTCTTGACTGCAATTGCCTCTTCTTCTTCTTCTTCTTCTTCCTTTTTGTTTTTTTTTTTTTTGCCTGCATTCACAATTACTTTTCAC AAAGGTGTTGGAAATGGCATTTTTATTTTACACAGATAGTTGCACATATATAGAGAAAATTAGGAAAGTCCATTTTTAACTCTAAAGGGA ACTTTTTTCTCTAAAACTTACTTGTAAAGCTACTGATACAGATGGGCCGCATGGTTAGGGGAAAGCTCCTCTGTGAAAAGAGCATGTTTA >35628_35628_2_HAT1-EXOC6B_HAT1_chr2_172844276_ENST00000392584_EXOC6B_chr2_72411316_ENST00000272427_length(amino acids)=369AA_BP=290 MLYYIAGSLSTMFRVEYASKVDENFDCVEADDVEGKIRQIIPPGFCTNTNDFLSLLEKEVDFKPFGTLLHTYSVLSPTGGENFTFQIYKA DMTCRGFREYHERLQTFLMWFIETASFIDVDDERWHYFLVFEKYNKDGATLFATVGYMTVYNYYVYPDKTRPRVSQMLILTPFQGQGHGA QLLETVHRYYTEFPTVLDITAEDPSKSYVKLRDFVLVKLCQDLPCFSREKLMQGFNEDMAIEAQQKFKINKQHARRVYEILRLLVTDMSD AEQYRSYRLDIKRRLISPYKLLDLFIQWDWSTYLADYGQPNCKYLRVNPVTALTLLEKMKDTSRKNNMFAQFRKNERDKQKLIDTVAKQL -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for HAT1-EXOC6B |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
| Hgene | HAT1 | chr2:172844276 | chr2:72411316 | ENST00000264108 | + | 10 | 11 | 62_64 | 364.0 | 420.0 | histone H4 N-terminus |
| Hgene | HAT1 | chr2:172844276 | chr2:72411316 | ENST00000392584 | + | 9 | 10 | 62_64 | 279.0 | 335.0 | histone H4 N-terminus |
| Hgene | HAT1 | chr2:172844276 | chr2:72411316 | ENST00000264108 | + | 10 | 11 | 225_227 | 364.0 | 420.0 | histone H4 N-terminus |
| Hgene | HAT1 | chr2:172844276 | chr2:72411316 | ENST00000392584 | + | 9 | 10 | 225_227 | 279.0 | 335.0 | histone H4 N-terminus |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for HAT1-EXOC6B |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for HAT1-EXOC6B |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
