
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:HGSNAT-FUT10 (FusionGDB2 ID:36253) |
Fusion Gene Summary for HGSNAT-FUT10 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: HGSNAT-FUT10 | Fusion gene ID: 36253 | Hgene | Tgene | Gene symbol | HGSNAT | FUT10 | Gene ID | 138050 | 84750 |
| Gene name | heparan-alpha-glucosaminide N-acetyltransferase | fucosyltransferase 10 | |
| Synonyms | HGNAT|MPS3C|RP73|TMEM76 | FUCTX | |
| Cytomap | 8p11.21-p11.1 | 8p12 | |
| Type of gene | protein-coding | protein-coding | |
| Description | heparan-alpha-glucosaminide N-acetyltransferasetransmembrane protein 76 | alpha-(1,3)-fucosyltransferase 10alpha (1,3) fucosyltransferasealpha 1,3-fucosyl transferasefuc-TXfucT-Xfucosyltransferase Xgalactoside 3-L-fucosyltransferase 10 | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | Q68CP4 | Q6P4F1 | |
| Ensembl transtripts involved in fusion gene | ENST00000297798, ENST00000379644, ENST00000458501, ENST00000521576, | ENST00000335589, ENST00000518076, ENST00000327671, ENST00000524021, ENST00000518672, | |
| Fusion gene scores | * DoF score | 9 X 8 X 7=504 | 7 X 6 X 5=210 |
| # samples | 9 | 8 | |
| ** MAII score | log2(9/504*10)=-2.48542682717024 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(8/210*10)=-1.39231742277876 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: HGSNAT [Title/Abstract] AND FUT10 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | HGSNAT(43037403)-FUT10(33230322), # samples:3 | ||
| Anticipated loss of major functional domain due to fusion event. | HGSNAT-FUT10 seems lost the major protein functional domain in Tgene partner, which is a essential gene due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | HGSNAT | GO:0007041 | lysosomal transport | 20650889 |
| Hgene | HGSNAT | GO:0051259 | protein complex oligomerization | 20650889 |
 Fusion gene breakpoints across HGSNAT (5'-gene) Fusion gene breakpoints across HGSNAT (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
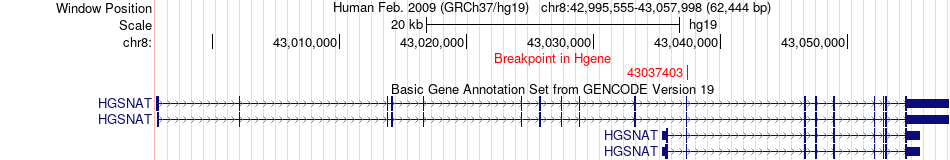 |
 Fusion gene breakpoints across FUT10 (3'-gene) Fusion gene breakpoints across FUT10 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
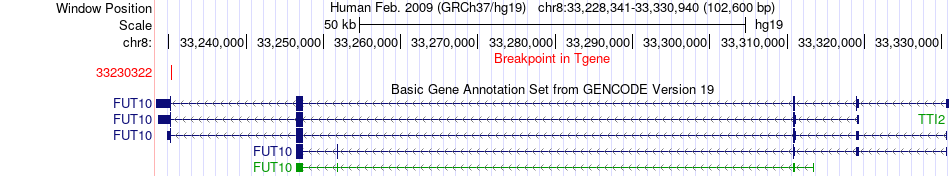 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | BLCA | TCGA-UY-A8OC-01A | HGSNAT | chr8 | 43037403 | - | FUT10 | chr8 | 33230322 | - |
| ChimerDB4 | BLCA | TCGA-UY-A8OC-01A | HGSNAT | chr8 | 43037403 | + | FUT10 | chr8 | 33230322 | - |
Top |
Fusion Gene ORF analysis for HGSNAT-FUT10 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-intron | ENST00000297798 | ENST00000335589 | HGSNAT | chr8 | 43037403 | + | FUT10 | chr8 | 33230322 | - |
| 5CDS-intron | ENST00000297798 | ENST00000518076 | HGSNAT | chr8 | 43037403 | + | FUT10 | chr8 | 33230322 | - |
| 5CDS-intron | ENST00000379644 | ENST00000335589 | HGSNAT | chr8 | 43037403 | + | FUT10 | chr8 | 33230322 | - |
| 5CDS-intron | ENST00000379644 | ENST00000518076 | HGSNAT | chr8 | 43037403 | + | FUT10 | chr8 | 33230322 | - |
| 5CDS-intron | ENST00000458501 | ENST00000335589 | HGSNAT | chr8 | 43037403 | + | FUT10 | chr8 | 33230322 | - |
| 5CDS-intron | ENST00000458501 | ENST00000518076 | HGSNAT | chr8 | 43037403 | + | FUT10 | chr8 | 33230322 | - |
| 5CDS-intron | ENST00000521576 | ENST00000335589 | HGSNAT | chr8 | 43037403 | + | FUT10 | chr8 | 33230322 | - |
| 5CDS-intron | ENST00000521576 | ENST00000518076 | HGSNAT | chr8 | 43037403 | + | FUT10 | chr8 | 33230322 | - |
| Frame-shift | ENST00000297798 | ENST00000327671 | HGSNAT | chr8 | 43037403 | + | FUT10 | chr8 | 33230322 | - |
| Frame-shift | ENST00000297798 | ENST00000524021 | HGSNAT | chr8 | 43037403 | + | FUT10 | chr8 | 33230322 | - |
| Frame-shift | ENST00000379644 | ENST00000327671 | HGSNAT | chr8 | 43037403 | + | FUT10 | chr8 | 33230322 | - |
| Frame-shift | ENST00000379644 | ENST00000524021 | HGSNAT | chr8 | 43037403 | + | FUT10 | chr8 | 33230322 | - |
| Frame-shift | ENST00000458501 | ENST00000327671 | HGSNAT | chr8 | 43037403 | + | FUT10 | chr8 | 33230322 | - |
| Frame-shift | ENST00000458501 | ENST00000524021 | HGSNAT | chr8 | 43037403 | + | FUT10 | chr8 | 33230322 | - |
| Frame-shift | ENST00000521576 | ENST00000327671 | HGSNAT | chr8 | 43037403 | + | FUT10 | chr8 | 33230322 | - |
| Frame-shift | ENST00000521576 | ENST00000524021 | HGSNAT | chr8 | 43037403 | + | FUT10 | chr8 | 33230322 | - |
| In-frame | ENST00000297798 | ENST00000518672 | HGSNAT | chr8 | 43037403 | + | FUT10 | chr8 | 33230322 | - |
| In-frame | ENST00000379644 | ENST00000518672 | HGSNAT | chr8 | 43037403 | + | FUT10 | chr8 | 33230322 | - |
| In-frame | ENST00000458501 | ENST00000518672 | HGSNAT | chr8 | 43037403 | + | FUT10 | chr8 | 33230322 | - |
| In-frame | ENST00000521576 | ENST00000518672 | HGSNAT | chr8 | 43037403 | + | FUT10 | chr8 | 33230322 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000458501 | HGSNAT | chr8 | 43037403 | + | ENST00000518672 | FUT10 | chr8 | 33230322 | - | 2956 | 1212 | 0 | 1439 | 479 |
| ENST00000379644 | HGSNAT | chr8 | 43037403 | + | ENST00000518672 | FUT10 | chr8 | 33230322 | - | 2914 | 1170 | 42 | 1397 | 451 |
| ENST00000297798 | HGSNAT | chr8 | 43037403 | + | ENST00000518672 | FUT10 | chr8 | 33230322 | - | 2310 | 566 | 242 | 793 | 183 |
| ENST00000521576 | HGSNAT | chr8 | 43037403 | + | ENST00000518672 | FUT10 | chr8 | 33230322 | - | 2310 | 566 | 242 | 793 | 183 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000458501 | ENST00000518672 | HGSNAT | chr8 | 43037403 | + | FUT10 | chr8 | 33230322 | - | 0.007155898 | 0.99284405 |
| ENST00000379644 | ENST00000518672 | HGSNAT | chr8 | 43037403 | + | FUT10 | chr8 | 33230322 | - | 0.001239408 | 0.9987606 |
| ENST00000297798 | ENST00000518672 | HGSNAT | chr8 | 43037403 | + | FUT10 | chr8 | 33230322 | - | 0.22953247 | 0.7704676 |
| ENST00000521576 | ENST00000518672 | HGSNAT | chr8 | 43037403 | + | FUT10 | chr8 | 33230322 | - | 0.22953247 | 0.7704676 |
Top |
Fusion Genomic Features for HGSNAT-FUT10 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
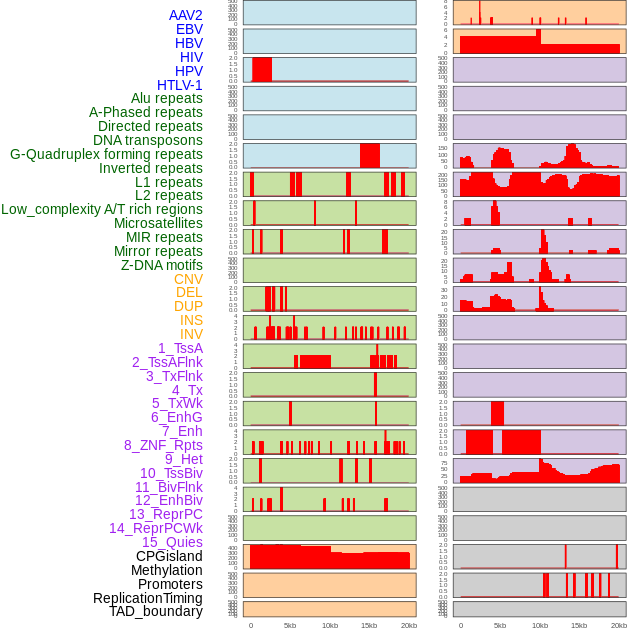 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for HGSNAT-FUT10 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr8:43037403/chr8:33230322) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| HGSNAT | FUT10 |
| FUNCTION: Lysosomal acetyltransferase that acetylates the non-reducing terminal alpha-glucosamine residue of intralysosomal heparin or heparan sulfate, converting it into a substrate for luminal alpha-N-acetyl glucosaminidase. {ECO:0000269|PubMed:16960811, ECO:0000269|PubMed:17033958, ECO:0000269|PubMed:19823584, ECO:0000269|PubMed:20650889}. | FUNCTION: Probable fucosyltransferase. {ECO:0000250}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | HGSNAT | chr8:43037403 | chr8:33230322 | ENST00000379644 | + | 11 | 18 | 4_64 | 376 | 636.0 | Compositional bias | Note=Ala-rich |
| Hgene | HGSNAT | chr8:43037403 | chr8:33230322 | ENST00000458501 | + | 11 | 18 | 4_64 | 404 | 664.0 | Compositional bias | Note=Ala-rich |
| Hgene | HGSNAT | chr8:43037403 | chr8:33230322 | ENST00000379644 | + | 11 | 18 | 1_190 | 376 | 636.0 | Topological domain | Lumenal%2C vesicle |
| Hgene | HGSNAT | chr8:43037403 | chr8:33230322 | ENST00000379644 | + | 11 | 18 | 212_275 | 376 | 636.0 | Topological domain | Cytoplasmic |
| Hgene | HGSNAT | chr8:43037403 | chr8:33230322 | ENST00000379644 | + | 11 | 18 | 297_302 | 376 | 636.0 | Topological domain | Lumenal%2C vesicle |
| Hgene | HGSNAT | chr8:43037403 | chr8:33230322 | ENST00000379644 | + | 11 | 18 | 324_345 | 376 | 636.0 | Topological domain | Cytoplasmic |
| Hgene | HGSNAT | chr8:43037403 | chr8:33230322 | ENST00000379644 | + | 11 | 18 | 367_374 | 376 | 636.0 | Topological domain | Lumenal%2C vesicle |
| Hgene | HGSNAT | chr8:43037403 | chr8:33230322 | ENST00000458501 | + | 11 | 18 | 1_190 | 404 | 664.0 | Topological domain | Lumenal%2C vesicle |
| Hgene | HGSNAT | chr8:43037403 | chr8:33230322 | ENST00000458501 | + | 11 | 18 | 212_275 | 404 | 664.0 | Topological domain | Cytoplasmic |
| Hgene | HGSNAT | chr8:43037403 | chr8:33230322 | ENST00000458501 | + | 11 | 18 | 297_302 | 404 | 664.0 | Topological domain | Lumenal%2C vesicle |
| Hgene | HGSNAT | chr8:43037403 | chr8:33230322 | ENST00000458501 | + | 11 | 18 | 324_345 | 404 | 664.0 | Topological domain | Cytoplasmic |
| Hgene | HGSNAT | chr8:43037403 | chr8:33230322 | ENST00000458501 | + | 11 | 18 | 367_374 | 404 | 664.0 | Topological domain | Lumenal%2C vesicle |
| Hgene | HGSNAT | chr8:43037403 | chr8:33230322 | ENST00000379644 | + | 11 | 18 | 191_211 | 376 | 636.0 | Transmembrane | Helical |
| Hgene | HGSNAT | chr8:43037403 | chr8:33230322 | ENST00000379644 | + | 11 | 18 | 276_296 | 376 | 636.0 | Transmembrane | Helical |
| Hgene | HGSNAT | chr8:43037403 | chr8:33230322 | ENST00000379644 | + | 11 | 18 | 303_323 | 376 | 636.0 | Transmembrane | Helical |
| Hgene | HGSNAT | chr8:43037403 | chr8:33230322 | ENST00000379644 | + | 11 | 18 | 346_366 | 376 | 636.0 | Transmembrane | Helical |
| Hgene | HGSNAT | chr8:43037403 | chr8:33230322 | ENST00000458501 | + | 11 | 18 | 191_211 | 404 | 664.0 | Transmembrane | Helical |
| Hgene | HGSNAT | chr8:43037403 | chr8:33230322 | ENST00000458501 | + | 11 | 18 | 276_296 | 404 | 664.0 | Transmembrane | Helical |
| Hgene | HGSNAT | chr8:43037403 | chr8:33230322 | ENST00000458501 | + | 11 | 18 | 303_323 | 404 | 664.0 | Transmembrane | Helical |
| Hgene | HGSNAT | chr8:43037403 | chr8:33230322 | ENST00000458501 | + | 11 | 18 | 346_366 | 404 | 664.0 | Transmembrane | Helical |
| Hgene | HGSNAT | chr8:43037403 | chr8:33230322 | ENST00000458501 | + | 11 | 18 | 375_395 | 404 | 664.0 | Transmembrane | Helical |
| Tgene | FUT10 | chr8:43037403 | chr8:33230322 | ENST00000335589 | 0 | 5 | 1_8 | 0 | 358.0 | Topological domain | Cytoplasmic | |
| Tgene | FUT10 | chr8:43037403 | chr8:33230322 | ENST00000335589 | 0 | 5 | 32_479 | 0 | 358.0 | Topological domain | Lumenal | |
| Tgene | FUT10 | chr8:43037403 | chr8:33230322 | ENST00000335589 | 0 | 5 | 9_31 | 0 | 358.0 | Transmembrane | Helical%3B Signal-anchor for type II membrane protein |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | HGSNAT | chr8:43037403 | chr8:33230322 | ENST00000379644 | + | 11 | 18 | 624_635 | 376 | 636.0 | Region | Note=Lysosomal targeting region |
| Hgene | HGSNAT | chr8:43037403 | chr8:33230322 | ENST00000458501 | + | 11 | 18 | 624_635 | 404 | 664.0 | Region | Note=Lysosomal targeting region |
| Hgene | HGSNAT | chr8:43037403 | chr8:33230322 | ENST00000379644 | + | 11 | 18 | 396_420 | 376 | 636.0 | Topological domain | Cytoplasmic |
| Hgene | HGSNAT | chr8:43037403 | chr8:33230322 | ENST00000379644 | + | 11 | 18 | 442_500 | 376 | 636.0 | Topological domain | Lumenal%2C vesicle |
| Hgene | HGSNAT | chr8:43037403 | chr8:33230322 | ENST00000379644 | + | 11 | 18 | 522_529 | 376 | 636.0 | Topological domain | Cytoplasmic |
| Hgene | HGSNAT | chr8:43037403 | chr8:33230322 | ENST00000379644 | + | 11 | 18 | 551_564 | 376 | 636.0 | Topological domain | Lumenal%2C vesicle |
| Hgene | HGSNAT | chr8:43037403 | chr8:33230322 | ENST00000379644 | + | 11 | 18 | 586_592 | 376 | 636.0 | Topological domain | Cytoplasmic |
| Hgene | HGSNAT | chr8:43037403 | chr8:33230322 | ENST00000379644 | + | 11 | 18 | 614_634 | 376 | 636.0 | Topological domain | Lumenal%2C vesicle |
| Hgene | HGSNAT | chr8:43037403 | chr8:33230322 | ENST00000379644 | + | 11 | 18 | 656_663 | 376 | 636.0 | Topological domain | Cytoplasmic |
| Hgene | HGSNAT | chr8:43037403 | chr8:33230322 | ENST00000458501 | + | 11 | 18 | 396_420 | 404 | 664.0 | Topological domain | Cytoplasmic |
| Hgene | HGSNAT | chr8:43037403 | chr8:33230322 | ENST00000458501 | + | 11 | 18 | 442_500 | 404 | 664.0 | Topological domain | Lumenal%2C vesicle |
| Hgene | HGSNAT | chr8:43037403 | chr8:33230322 | ENST00000458501 | + | 11 | 18 | 522_529 | 404 | 664.0 | Topological domain | Cytoplasmic |
| Hgene | HGSNAT | chr8:43037403 | chr8:33230322 | ENST00000458501 | + | 11 | 18 | 551_564 | 404 | 664.0 | Topological domain | Lumenal%2C vesicle |
| Hgene | HGSNAT | chr8:43037403 | chr8:33230322 | ENST00000458501 | + | 11 | 18 | 586_592 | 404 | 664.0 | Topological domain | Cytoplasmic |
| Hgene | HGSNAT | chr8:43037403 | chr8:33230322 | ENST00000458501 | + | 11 | 18 | 614_634 | 404 | 664.0 | Topological domain | Lumenal%2C vesicle |
| Hgene | HGSNAT | chr8:43037403 | chr8:33230322 | ENST00000458501 | + | 11 | 18 | 656_663 | 404 | 664.0 | Topological domain | Cytoplasmic |
| Hgene | HGSNAT | chr8:43037403 | chr8:33230322 | ENST00000379644 | + | 11 | 18 | 375_395 | 376 | 636.0 | Transmembrane | Helical |
| Hgene | HGSNAT | chr8:43037403 | chr8:33230322 | ENST00000379644 | + | 11 | 18 | 421_441 | 376 | 636.0 | Transmembrane | Helical |
| Hgene | HGSNAT | chr8:43037403 | chr8:33230322 | ENST00000379644 | + | 11 | 18 | 501_521 | 376 | 636.0 | Transmembrane | Helical |
| Hgene | HGSNAT | chr8:43037403 | chr8:33230322 | ENST00000379644 | + | 11 | 18 | 530_550 | 376 | 636.0 | Transmembrane | Helical |
| Hgene | HGSNAT | chr8:43037403 | chr8:33230322 | ENST00000379644 | + | 11 | 18 | 565_585 | 376 | 636.0 | Transmembrane | Helical |
| Hgene | HGSNAT | chr8:43037403 | chr8:33230322 | ENST00000379644 | + | 11 | 18 | 593_613 | 376 | 636.0 | Transmembrane | Helical |
| Hgene | HGSNAT | chr8:43037403 | chr8:33230322 | ENST00000379644 | + | 11 | 18 | 635_655 | 376 | 636.0 | Transmembrane | Helical |
| Hgene | HGSNAT | chr8:43037403 | chr8:33230322 | ENST00000458501 | + | 11 | 18 | 421_441 | 404 | 664.0 | Transmembrane | Helical |
| Hgene | HGSNAT | chr8:43037403 | chr8:33230322 | ENST00000458501 | + | 11 | 18 | 501_521 | 404 | 664.0 | Transmembrane | Helical |
| Hgene | HGSNAT | chr8:43037403 | chr8:33230322 | ENST00000458501 | + | 11 | 18 | 530_550 | 404 | 664.0 | Transmembrane | Helical |
| Hgene | HGSNAT | chr8:43037403 | chr8:33230322 | ENST00000458501 | + | 11 | 18 | 565_585 | 404 | 664.0 | Transmembrane | Helical |
| Hgene | HGSNAT | chr8:43037403 | chr8:33230322 | ENST00000458501 | + | 11 | 18 | 593_613 | 404 | 664.0 | Transmembrane | Helical |
| Hgene | HGSNAT | chr8:43037403 | chr8:33230322 | ENST00000458501 | + | 11 | 18 | 635_655 | 404 | 664.0 | Transmembrane | Helical |
| Tgene | FUT10 | chr8:43037403 | chr8:33230322 | ENST00000327671 | 3 | 5 | 1_8 | 404 | 480.0 | Topological domain | Cytoplasmic | |
| Tgene | FUT10 | chr8:43037403 | chr8:33230322 | ENST00000327671 | 3 | 5 | 32_479 | 404 | 480.0 | Topological domain | Lumenal | |
| Tgene | FUT10 | chr8:43037403 | chr8:33230322 | ENST00000518672 | 2 | 4 | 1_8 | 376 | 452.0 | Topological domain | Cytoplasmic | |
| Tgene | FUT10 | chr8:43037403 | chr8:33230322 | ENST00000518672 | 2 | 4 | 32_479 | 376 | 452.0 | Topological domain | Lumenal | |
| Tgene | FUT10 | chr8:43037403 | chr8:33230322 | ENST00000524021 | 3 | 5 | 1_8 | 376 | 452.0 | Topological domain | Cytoplasmic | |
| Tgene | FUT10 | chr8:43037403 | chr8:33230322 | ENST00000524021 | 3 | 5 | 32_479 | 376 | 452.0 | Topological domain | Lumenal | |
| Tgene | FUT10 | chr8:43037403 | chr8:33230322 | ENST00000327671 | 3 | 5 | 9_31 | 404 | 480.0 | Transmembrane | Helical%3B Signal-anchor for type II membrane protein | |
| Tgene | FUT10 | chr8:43037403 | chr8:33230322 | ENST00000518672 | 2 | 4 | 9_31 | 376 | 452.0 | Transmembrane | Helical%3B Signal-anchor for type II membrane protein | |
| Tgene | FUT10 | chr8:43037403 | chr8:33230322 | ENST00000524021 | 3 | 5 | 9_31 | 376 | 452.0 | Transmembrane | Helical%3B Signal-anchor for type II membrane protein |
Top |
Fusion Gene Sequence for HGSNAT-FUT10 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >36253_36253_1_HGSNAT-FUT10_HGSNAT_chr8_43037403_ENST00000297798_FUT10_chr8_33230322_ENST00000518672_length(transcript)=2310nt_BP=566nt ATTTCCCAGACGGGGTGGTTGCCGGACGGAGGGGCTCCTCACTTCTCAGACGGGGCGGTTGCCAGGCAGAGGGTTTCCTCACTTCTCAGA CGGAGCGGCCGGGCAGAGACGCTCCCCACCTCCCAGACAGGGCTGCGGCCCAGCAGAGGCACTCCTCACATCCCAGACAGGGCGGCGGGG CAGAGGTGCTCCCCACATCTCAGACGATGGGCGGCCGGGCAGAGACGCTCCTCACTTCCTAGATGGGATGGCGGCGGGGAAGAGGCGCTC CTCGCTTCCCAGATGGGATGGCGGCCGGGCAGAGACGCTCCTCACTTTCCAGACTGGGCAGCCAGGCAGAGGGGCTCCTCACATCCCAGA CGATGGGTGGCCAAGCAGAGACGCTCCTCACTTCCCAGACGGGGTGGCTGCTGGGCAGAGGCTGCAATCTCGGCTCTCCGGGAGGCCAAG TGTCTTGGGACAAGGTGCGCATTCCTGGTGTGCTGCAGCGATTGGGAGTGACATACTTTGTGGTTGCTGTGTTGGAGCTCCTCTTTGCTA AACCTGTGCCTGAACATTGTGCCTCGGGCTTACCACCCAAAAGATGGGAGGCAGAAGATACCCACCTGAGTTGCCCAGAGCCCACAGTGT TTGCTTTCTCACCACTCCGGACTCCACCTTTGAGCTCTTTGCGAGAGATGTGGATTTCCAGCTTTGAACAATCCAAGAAAGAAGCCCAGG CACTAAGGTGGCTGGTTGATAGGAATCAAAACTTTTCATCTCAAGAGTTTTGGGGCCTAGTATTCAAGGACTGATTTCAAAAATGATCAG AATGAAACAGACTAGAGCCTTCTTGAGGTTTATTTTCTAGGTTGCCTTAATATTTGAACATAATAGCTATTCTGTTGACTATCCATCAGG ATAATAATTAGTTGCTGCAGTACTCATAATGAGCCCTTTCAAGGAATAGATGTAAATTACCCTTAGGGGTAGCCACTATATTTTTTATAC TAAAAAAGGAAATATTTCTTATGGACACTGGTTATCTTCCATGTCCGTGTCTGTGGACCCACATACTTACTGCCTTTTGTGGAAATTCAC CTAGAGTGAGTACTTTCCTTGTGCAATGGAGGGAGACGATCTAGCTATAGATTCTTGATACTCACCTCACTCTCAACAGCTTATTGGTAT TTGGCAAACCTGGTATCCTGCAGCCGAGCATGAACATGAAATACATTAGAACTTGGCAATGAGAGATCCCATCTGTTGGTTTCCAGGGAT GTAAGTGAGTAATAAAAGCTGTTTCAAGTTTATCCACTATCCACTCTTCAAGTGAAAAGAACTTATGTGTGTCTACCTTCTTCTCCATAC CCTGAGCCACCTCTCTCACTTTCTTTAGTTGTGCCTTGCCCTATCGCCTCCCATTTTCCCTTCAAAGTAGAATGTCTTGTAAAGACACAG TAACTCCTGTGTGTTTTGATTAGTGAGATGTTACCACTTACCCCTTCTGCTGCGCAGAATAATATTAAGAATTGCTATGCTTATGATTCA GGCTGGACAGCCTGGACATTTTGAGCAGCGCGAGAGTAGAAGTGAATACCCCCAGGAACTCCAATTGTGCTGAAGCTGATTTGTCTGTTC GTAGGATGATGTGCAGCCTGTCACAGTGGCAGCCACAGGCATTCAGTTTGTGGGCATTTCCTGTCATTTCGGATCCTGCTATAGGGAAGG TTCAGCTCTCCTGTCTGGGGCATCTACTGGAGCACCTCAGAAGACAAGGAGGAAAACACTGGCTCATGTTTTCTTTCCATGTGAGGGAGG GGAGGTTTCTAGATGATACTAAAAGCCTGTTGCCACTGTTGTGTCATGATTTCCATTCTTCATCATGGAAAGCCACAAGCCTCAGAAGGC AGCCAGAGGACAGGAAGAGTCTGTGATGCTCATGTCTGTGTCATGGCATGAGGGGGAATTGACGAGTGCTTGCTTAGCATCAGCTCTCAT TGTATTGAAGGCTGTGCCCTTGGTGTGCTTAGCAGTGAGCAGCGTCTTCATTAGGTAGCTCTGGCTGGGCTTTGTCAGCCAGCGTGTGAG AAAGGAGAACCAAAACCAATACCTCCTTATAGAAGCCAGAAGGATCACATTGGATGAGAGAAGGTGTCTATAACTGGGTTCTTCCCAATC CCAGCTCTTTCATTAACTAAATATGATCTTGGACATATCTCTTAACCTCCATCGGCCTTAGTTTTTGCATCCAAAAAGAAGTGATTGGGC >36253_36253_1_HGSNAT-FUT10_HGSNAT_chr8_43037403_ENST00000297798_FUT10_chr8_33230322_ENST00000518672_length(amino acids)=183AA_BP=108 MGWRRGRGAPRFPDGMAAGQRRSSLSRLGSQAEGLLTSQTMGGQAETLLTSQTGWLLGRGCNLGSPGGQVSWDKVRIPGVLQRLGVTYFV VAVLELLFAKPVPEHCASGLPPKRWEAEDTHLSCPEPTVFAFSPLRTPPLSSLREMWISSFEQSKKEAQALRWLVDRNQNFSSQEFWGLV -------------------------------------------------------------- >36253_36253_2_HGSNAT-FUT10_HGSNAT_chr8_43037403_ENST00000379644_FUT10_chr8_33230322_ENST00000518672_length(transcript)=2914nt_BP=1170nt GGGCGCAGCGGGCAGGCAAGGGCGGCCGAGCGGGCGGCGGGCATGAGCGGGGCGGGCAGGGCGCTGGCCGCGCTGCTGCTGGCCGCGTCC GTGCTGAGCGCCGCGCTGCTGGCCCCCGGCGGCTCTTCGGGGCGCGATGCCCAGGCCGCGCCGCCACGAGACTTAGACAAAAAAAGACAT GCAGAGCTGAAGATGGATCAGGCTTTGCTACTCATCCATAATGAACTTCTCTGGACCAACTTGACCGTCTACTGGAAATCTGAATGCTGT TATCACTGCTTGTTTCAGGTTCTGGTAAACGTTCCTCAGAGTCCAAAAGCAGGGAAGCCTAGTGCTGCAGCTGCCTCTGTCAGCACCCAG CACGGATCTATCCTGCAGCTGAACGACACCTTGGAAGAGAAAGAAGTTTGTAGGTTGGAATACAGATTTGGAGAATTTGGAAACTATTCT CTCTTGGTAAAGAACATCCATAATGGAGTTAGTGAAATTGCCTGTGACCTGGCTGTGAACGAGGATCCAGTTGATAGTAACCTTCCTGTG AGCATTGCATTCCTTATTGGTCTTGCTGTCATCATTGTGATATCCTTTCTGAGGCTCTTGTTGAGTTTGGATGACTTTAACAATTGGATT TCTAAAGCCATAAGTTCTCGAGAAACTGATCGCCTCATCAATTCTGAGCTGGGATCTCCCAGCAGGACAGACCCTCTCGATGGTGATGTT CAGCCAGCAACGTGGCGTCTATCTGCCCTGCCGCCCCGCCTCCGCAGCGTGGACACCTTCAGGGGGATTGCTCTTATACTCATGGTCTTT GTCAATTATGGAGGAGGAAAATATTGGTACTTCAAACATGCAAGTTGGAATGGGCTGACAGTGGCTGACCTCGTGTTCCCGTGGTTTGTA TTTATTATGGGATCTTCCATTTTTCTATCGATGACTTCTATACTGCAACGGGGGTGTTCAAAATTCAGATTGCTGGGGAAGATTGCATGG AGGAGTTTCCTGTTAATCTGCATAGGAATTATCATTGTGAATCCCAATTATTGCCTTGGTCCATTGTCTTGGGACAAGGTGCGCATTCCT GGTGTGCTGCAGCGATTGGGAGTGACATACTTTGTGGTTGCTGTGTTGGAGCTCCTCTTTGCTAAACCTGTGCCTGAACATTGTGCCTCG GGCTTACCACCCAAAAGATGGGAGGCAGAAGATACCCACCTGAGTTGCCCAGAGCCCACAGTGTTTGCTTTCTCACCACTCCGGACTCCA CCTTTGAGCTCTTTGCGAGAGATGTGGATTTCCAGCTTTGAACAATCCAAGAAAGAAGCCCAGGCACTAAGGTGGCTGGTTGATAGGAAT CAAAACTTTTCATCTCAAGAGTTTTGGGGCCTAGTATTCAAGGACTGATTTCAAAAATGATCAGAATGAAACAGACTAGAGCCTTCTTGA GGTTTATTTTCTAGGTTGCCTTAATATTTGAACATAATAGCTATTCTGTTGACTATCCATCAGGATAATAATTAGTTGCTGCAGTACTCA TAATGAGCCCTTTCAAGGAATAGATGTAAATTACCCTTAGGGGTAGCCACTATATTTTTTATACTAAAAAAGGAAATATTTCTTATGGAC ACTGGTTATCTTCCATGTCCGTGTCTGTGGACCCACATACTTACTGCCTTTTGTGGAAATTCACCTAGAGTGAGTACTTTCCTTGTGCAA TGGAGGGAGACGATCTAGCTATAGATTCTTGATACTCACCTCACTCTCAACAGCTTATTGGTATTTGGCAAACCTGGTATCCTGCAGCCG AGCATGAACATGAAATACATTAGAACTTGGCAATGAGAGATCCCATCTGTTGGTTTCCAGGGATGTAAGTGAGTAATAAAAGCTGTTTCA AGTTTATCCACTATCCACTCTTCAAGTGAAAAGAACTTATGTGTGTCTACCTTCTTCTCCATACCCTGAGCCACCTCTCTCACTTTCTTT AGTTGTGCCTTGCCCTATCGCCTCCCATTTTCCCTTCAAAGTAGAATGTCTTGTAAAGACACAGTAACTCCTGTGTGTTTTGATTAGTGA GATGTTACCACTTACCCCTTCTGCTGCGCAGAATAATATTAAGAATTGCTATGCTTATGATTCAGGCTGGACAGCCTGGACATTTTGAGC AGCGCGAGAGTAGAAGTGAATACCCCCAGGAACTCCAATTGTGCTGAAGCTGATTTGTCTGTTCGTAGGATGATGTGCAGCCTGTCACAG TGGCAGCCACAGGCATTCAGTTTGTGGGCATTTCCTGTCATTTCGGATCCTGCTATAGGGAAGGTTCAGCTCTCCTGTCTGGGGCATCTA CTGGAGCACCTCAGAAGACAAGGAGGAAAACACTGGCTCATGTTTTCTTTCCATGTGAGGGAGGGGAGGTTTCTAGATGATACTAAAAGC CTGTTGCCACTGTTGTGTCATGATTTCCATTCTTCATCATGGAAAGCCACAAGCCTCAGAAGGCAGCCAGAGGACAGGAAGAGTCTGTGA TGCTCATGTCTGTGTCATGGCATGAGGGGGAATTGACGAGTGCTTGCTTAGCATCAGCTCTCATTGTATTGAAGGCTGTGCCCTTGGTGT GCTTAGCAGTGAGCAGCGTCTTCATTAGGTAGCTCTGGCTGGGCTTTGTCAGCCAGCGTGTGAGAAAGGAGAACCAAAACCAATACCTCC TTATAGAAGCCAGAAGGATCACATTGGATGAGAGAAGGTGTCTATAACTGGGTTCTTCCCAATCCCAGCTCTTTCATTAACTAAATATGA TCTTGGACATATCTCTTAACCTCCATCGGCCTTAGTTTTTGCATCCAAAAAGAAGTGATTGGGCTGGTCTTGGAAGGTTTTTTTCTTTTT >36253_36253_2_HGSNAT-FUT10_HGSNAT_chr8_43037403_ENST00000379644_FUT10_chr8_33230322_ENST00000518672_length(amino acids)=451AA_BP=376 MSGAGRALAALLLAASVLSAALLAPGGSSGRDAQAAPPRDLDKKRHAELKMDQALLLIHNELLWTNLTVYWKSECCYHCLFQVLVNVPQS PKAGKPSAAAASVSTQHGSILQLNDTLEEKEVCRLEYRFGEFGNYSLLVKNIHNGVSEIACDLAVNEDPVDSNLPVSIAFLIGLAVIIVI SFLRLLLSLDDFNNWISKAISSRETDRLINSELGSPSRTDPLDGDVQPATWRLSALPPRLRSVDTFRGIALILMVFVNYGGGKYWYFKHA SWNGLTVADLVFPWFVFIMGSSIFLSMTSILQRGCSKFRLLGKIAWRSFLLICIGIIIVNPNYCLGPLSWDKVRIPGVLQRLGVTYFVVA VLELLFAKPVPEHCASGLPPKRWEAEDTHLSCPEPTVFAFSPLRTPPLSSLREMWISSFEQSKKEAQALRWLVDRNQNFSSQEFWGLVFK -------------------------------------------------------------- >36253_36253_3_HGSNAT-FUT10_HGSNAT_chr8_43037403_ENST00000458501_FUT10_chr8_33230322_ENST00000518672_length(transcript)=2956nt_BP=1212nt ATGACGGGCGCGCGGGCGTCCGCGGCGGAGCAGCGCAGGGCGGGGCGCAGCGGGCAGGCAAGGGCGGCCGAGCGGGCGGCGGGCATGAGC GGGGCGGGCAGGGCGCTGGCCGCGCTGCTGCTGGCCGCGTCCGTGCTGAGCGCCGCGCTGCTGGCCCCCGGCGGCTCTTCGGGGCGCGAT GCCCAGGCCGCGCCGCCACGAGACTTAGACAAAAAAAGACATGCAGAGCTGAAGATGGATCAGGCTTTGCTACTCATCCATAATGAACTT CTCTGGACCAACTTGACCGTCTACTGGAAATCTGAATGCTGTTATCACTGCTTGTTTCAGGTTCTGGTAAACGTTCCTCAGAGTCCAAAA GCAGGGAAGCCTAGTGCTGCAGCTGCCTCTGTCAGCACCCAGCACGGATCTATCCTGCAGCTGAACGACACCTTGGAAGAGAAAGAAGTT TGTAGGTTGGAATACAGATTTGGAGAATTTGGAAACTATTCTCTCTTGGTAAAGAACATCCATAATGGAGTTAGTGAAATTGCCTGTGAC CTGGCTGTGAACGAGGATCCAGTTGATAGTAACCTTCCTGTGAGCATTGCATTCCTTATTGGTCTTGCTGTCATCATTGTGATATCCTTT CTGAGGCTCTTGTTGAGTTTGGATGACTTTAACAATTGGATTTCTAAAGCCATAAGTTCTCGAGAAACTGATCGCCTCATCAATTCTGAG CTGGGATCTCCCAGCAGGACAGACCCTCTCGATGGTGATGTTCAGCCAGCAACGTGGCGTCTATCTGCCCTGCCGCCCCGCCTCCGCAGC GTGGACACCTTCAGGGGGATTGCTCTTATACTCATGGTCTTTGTCAATTATGGAGGAGGAAAATATTGGTACTTCAAACATGCAAGTTGG AATGGGCTGACAGTGGCTGACCTCGTGTTCCCGTGGTTTGTATTTATTATGGGATCTTCCATTTTTCTATCGATGACTTCTATACTGCAA CGGGGGTGTTCAAAATTCAGATTGCTGGGGAAGATTGCATGGAGGAGTTTCCTGTTAATCTGCATAGGAATTATCATTGTGAATCCCAAT TATTGCCTTGGTCCATTGTCTTGGGACAAGGTGCGCATTCCTGGTGTGCTGCAGCGATTGGGAGTGACATACTTTGTGGTTGCTGTGTTG GAGCTCCTCTTTGCTAAACCTGTGCCTGAACATTGTGCCTCGGGCTTACCACCCAAAAGATGGGAGGCAGAAGATACCCACCTGAGTTGC CCAGAGCCCACAGTGTTTGCTTTCTCACCACTCCGGACTCCACCTTTGAGCTCTTTGCGAGAGATGTGGATTTCCAGCTTTGAACAATCC AAGAAAGAAGCCCAGGCACTAAGGTGGCTGGTTGATAGGAATCAAAACTTTTCATCTCAAGAGTTTTGGGGCCTAGTATTCAAGGACTGA TTTCAAAAATGATCAGAATGAAACAGACTAGAGCCTTCTTGAGGTTTATTTTCTAGGTTGCCTTAATATTTGAACATAATAGCTATTCTG TTGACTATCCATCAGGATAATAATTAGTTGCTGCAGTACTCATAATGAGCCCTTTCAAGGAATAGATGTAAATTACCCTTAGGGGTAGCC ACTATATTTTTTATACTAAAAAAGGAAATATTTCTTATGGACACTGGTTATCTTCCATGTCCGTGTCTGTGGACCCACATACTTACTGCC TTTTGTGGAAATTCACCTAGAGTGAGTACTTTCCTTGTGCAATGGAGGGAGACGATCTAGCTATAGATTCTTGATACTCACCTCACTCTC AACAGCTTATTGGTATTTGGCAAACCTGGTATCCTGCAGCCGAGCATGAACATGAAATACATTAGAACTTGGCAATGAGAGATCCCATCT GTTGGTTTCCAGGGATGTAAGTGAGTAATAAAAGCTGTTTCAAGTTTATCCACTATCCACTCTTCAAGTGAAAAGAACTTATGTGTGTCT ACCTTCTTCTCCATACCCTGAGCCACCTCTCTCACTTTCTTTAGTTGTGCCTTGCCCTATCGCCTCCCATTTTCCCTTCAAAGTAGAATG TCTTGTAAAGACACAGTAACTCCTGTGTGTTTTGATTAGTGAGATGTTACCACTTACCCCTTCTGCTGCGCAGAATAATATTAAGAATTG CTATGCTTATGATTCAGGCTGGACAGCCTGGACATTTTGAGCAGCGCGAGAGTAGAAGTGAATACCCCCAGGAACTCCAATTGTGCTGAA GCTGATTTGTCTGTTCGTAGGATGATGTGCAGCCTGTCACAGTGGCAGCCACAGGCATTCAGTTTGTGGGCATTTCCTGTCATTTCGGAT CCTGCTATAGGGAAGGTTCAGCTCTCCTGTCTGGGGCATCTACTGGAGCACCTCAGAAGACAAGGAGGAAAACACTGGCTCATGTTTTCT TTCCATGTGAGGGAGGGGAGGTTTCTAGATGATACTAAAAGCCTGTTGCCACTGTTGTGTCATGATTTCCATTCTTCATCATGGAAAGCC ACAAGCCTCAGAAGGCAGCCAGAGGACAGGAAGAGTCTGTGATGCTCATGTCTGTGTCATGGCATGAGGGGGAATTGACGAGTGCTTGCT TAGCATCAGCTCTCATTGTATTGAAGGCTGTGCCCTTGGTGTGCTTAGCAGTGAGCAGCGTCTTCATTAGGTAGCTCTGGCTGGGCTTTG TCAGCCAGCGTGTGAGAAAGGAGAACCAAAACCAATACCTCCTTATAGAAGCCAGAAGGATCACATTGGATGAGAGAAGGTGTCTATAAC TGGGTTCTTCCCAATCCCAGCTCTTTCATTAACTAAATATGATCTTGGACATATCTCTTAACCTCCATCGGCCTTAGTTTTTGCATCCAA >36253_36253_3_HGSNAT-FUT10_HGSNAT_chr8_43037403_ENST00000458501_FUT10_chr8_33230322_ENST00000518672_length(amino acids)=479AA_BP=404 MTGARASAAEQRRAGRSGQARAAERAAGMSGAGRALAALLLAASVLSAALLAPGGSSGRDAQAAPPRDLDKKRHAELKMDQALLLIHNEL LWTNLTVYWKSECCYHCLFQVLVNVPQSPKAGKPSAAAASVSTQHGSILQLNDTLEEKEVCRLEYRFGEFGNYSLLVKNIHNGVSEIACD LAVNEDPVDSNLPVSIAFLIGLAVIIVISFLRLLLSLDDFNNWISKAISSRETDRLINSELGSPSRTDPLDGDVQPATWRLSALPPRLRS VDTFRGIALILMVFVNYGGGKYWYFKHASWNGLTVADLVFPWFVFIMGSSIFLSMTSILQRGCSKFRLLGKIAWRSFLLICIGIIIVNPN YCLGPLSWDKVRIPGVLQRLGVTYFVVAVLELLFAKPVPEHCASGLPPKRWEAEDTHLSCPEPTVFAFSPLRTPPLSSLREMWISSFEQS -------------------------------------------------------------- >36253_36253_4_HGSNAT-FUT10_HGSNAT_chr8_43037403_ENST00000521576_FUT10_chr8_33230322_ENST00000518672_length(transcript)=2310nt_BP=566nt ATTTCCCAGACGGGGTGGTTGCCGGACGGAGGGGCTCCTCACTTCTCAGACGGGGCGGTTGCCAGGCAGAGGGTTTCCTCACTTCTCAGA CGGAGCGGCCGGGCAGAGACGCTCCCCACCTCCCAGACAGGGCTGCGGCCCAGCAGAGGCACTCCTCACATCCCAGACAGGGCGGCGGGG CAGAGGTGCTCCCCACATCTCAGACGATGGGCGGCCGGGCAGAGACGCTCCTCACTTCCTAGATGGGATGGCGGCGGGGAAGAGGCGCTC CTCGCTTCCCAGATGGGATGGCGGCCGGGCAGAGACGCTCCTCACTTTCCAGACTGGGCAGCCAGGCAGAGGGGCTCCTCACATCCCAGA CGATGGGTGGCCAAGCAGAGACGCTCCTCACTTCCCAGACGGGGTGGCTGCTGGGCAGAGGCTGCAATCTCGGCTCTCCGGGAGGCCAAG TGTCTTGGGACAAGGTGCGCATTCCTGGTGTGCTGCAGCGATTGGGAGTGACATACTTTGTGGTTGCTGTGTTGGAGCTCCTCTTTGCTA AACCTGTGCCTGAACATTGTGCCTCGGGCTTACCACCCAAAAGATGGGAGGCAGAAGATACCCACCTGAGTTGCCCAGAGCCCACAGTGT TTGCTTTCTCACCACTCCGGACTCCACCTTTGAGCTCTTTGCGAGAGATGTGGATTTCCAGCTTTGAACAATCCAAGAAAGAAGCCCAGG CACTAAGGTGGCTGGTTGATAGGAATCAAAACTTTTCATCTCAAGAGTTTTGGGGCCTAGTATTCAAGGACTGATTTCAAAAATGATCAG AATGAAACAGACTAGAGCCTTCTTGAGGTTTATTTTCTAGGTTGCCTTAATATTTGAACATAATAGCTATTCTGTTGACTATCCATCAGG ATAATAATTAGTTGCTGCAGTACTCATAATGAGCCCTTTCAAGGAATAGATGTAAATTACCCTTAGGGGTAGCCACTATATTTTTTATAC TAAAAAAGGAAATATTTCTTATGGACACTGGTTATCTTCCATGTCCGTGTCTGTGGACCCACATACTTACTGCCTTTTGTGGAAATTCAC CTAGAGTGAGTACTTTCCTTGTGCAATGGAGGGAGACGATCTAGCTATAGATTCTTGATACTCACCTCACTCTCAACAGCTTATTGGTAT TTGGCAAACCTGGTATCCTGCAGCCGAGCATGAACATGAAATACATTAGAACTTGGCAATGAGAGATCCCATCTGTTGGTTTCCAGGGAT GTAAGTGAGTAATAAAAGCTGTTTCAAGTTTATCCACTATCCACTCTTCAAGTGAAAAGAACTTATGTGTGTCTACCTTCTTCTCCATAC CCTGAGCCACCTCTCTCACTTTCTTTAGTTGTGCCTTGCCCTATCGCCTCCCATTTTCCCTTCAAAGTAGAATGTCTTGTAAAGACACAG TAACTCCTGTGTGTTTTGATTAGTGAGATGTTACCACTTACCCCTTCTGCTGCGCAGAATAATATTAAGAATTGCTATGCTTATGATTCA GGCTGGACAGCCTGGACATTTTGAGCAGCGCGAGAGTAGAAGTGAATACCCCCAGGAACTCCAATTGTGCTGAAGCTGATTTGTCTGTTC GTAGGATGATGTGCAGCCTGTCACAGTGGCAGCCACAGGCATTCAGTTTGTGGGCATTTCCTGTCATTTCGGATCCTGCTATAGGGAAGG TTCAGCTCTCCTGTCTGGGGCATCTACTGGAGCACCTCAGAAGACAAGGAGGAAAACACTGGCTCATGTTTTCTTTCCATGTGAGGGAGG GGAGGTTTCTAGATGATACTAAAAGCCTGTTGCCACTGTTGTGTCATGATTTCCATTCTTCATCATGGAAAGCCACAAGCCTCAGAAGGC AGCCAGAGGACAGGAAGAGTCTGTGATGCTCATGTCTGTGTCATGGCATGAGGGGGAATTGACGAGTGCTTGCTTAGCATCAGCTCTCAT TGTATTGAAGGCTGTGCCCTTGGTGTGCTTAGCAGTGAGCAGCGTCTTCATTAGGTAGCTCTGGCTGGGCTTTGTCAGCCAGCGTGTGAG AAAGGAGAACCAAAACCAATACCTCCTTATAGAAGCCAGAAGGATCACATTGGATGAGAGAAGGTGTCTATAACTGGGTTCTTCCCAATC CCAGCTCTTTCATTAACTAAATATGATCTTGGACATATCTCTTAACCTCCATCGGCCTTAGTTTTTGCATCCAAAAAGAAGTGATTGGGC >36253_36253_4_HGSNAT-FUT10_HGSNAT_chr8_43037403_ENST00000521576_FUT10_chr8_33230322_ENST00000518672_length(amino acids)=183AA_BP=108 MGWRRGRGAPRFPDGMAAGQRRSSLSRLGSQAEGLLTSQTMGGQAETLLTSQTGWLLGRGCNLGSPGGQVSWDKVRIPGVLQRLGVTYFV VAVLELLFAKPVPEHCASGLPPKRWEAEDTHLSCPEPTVFAFSPLRTPPLSSLREMWISSFEQSKKEAQALRWLVDRNQNFSSQEFWGLV -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for HGSNAT-FUT10 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for HGSNAT-FUT10 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for HGSNAT-FUT10 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
