
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:HK1-UTP23 (FusionGDB2 ID:36585) |
Fusion Gene Summary for HK1-UTP23 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: HK1-UTP23 | Fusion gene ID: 36585 | Hgene | Tgene | Gene symbol | HK1 | UTP23 | Gene ID | 255061 | 84294 |
| Gene name | tachykinin precursor 4 | UTP23 small subunit processome component | |
| Synonyms | EK|HK-1|HK1|PPT-C | C8orf53 | |
| Cytomap | 17q21.33 | 8q24.11 | |
| Type of gene | protein-coding | protein-coding | |
| Description | tachykinin-4endokininpreprotachykinin-Ctachykinin 4 (hemokinin) | rRNA-processing protein UTP23 homologUTP23, small subunit (SSU) processome component, homolog | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | . | . | |
| Ensembl transtripts involved in fusion gene | ENST00000494253, ENST00000298649, ENST00000359426, ENST00000360289, ENST00000404387, ENST00000448642, | ENST00000517820, ENST00000520733, ENST00000309822, ENST00000357148, | |
| Fusion gene scores | * DoF score | 15 X 16 X 5=1200 | 3 X 2 X 3=18 |
| # samples | 21 | 3 | |
| ** MAII score | log2(21/1200*10)=-2.51457317282976 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(3/18*10)=0.736965594166206 effective Gene in Pan-Cancer Fusion Genes (eGinPCFGs). DoF>8 and MAII>0 | |
| Context | PubMed: HK1 [Title/Abstract] AND UTP23 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | HK1(71146174)-UTP23(117861127), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | HK1-UTP23 seems lost the major protein functional domain in Hgene partner, which is a cell metabolism gene due to the frame-shifted ORF. HK1-UTP23 seems lost the major protein functional domain in Tgene partner, which is a essential gene due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | HK1 | GO:1902093 | positive regulation of flagellated sperm motility | 17437961 |
 Fusion gene breakpoints across HK1 (5'-gene) Fusion gene breakpoints across HK1 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
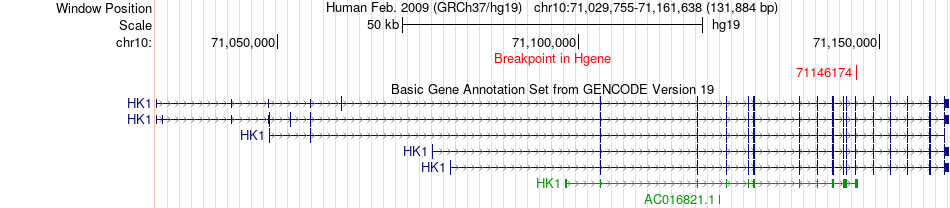 |
 Fusion gene breakpoints across UTP23 (3'-gene) Fusion gene breakpoints across UTP23 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
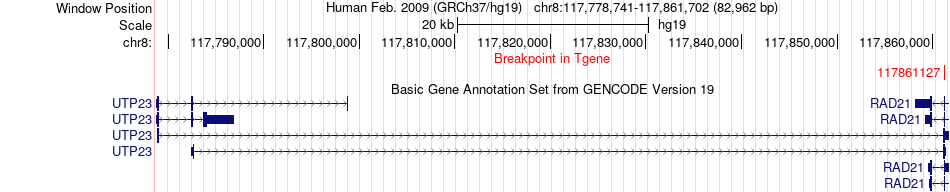 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | BRCA | TCGA-E9-A1RH-01A | HK1 | chr10 | 71146174 | + | UTP23 | chr8 | 117861127 | + |
Top |
Fusion Gene ORF analysis for HK1-UTP23 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 3UTR-3CDS | ENST00000494253 | ENST00000517820 | HK1 | chr10 | 71146174 | + | UTP23 | chr8 | 117861127 | + |
| 3UTR-3CDS | ENST00000494253 | ENST00000520733 | HK1 | chr10 | 71146174 | + | UTP23 | chr8 | 117861127 | + |
| 3UTR-intron | ENST00000494253 | ENST00000309822 | HK1 | chr10 | 71146174 | + | UTP23 | chr8 | 117861127 | + |
| 3UTR-intron | ENST00000494253 | ENST00000357148 | HK1 | chr10 | 71146174 | + | UTP23 | chr8 | 117861127 | + |
| 5CDS-intron | ENST00000298649 | ENST00000309822 | HK1 | chr10 | 71146174 | + | UTP23 | chr8 | 117861127 | + |
| 5CDS-intron | ENST00000298649 | ENST00000357148 | HK1 | chr10 | 71146174 | + | UTP23 | chr8 | 117861127 | + |
| 5CDS-intron | ENST00000359426 | ENST00000309822 | HK1 | chr10 | 71146174 | + | UTP23 | chr8 | 117861127 | + |
| 5CDS-intron | ENST00000359426 | ENST00000357148 | HK1 | chr10 | 71146174 | + | UTP23 | chr8 | 117861127 | + |
| 5CDS-intron | ENST00000360289 | ENST00000309822 | HK1 | chr10 | 71146174 | + | UTP23 | chr8 | 117861127 | + |
| 5CDS-intron | ENST00000360289 | ENST00000357148 | HK1 | chr10 | 71146174 | + | UTP23 | chr8 | 117861127 | + |
| 5CDS-intron | ENST00000404387 | ENST00000309822 | HK1 | chr10 | 71146174 | + | UTP23 | chr8 | 117861127 | + |
| 5CDS-intron | ENST00000404387 | ENST00000357148 | HK1 | chr10 | 71146174 | + | UTP23 | chr8 | 117861127 | + |
| 5CDS-intron | ENST00000448642 | ENST00000309822 | HK1 | chr10 | 71146174 | + | UTP23 | chr8 | 117861127 | + |
| 5CDS-intron | ENST00000448642 | ENST00000357148 | HK1 | chr10 | 71146174 | + | UTP23 | chr8 | 117861127 | + |
| Frame-shift | ENST00000298649 | ENST00000517820 | HK1 | chr10 | 71146174 | + | UTP23 | chr8 | 117861127 | + |
| Frame-shift | ENST00000359426 | ENST00000517820 | HK1 | chr10 | 71146174 | + | UTP23 | chr8 | 117861127 | + |
| Frame-shift | ENST00000360289 | ENST00000517820 | HK1 | chr10 | 71146174 | + | UTP23 | chr8 | 117861127 | + |
| Frame-shift | ENST00000404387 | ENST00000517820 | HK1 | chr10 | 71146174 | + | UTP23 | chr8 | 117861127 | + |
| Frame-shift | ENST00000448642 | ENST00000517820 | HK1 | chr10 | 71146174 | + | UTP23 | chr8 | 117861127 | + |
| In-frame | ENST00000298649 | ENST00000520733 | HK1 | chr10 | 71146174 | + | UTP23 | chr8 | 117861127 | + |
| In-frame | ENST00000359426 | ENST00000520733 | HK1 | chr10 | 71146174 | + | UTP23 | chr8 | 117861127 | + |
| In-frame | ENST00000360289 | ENST00000520733 | HK1 | chr10 | 71146174 | + | UTP23 | chr8 | 117861127 | + |
| In-frame | ENST00000404387 | ENST00000520733 | HK1 | chr10 | 71146174 | + | UTP23 | chr8 | 117861127 | + |
| In-frame | ENST00000448642 | ENST00000520733 | HK1 | chr10 | 71146174 | + | UTP23 | chr8 | 117861127 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000360289 | HK1 | chr10 | 71146174 | + | ENST00000520733 | UTP23 | chr8 | 117861127 | + | 2590 | 2398 | 481 | 2547 | 688 |
| ENST00000448642 | HK1 | chr10 | 71146174 | + | ENST00000520733 | UTP23 | chr8 | 117861127 | + | 2621 | 2429 | 389 | 2578 | 729 |
| ENST00000404387 | HK1 | chr10 | 71146174 | + | ENST00000520733 | UTP23 | chr8 | 117861127 | + | 2139 | 1947 | 0 | 2096 | 698 |
| ENST00000298649 | HK1 | chr10 | 71146174 | + | ENST00000520733 | UTP23 | chr8 | 117861127 | + | 2225 | 2033 | 101 | 2182 | 693 |
| ENST00000359426 | HK1 | chr10 | 71146174 | + | ENST00000520733 | UTP23 | chr8 | 117861127 | + | 2231 | 2039 | 50 | 2188 | 712 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000360289 | ENST00000520733 | HK1 | chr10 | 71146174 | + | UTP23 | chr8 | 117861127 | + | 0.002341894 | 0.99765813 |
| ENST00000448642 | ENST00000520733 | HK1 | chr10 | 71146174 | + | UTP23 | chr8 | 117861127 | + | 0.00370185 | 0.99629813 |
| ENST00000404387 | ENST00000520733 | HK1 | chr10 | 71146174 | + | UTP23 | chr8 | 117861127 | + | 0.003036913 | 0.9969631 |
| ENST00000298649 | ENST00000520733 | HK1 | chr10 | 71146174 | + | UTP23 | chr8 | 117861127 | + | 0.003060782 | 0.9969392 |
| ENST00000359426 | ENST00000520733 | HK1 | chr10 | 71146174 | + | UTP23 | chr8 | 117861127 | + | 0.00295943 | 0.99704057 |
Top |
Fusion Genomic Features for HK1-UTP23 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| HK1 | chr10 | 71146174 | + | UTP23 | chr8 | 117861126 | + | 0.000340856 | 0.9996592 |
| HK1 | chr10 | 71146174 | + | UTP23 | chr8 | 117861126 | + | 0.000340856 | 0.9996592 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
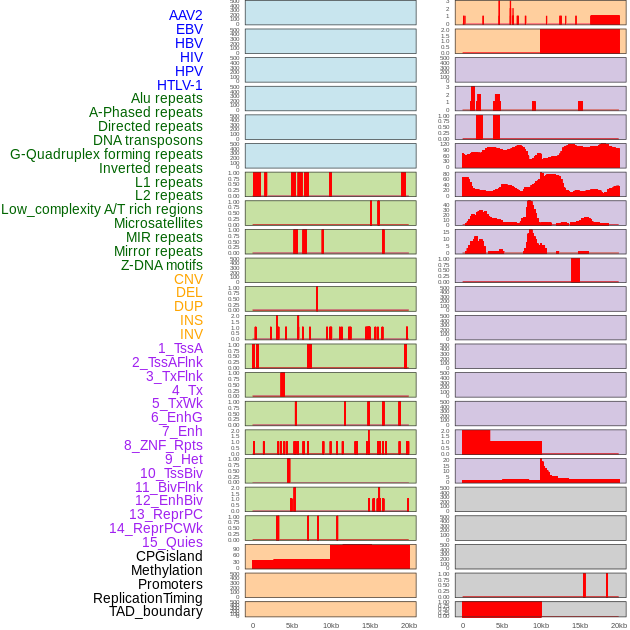 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
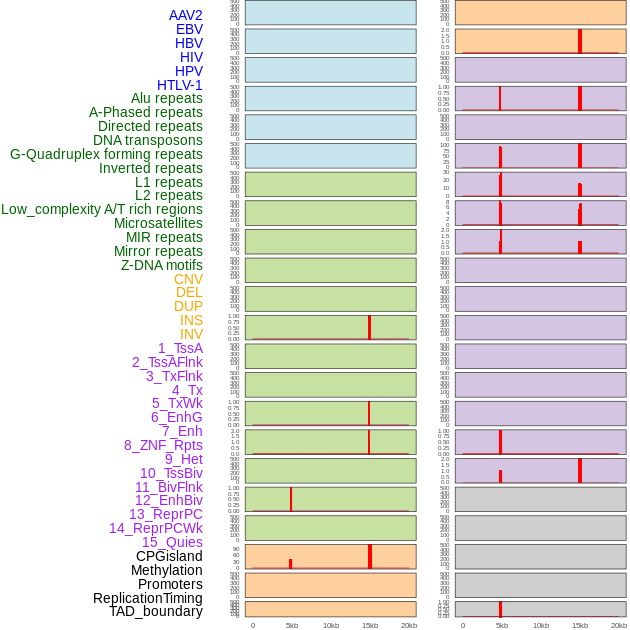 |
Top |
Fusion Protein Features for HK1-UTP23 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr10:71146174/chr8:117861127) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| . | . |
| FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. | FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | HK1 | chr10:71146174 | chr8:117861127 | ENST00000298649 | + | 13 | 18 | 16_458 | 644 | 917.0 | Domain | Hexokinase 1 |
| Hgene | HK1 | chr10:71146174 | chr8:117861127 | ENST00000359426 | + | 13 | 18 | 16_458 | 645 | 918.0 | Domain | Hexokinase 1 |
| Hgene | HK1 | chr10:71146174 | chr8:117861127 | ENST00000360289 | + | 17 | 22 | 16_458 | 633 | 906.0 | Domain | Hexokinase 1 |
| Hgene | HK1 | chr10:71146174 | chr8:117861127 | ENST00000404387 | + | 14 | 19 | 16_458 | 649 | 922.0 | Domain | Hexokinase 1 |
| Hgene | HK1 | chr10:71146174 | chr8:117861127 | ENST00000298649 | + | 13 | 18 | 425_426 | 644 | 917.0 | Nucleotide binding | ATP 1 |
| Hgene | HK1 | chr10:71146174 | chr8:117861127 | ENST00000298649 | + | 13 | 18 | 532_537 | 644 | 917.0 | Nucleotide binding | ATP 2 |
| Hgene | HK1 | chr10:71146174 | chr8:117861127 | ENST00000298649 | + | 13 | 18 | 84_89 | 644 | 917.0 | Nucleotide binding | ATP 1 |
| Hgene | HK1 | chr10:71146174 | chr8:117861127 | ENST00000359426 | + | 13 | 18 | 425_426 | 645 | 918.0 | Nucleotide binding | ATP 1 |
| Hgene | HK1 | chr10:71146174 | chr8:117861127 | ENST00000359426 | + | 13 | 18 | 532_537 | 645 | 918.0 | Nucleotide binding | ATP 2 |
| Hgene | HK1 | chr10:71146174 | chr8:117861127 | ENST00000359426 | + | 13 | 18 | 84_89 | 645 | 918.0 | Nucleotide binding | ATP 1 |
| Hgene | HK1 | chr10:71146174 | chr8:117861127 | ENST00000360289 | + | 17 | 22 | 425_426 | 633 | 906.0 | Nucleotide binding | ATP 1 |
| Hgene | HK1 | chr10:71146174 | chr8:117861127 | ENST00000360289 | + | 17 | 22 | 532_537 | 633 | 906.0 | Nucleotide binding | ATP 2 |
| Hgene | HK1 | chr10:71146174 | chr8:117861127 | ENST00000360289 | + | 17 | 22 | 84_89 | 633 | 906.0 | Nucleotide binding | ATP 1 |
| Hgene | HK1 | chr10:71146174 | chr8:117861127 | ENST00000404387 | + | 14 | 19 | 425_426 | 649 | 922.0 | Nucleotide binding | ATP 1 |
| Hgene | HK1 | chr10:71146174 | chr8:117861127 | ENST00000404387 | + | 14 | 19 | 532_537 | 649 | 922.0 | Nucleotide binding | ATP 2 |
| Hgene | HK1 | chr10:71146174 | chr8:117861127 | ENST00000404387 | + | 14 | 19 | 84_89 | 649 | 922.0 | Nucleotide binding | ATP 1 |
| Hgene | HK1 | chr10:71146174 | chr8:117861127 | ENST00000298649 | + | 13 | 18 | 172_173 | 644 | 917.0 | Region | Substrate 1 binding |
| Hgene | HK1 | chr10:71146174 | chr8:117861127 | ENST00000298649 | + | 13 | 18 | 1_10 | 644 | 917.0 | Region | Mitochondrial-binding peptide (MBP) |
| Hgene | HK1 | chr10:71146174 | chr8:117861127 | ENST00000298649 | + | 13 | 18 | 208_209 | 644 | 917.0 | Region | Substrate 1 binding |
| Hgene | HK1 | chr10:71146174 | chr8:117861127 | ENST00000298649 | + | 13 | 18 | 208_447 | 644 | 917.0 | Region | Hexokinase large subdomain 1 |
| Hgene | HK1 | chr10:71146174 | chr8:117861127 | ENST00000298649 | + | 13 | 18 | 291_294 | 644 | 917.0 | Region | Substrate 1 binding |
| Hgene | HK1 | chr10:71146174 | chr8:117861127 | ENST00000298649 | + | 13 | 18 | 413_415 | 644 | 917.0 | Region | Glucose-6-phosphate 1 binding |
| Hgene | HK1 | chr10:71146174 | chr8:117861127 | ENST00000298649 | + | 13 | 18 | 532_536 | 644 | 917.0 | Region | Glucose-6-phosphate 2 binding |
| Hgene | HK1 | chr10:71146174 | chr8:117861127 | ENST00000298649 | + | 13 | 18 | 603_604 | 644 | 917.0 | Region | Substrate 2 binding |
| Hgene | HK1 | chr10:71146174 | chr8:117861127 | ENST00000298649 | + | 13 | 18 | 620_621 | 644 | 917.0 | Region | Substrate 2 binding |
| Hgene | HK1 | chr10:71146174 | chr8:117861127 | ENST00000298649 | + | 13 | 18 | 73_207 | 644 | 917.0 | Region | Hexokinase small subdomain 1 |
| Hgene | HK1 | chr10:71146174 | chr8:117861127 | ENST00000298649 | + | 13 | 18 | 84_91 | 644 | 917.0 | Region | Glucose-6-phosphate 1 binding |
| Hgene | HK1 | chr10:71146174 | chr8:117861127 | ENST00000359426 | + | 13 | 18 | 172_173 | 645 | 918.0 | Region | Substrate 1 binding |
| Hgene | HK1 | chr10:71146174 | chr8:117861127 | ENST00000359426 | + | 13 | 18 | 1_10 | 645 | 918.0 | Region | Mitochondrial-binding peptide (MBP) |
| Hgene | HK1 | chr10:71146174 | chr8:117861127 | ENST00000359426 | + | 13 | 18 | 208_209 | 645 | 918.0 | Region | Substrate 1 binding |
| Hgene | HK1 | chr10:71146174 | chr8:117861127 | ENST00000359426 | + | 13 | 18 | 208_447 | 645 | 918.0 | Region | Hexokinase large subdomain 1 |
| Hgene | HK1 | chr10:71146174 | chr8:117861127 | ENST00000359426 | + | 13 | 18 | 291_294 | 645 | 918.0 | Region | Substrate 1 binding |
| Hgene | HK1 | chr10:71146174 | chr8:117861127 | ENST00000359426 | + | 13 | 18 | 413_415 | 645 | 918.0 | Region | Glucose-6-phosphate 1 binding |
| Hgene | HK1 | chr10:71146174 | chr8:117861127 | ENST00000359426 | + | 13 | 18 | 532_536 | 645 | 918.0 | Region | Glucose-6-phosphate 2 binding |
| Hgene | HK1 | chr10:71146174 | chr8:117861127 | ENST00000359426 | + | 13 | 18 | 603_604 | 645 | 918.0 | Region | Substrate 2 binding |
| Hgene | HK1 | chr10:71146174 | chr8:117861127 | ENST00000359426 | + | 13 | 18 | 620_621 | 645 | 918.0 | Region | Substrate 2 binding |
| Hgene | HK1 | chr10:71146174 | chr8:117861127 | ENST00000359426 | + | 13 | 18 | 73_207 | 645 | 918.0 | Region | Hexokinase small subdomain 1 |
| Hgene | HK1 | chr10:71146174 | chr8:117861127 | ENST00000359426 | + | 13 | 18 | 84_91 | 645 | 918.0 | Region | Glucose-6-phosphate 1 binding |
| Hgene | HK1 | chr10:71146174 | chr8:117861127 | ENST00000360289 | + | 17 | 22 | 172_173 | 633 | 906.0 | Region | Substrate 1 binding |
| Hgene | HK1 | chr10:71146174 | chr8:117861127 | ENST00000360289 | + | 17 | 22 | 1_10 | 633 | 906.0 | Region | Mitochondrial-binding peptide (MBP) |
| Hgene | HK1 | chr10:71146174 | chr8:117861127 | ENST00000360289 | + | 17 | 22 | 208_209 | 633 | 906.0 | Region | Substrate 1 binding |
| Hgene | HK1 | chr10:71146174 | chr8:117861127 | ENST00000360289 | + | 17 | 22 | 208_447 | 633 | 906.0 | Region | Hexokinase large subdomain 1 |
| Hgene | HK1 | chr10:71146174 | chr8:117861127 | ENST00000360289 | + | 17 | 22 | 291_294 | 633 | 906.0 | Region | Substrate 1 binding |
| Hgene | HK1 | chr10:71146174 | chr8:117861127 | ENST00000360289 | + | 17 | 22 | 413_415 | 633 | 906.0 | Region | Glucose-6-phosphate 1 binding |
| Hgene | HK1 | chr10:71146174 | chr8:117861127 | ENST00000360289 | + | 17 | 22 | 532_536 | 633 | 906.0 | Region | Glucose-6-phosphate 2 binding |
| Hgene | HK1 | chr10:71146174 | chr8:117861127 | ENST00000360289 | + | 17 | 22 | 603_604 | 633 | 906.0 | Region | Substrate 2 binding |
| Hgene | HK1 | chr10:71146174 | chr8:117861127 | ENST00000360289 | + | 17 | 22 | 620_621 | 633 | 906.0 | Region | Substrate 2 binding |
| Hgene | HK1 | chr10:71146174 | chr8:117861127 | ENST00000360289 | + | 17 | 22 | 73_207 | 633 | 906.0 | Region | Hexokinase small subdomain 1 |
| Hgene | HK1 | chr10:71146174 | chr8:117861127 | ENST00000360289 | + | 17 | 22 | 84_91 | 633 | 906.0 | Region | Glucose-6-phosphate 1 binding |
| Hgene | HK1 | chr10:71146174 | chr8:117861127 | ENST00000404387 | + | 14 | 19 | 172_173 | 649 | 922.0 | Region | Substrate 1 binding |
| Hgene | HK1 | chr10:71146174 | chr8:117861127 | ENST00000404387 | + | 14 | 19 | 1_10 | 649 | 922.0 | Region | Mitochondrial-binding peptide (MBP) |
| Hgene | HK1 | chr10:71146174 | chr8:117861127 | ENST00000404387 | + | 14 | 19 | 208_209 | 649 | 922.0 | Region | Substrate 1 binding |
| Hgene | HK1 | chr10:71146174 | chr8:117861127 | ENST00000404387 | + | 14 | 19 | 208_447 | 649 | 922.0 | Region | Hexokinase large subdomain 1 |
| Hgene | HK1 | chr10:71146174 | chr8:117861127 | ENST00000404387 | + | 14 | 19 | 291_294 | 649 | 922.0 | Region | Substrate 1 binding |
| Hgene | HK1 | chr10:71146174 | chr8:117861127 | ENST00000404387 | + | 14 | 19 | 413_415 | 649 | 922.0 | Region | Glucose-6-phosphate 1 binding |
| Hgene | HK1 | chr10:71146174 | chr8:117861127 | ENST00000404387 | + | 14 | 19 | 532_536 | 649 | 922.0 | Region | Glucose-6-phosphate 2 binding |
| Hgene | HK1 | chr10:71146174 | chr8:117861127 | ENST00000404387 | + | 14 | 19 | 603_604 | 649 | 922.0 | Region | Substrate 2 binding |
| Hgene | HK1 | chr10:71146174 | chr8:117861127 | ENST00000404387 | + | 14 | 19 | 620_621 | 649 | 922.0 | Region | Substrate 2 binding |
| Hgene | HK1 | chr10:71146174 | chr8:117861127 | ENST00000404387 | + | 14 | 19 | 73_207 | 649 | 922.0 | Region | Hexokinase small subdomain 1 |
| Hgene | HK1 | chr10:71146174 | chr8:117861127 | ENST00000404387 | + | 14 | 19 | 84_91 | 649 | 922.0 | Region | Glucose-6-phosphate 1 binding |
| Tgene | UTP23 | chr10:71146174 | chr8:117861127 | ENST00000309822 | 0 | 3 | 209_213 | 0 | 250.0 | Compositional bias | Note=Poly-Lys | |
| Tgene | UTP23 | chr10:71146174 | chr8:117861127 | ENST00000357148 | 0 | 3 | 209_213 | 0 | 146.0 | Compositional bias | Note=Poly-Lys |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | HK1 | chr10:71146174 | chr8:117861127 | ENST00000298649 | + | 13 | 18 | 464_906 | 644 | 917.0 | Domain | Hexokinase 2 |
| Hgene | HK1 | chr10:71146174 | chr8:117861127 | ENST00000359426 | + | 13 | 18 | 464_906 | 645 | 918.0 | Domain | Hexokinase 2 |
| Hgene | HK1 | chr10:71146174 | chr8:117861127 | ENST00000360289 | + | 17 | 22 | 464_906 | 633 | 906.0 | Domain | Hexokinase 2 |
| Hgene | HK1 | chr10:71146174 | chr8:117861127 | ENST00000404387 | + | 14 | 19 | 464_906 | 649 | 922.0 | Domain | Hexokinase 2 |
| Hgene | HK1 | chr10:71146174 | chr8:117861127 | ENST00000298649 | + | 13 | 18 | 747_748 | 644 | 917.0 | Nucleotide binding | ATP 2 |
| Hgene | HK1 | chr10:71146174 | chr8:117861127 | ENST00000298649 | + | 13 | 18 | 784_788 | 644 | 917.0 | Nucleotide binding | ATP 2 |
| Hgene | HK1 | chr10:71146174 | chr8:117861127 | ENST00000359426 | + | 13 | 18 | 747_748 | 645 | 918.0 | Nucleotide binding | ATP 2 |
| Hgene | HK1 | chr10:71146174 | chr8:117861127 | ENST00000359426 | + | 13 | 18 | 784_788 | 645 | 918.0 | Nucleotide binding | ATP 2 |
| Hgene | HK1 | chr10:71146174 | chr8:117861127 | ENST00000360289 | + | 17 | 22 | 747_748 | 633 | 906.0 | Nucleotide binding | ATP 2 |
| Hgene | HK1 | chr10:71146174 | chr8:117861127 | ENST00000360289 | + | 17 | 22 | 784_788 | 633 | 906.0 | Nucleotide binding | ATP 2 |
| Hgene | HK1 | chr10:71146174 | chr8:117861127 | ENST00000404387 | + | 14 | 19 | 747_748 | 649 | 922.0 | Nucleotide binding | ATP 2 |
| Hgene | HK1 | chr10:71146174 | chr8:117861127 | ENST00000404387 | + | 14 | 19 | 784_788 | 649 | 922.0 | Nucleotide binding | ATP 2 |
| Hgene | HK1 | chr10:71146174 | chr8:117861127 | ENST00000298649 | + | 13 | 18 | 521_655 | 644 | 917.0 | Region | Hexokinase small subdomain 2 |
| Hgene | HK1 | chr10:71146174 | chr8:117861127 | ENST00000298649 | + | 13 | 18 | 656_657 | 644 | 917.0 | Region | Substrate 2 binding |
| Hgene | HK1 | chr10:71146174 | chr8:117861127 | ENST00000298649 | + | 13 | 18 | 656_895 | 644 | 917.0 | Region | Hexokinase large subdomain 2 |
| Hgene | HK1 | chr10:71146174 | chr8:117861127 | ENST00000298649 | + | 13 | 18 | 682_683 | 644 | 917.0 | Region | Substrate 2 binding |
| Hgene | HK1 | chr10:71146174 | chr8:117861127 | ENST00000298649 | + | 13 | 18 | 861_863 | 644 | 917.0 | Region | Glucose-6-phosphate 2 binding |
| Hgene | HK1 | chr10:71146174 | chr8:117861127 | ENST00000359426 | + | 13 | 18 | 521_655 | 645 | 918.0 | Region | Hexokinase small subdomain 2 |
| Hgene | HK1 | chr10:71146174 | chr8:117861127 | ENST00000359426 | + | 13 | 18 | 656_657 | 645 | 918.0 | Region | Substrate 2 binding |
| Hgene | HK1 | chr10:71146174 | chr8:117861127 | ENST00000359426 | + | 13 | 18 | 656_895 | 645 | 918.0 | Region | Hexokinase large subdomain 2 |
| Hgene | HK1 | chr10:71146174 | chr8:117861127 | ENST00000359426 | + | 13 | 18 | 682_683 | 645 | 918.0 | Region | Substrate 2 binding |
| Hgene | HK1 | chr10:71146174 | chr8:117861127 | ENST00000359426 | + | 13 | 18 | 861_863 | 645 | 918.0 | Region | Glucose-6-phosphate 2 binding |
| Hgene | HK1 | chr10:71146174 | chr8:117861127 | ENST00000360289 | + | 17 | 22 | 521_655 | 633 | 906.0 | Region | Hexokinase small subdomain 2 |
| Hgene | HK1 | chr10:71146174 | chr8:117861127 | ENST00000360289 | + | 17 | 22 | 656_657 | 633 | 906.0 | Region | Substrate 2 binding |
| Hgene | HK1 | chr10:71146174 | chr8:117861127 | ENST00000360289 | + | 17 | 22 | 656_895 | 633 | 906.0 | Region | Hexokinase large subdomain 2 |
| Hgene | HK1 | chr10:71146174 | chr8:117861127 | ENST00000360289 | + | 17 | 22 | 682_683 | 633 | 906.0 | Region | Substrate 2 binding |
| Hgene | HK1 | chr10:71146174 | chr8:117861127 | ENST00000360289 | + | 17 | 22 | 861_863 | 633 | 906.0 | Region | Glucose-6-phosphate 2 binding |
| Hgene | HK1 | chr10:71146174 | chr8:117861127 | ENST00000404387 | + | 14 | 19 | 521_655 | 649 | 922.0 | Region | Hexokinase small subdomain 2 |
| Hgene | HK1 | chr10:71146174 | chr8:117861127 | ENST00000404387 | + | 14 | 19 | 656_657 | 649 | 922.0 | Region | Substrate 2 binding |
| Hgene | HK1 | chr10:71146174 | chr8:117861127 | ENST00000404387 | + | 14 | 19 | 656_895 | 649 | 922.0 | Region | Hexokinase large subdomain 2 |
| Hgene | HK1 | chr10:71146174 | chr8:117861127 | ENST00000404387 | + | 14 | 19 | 682_683 | 649 | 922.0 | Region | Substrate 2 binding |
| Hgene | HK1 | chr10:71146174 | chr8:117861127 | ENST00000404387 | + | 14 | 19 | 861_863 | 649 | 922.0 | Region | Glucose-6-phosphate 2 binding |
Top |
Fusion Gene Sequence for HK1-UTP23 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >36585_36585_1_HK1-UTP23_HK1_chr10_71146174_ENST00000298649_UTP23_chr8_117861127_ENST00000520733_length(transcript)=2225nt_BP=2033nt CCTGGCTCAAAACTCTACCACAACCTGACACTGGGCAAGATGGCCCACCTGTGGCATCCCAGGTTCCTGCCCTGTAAAGAGGAGTCTGGG TGTATATCCAGATGGACTGTGAGCACAGCCTGAGTTTGCCCTGTCGAGGTGCTGAGGCCTGGGAGATTGGGATTGACAAGTATCTCTATG CCATGCGGCTCTCCGATGAAACTCTCATAGATATCATGACTCGCTTCAGGAAGGAGATGAAGAATGGCCTCTCCCGGGATTTTAATCCAA CAGCCACAGTCAAGATGTTGCCAACATTCGTAAGGTCCATTCCTGATGGCTCTGAAAAGGGAGATTTCATTGCCCTGGATCTTGGTGGGT CTTCCTTTCGAATTCTGCGGGTGCAAGTGAATCATGAGAAAAACCAGAATGTTCACATGGAGTCCGAGGTTTATGACACCCCAGAGAACA TCGTGCACGGCAGTGGAAGCCAGCTTTTTGATCATGTTGCTGAGTGCCTGGGAGATTTCATGGAGAAAAGGAAGATCAAGGACAAGAAGT TACCTGTGGGATTCACGTTTTCTTTTCCTTGCCAACAATCCAAAATAGATGAGGCCATCCTGATCACCTGGACAAAGCGATTTAAAGCGA GCGGAGTGGAAGGAGCAGATGTGGTCAAACTGCTTAACAAAGCCATCAAAAAGCGAGGGGACTATGATGCCAACATCGTAGCTGTGGTGA ATGACACAGTGGGCACCATGATGACCTGTGGCTATGACGACCAGCACTGTGAAGTCGGCCTGATCATCGGCACTGGCACCAATGCTTGCT ACATGGAGGAACTGAGGCACATTGATCTGGTGGAAGGAGACGAGGGGAGGATGTGTATCAATACAGAATGGGGAGCCTTTGGAGACGATG GATCATTAGAAGACATCCGGACAGAGTTTGACAGGGAGATAGACCGGGGATCCCTCAACCCTGGAAAACAGCTGTTTGAGAAGATGGTCA GTGGCATGTACTTGGGAGAGCTGGTTCGACTGATCCTAGTCAAGATGGCCAAGGAGGGCCTCTTATTTGAAGGGCGGATCACCCCGGAGC TGCTCACCCGAGGGAAGTTTAACACCAGTGATGTGTCAGCCATCGAAAAGAATAAGGAAGGCCTCCACAATGCCAAAGAAATCCTGACCC GCCTGGGAGTGGAGCCGTCCGATGATGACTGTGTCTCAGTCCAGCACGTTTGCACCATTGTCTCATTTCGCTCAGCCAACTTGGTGGCTG CCACACTGGGCGCCATCTTGAACCGCCTGCGTGATAACAAGGGCACACCCAGGCTGCGGACCACGGTTGGTGTCGACGGATCTCTTTACA AGACGCACCCACAGTATTCCCGGCGTTTCCACAAGACTCTAAGGCGCTTGGTGCCAGACTCCGATGTGCGCTTCCTCCTCTCGGAGAGTG GCAGCGGCAAGGGGGCTGCCATGGTGACGGCGGTGGCCTACCGCTTGGCCGAGCAGCACCGGCAGATAGAGGAGACCCTGGCTCATTTCC ACCTCACCAAGGACATGCTGCTGGAGGTGAAGAAGAGGATGCGGGCCGAGATGGAGCTGGGGCTGAGGAAGCAGACGCACAACAATGCCG TGGTTAAGATGCTGCCCTCCTTCGTCCGGAGAACTCCCGACGGGACCGAGAATGGTGACTTCTTGGCCCTGGATCTTGGAGGAACCAATT TCCGTGTGCTGCTGGTGAAAATCCGTAGTGGGAAAAAGAGAACGGTGGAAATGCACAACAAGATCTACGCCATTCCTATTGAAATCATGC AGGGCACTGGGGAAGAGCTGTTTGATCACATTGTCTCCTGCATCTCTGACTTCTTGGACTACATGGGGATCAAAGGCCCCAGGATGCCTC TGGGCTTCACGTTCTCATTTCCCTGCCAGCAGACGAGTCTGGACGCGGGAATCTTGATCACGTGGACAAAGGGTTTTAAGGCAACAGACT GCGTGGGCCACGATGTAGTCACCTTACTAAGGGATGCGATAAAAAGGAGAGAGGGAACAATGGCAAAAGCCTTTGATGAAGCATTTTCCT CTGAGACAACAGCGGCAATACCTGAAGACCATGAAGCATCTGCTGAGTCCTTTTGTTCCATCTTCTTTCTTCCTGATCTTGATCGCCCCC >36585_36585_1_HK1-UTP23_HK1_chr10_71146174_ENST00000298649_UTP23_chr8_117861127_ENST00000520733_length(amino acids)=693AA_BP=644 MDCEHSLSLPCRGAEAWEIGIDKYLYAMRLSDETLIDIMTRFRKEMKNGLSRDFNPTATVKMLPTFVRSIPDGSEKGDFIALDLGGSSFR ILRVQVNHEKNQNVHMESEVYDTPENIVHGSGSQLFDHVAECLGDFMEKRKIKDKKLPVGFTFSFPCQQSKIDEAILITWTKRFKASGVE GADVVKLLNKAIKKRGDYDANIVAVVNDTVGTMMTCGYDDQHCEVGLIIGTGTNACYMEELRHIDLVEGDEGRMCINTEWGAFGDDGSLE DIRTEFDREIDRGSLNPGKQLFEKMVSGMYLGELVRLILVKMAKEGLLFEGRITPELLTRGKFNTSDVSAIEKNKEGLHNAKEILTRLGV EPSDDDCVSVQHVCTIVSFRSANLVAATLGAILNRLRDNKGTPRLRTTVGVDGSLYKTHPQYSRRFHKTLRRLVPDSDVRFLLSESGSGK GAAMVTAVAYRLAEQHRQIEETLAHFHLTKDMLLEVKKRMRAEMELGLRKQTHNNAVVKMLPSFVRRTPDGTENGDFLALDLGGTNFRVL LVKIRSGKKRTVEMHNKIYAIPIEIMQGTGEELFDHIVSCISDFLDYMGIKGPRMPLGFTFSFPCQQTSLDAGILITWTKGFKATDCVGH -------------------------------------------------------------- >36585_36585_2_HK1-UTP23_HK1_chr10_71146174_ENST00000359426_UTP23_chr8_117861127_ENST00000520733_length(transcript)=2231nt_BP=2039nt GAGGAGGAGGAGCCGCCGAGCAGCCGCCGGAGGACCACGGCTCGCCAGGGCTGCGGAGGACCGACCGTCCCCACGCCTGCCGCCCCGCGA CCCCGACCGCCAGCATGATCGCCGCGCAGCTCCTGGCCTATTACTTCACGGAGCTGAAGGATGACCAGGTCAAAAAGATTGACAAGTATC TCTATGCCATGCGGCTCTCCGATGAAACTCTCATAGATATCATGACTCGCTTCAGGAAGGAGATGAAGAATGGCCTCTCCCGGGATTTTA ATCCAACAGCCACAGTCAAGATGTTGCCAACATTCGTAAGGTCCATTCCTGATGGCTCTGAAAAGGGAGATTTCATTGCCCTGGATCTTG GTGGGTCTTCCTTTCGAATTCTGCGGGTGCAAGTGAATCATGAGAAAAACCAGAATGTTCACATGGAGTCCGAGGTTTATGACACCCCAG AGAACATCGTGCACGGCAGTGGAAGCCAGCTTTTTGATCATGTTGCTGAGTGCCTGGGAGATTTCATGGAGAAAAGGAAGATCAAGGACA AGAAGTTACCTGTGGGATTCACGTTTTCTTTTCCTTGCCAACAATCCAAAATAGATGAGGCCATCCTGATCACCTGGACAAAGCGATTTA AAGCGAGCGGAGTGGAAGGAGCAGATGTGGTCAAACTGCTTAACAAAGCCATCAAAAAGCGAGGGGACTATGATGCCAACATCGTAGCTG TGGTGAATGACACAGTGGGCACCATGATGACCTGTGGCTATGACGACCAGCACTGTGAAGTCGGCCTGATCATCGGCACTGGCACCAATG CTTGCTACATGGAGGAACTGAGGCACATTGATCTGGTGGAAGGAGACGAGGGGAGGATGTGTATCAATACAGAATGGGGAGCCTTTGGAG ACGATGGATCATTAGAAGACATCCGGACAGAGTTTGACAGGGAGATAGACCGGGGATCCCTCAACCCTGGAAAACAGCTGTTTGAGAAGA TGGTCAGTGGCATGTACTTGGGAGAGCTGGTTCGACTGATCCTAGTCAAGATGGCCAAGGAGGGCCTCTTATTTGAAGGGCGGATCACCC CGGAGCTGCTCACCCGAGGGAAGTTTAACACCAGTGATGTGTCAGCCATCGAAAAGAATAAGGAAGGCCTCCACAATGCCAAAGAAATCC TGACCCGCCTGGGAGTGGAGCCGTCCGATGATGACTGTGTCTCAGTCCAGCACGTTTGCACCATTGTCTCATTTCGCTCAGCCAACTTGG TGGCTGCCACACTGGGCGCCATCTTGAACCGCCTGCGTGATAACAAGGGCACACCCAGGCTGCGGACCACGGTTGGTGTCGACGGATCTC TTTACAAGACGCACCCACAGTATTCCCGGCGTTTCCACAAGACTCTAAGGCGCTTGGTGCCAGACTCCGATGTGCGCTTCCTCCTCTCGG AGAGTGGCAGCGGCAAGGGGGCTGCCATGGTGACGGCGGTGGCCTACCGCTTGGCCGAGCAGCACCGGCAGATAGAGGAGACCCTGGCTC ATTTCCACCTCACCAAGGACATGCTGCTGGAGGTGAAGAAGAGGATGCGGGCCGAGATGGAGCTGGGGCTGAGGAAGCAGACGCACAACA ATGCCGTGGTTAAGATGCTGCCCTCCTTCGTCCGGAGAACTCCCGACGGGACCGAGAATGGTGACTTCTTGGCCCTGGATCTTGGAGGAA CCAATTTCCGTGTGCTGCTGGTGAAAATCCGTAGTGGGAAAAAGAGAACGGTGGAAATGCACAACAAGATCTACGCCATTCCTATTGAAA TCATGCAGGGCACTGGGGAAGAGCTGTTTGATCACATTGTCTCCTGCATCTCTGACTTCTTGGACTACATGGGGATCAAAGGCCCCAGGA TGCCTCTGGGCTTCACGTTCTCATTTCCCTGCCAGCAGACGAGTCTGGACGCGGGAATCTTGATCACGTGGACAAAGGGTTTTAAGGCAA CAGACTGCGTGGGCCACGATGTAGTCACCTTACTAAGGGATGCGATAAAAAGGAGAGAGGGAACAATGGCAAAAGCCTTTGATGAAGCAT TTTCCTCTGAGACAACAGCGGCAATACCTGAAGACCATGAAGCATCTGCTGAGTCCTTTTGTTCCATCTTCTTTCTTCCTGATCTTGATC >36585_36585_2_HK1-UTP23_HK1_chr10_71146174_ENST00000359426_UTP23_chr8_117861127_ENST00000520733_length(amino acids)=712AA_BP=663 MRRTDRPHACRPATPTASMIAAQLLAYYFTELKDDQVKKIDKYLYAMRLSDETLIDIMTRFRKEMKNGLSRDFNPTATVKMLPTFVRSIP DGSEKGDFIALDLGGSSFRILRVQVNHEKNQNVHMESEVYDTPENIVHGSGSQLFDHVAECLGDFMEKRKIKDKKLPVGFTFSFPCQQSK IDEAILITWTKRFKASGVEGADVVKLLNKAIKKRGDYDANIVAVVNDTVGTMMTCGYDDQHCEVGLIIGTGTNACYMEELRHIDLVEGDE GRMCINTEWGAFGDDGSLEDIRTEFDREIDRGSLNPGKQLFEKMVSGMYLGELVRLILVKMAKEGLLFEGRITPELLTRGKFNTSDVSAI EKNKEGLHNAKEILTRLGVEPSDDDCVSVQHVCTIVSFRSANLVAATLGAILNRLRDNKGTPRLRTTVGVDGSLYKTHPQYSRRFHKTLR RLVPDSDVRFLLSESGSGKGAAMVTAVAYRLAEQHRQIEETLAHFHLTKDMLLEVKKRMRAEMELGLRKQTHNNAVVKMLPSFVRRTPDG TENGDFLALDLGGTNFRVLLVKIRSGKKRTVEMHNKIYAIPIEIMQGTGEELFDHIVSCISDFLDYMGIKGPRMPLGFTFSFPCQQTSLD -------------------------------------------------------------- >36585_36585_3_HK1-UTP23_HK1_chr10_71146174_ENST00000360289_UTP23_chr8_117861127_ENST00000520733_length(transcript)=2590nt_BP=2398nt AAAACATCTATCTTGCTGTGTTTGGACAGGCCAGCCCCTGAAACATCTTGGGCAATGGAGGGTTAACTTCTCAAAGTTTAATAGGCAAGA CCAGCAACCATGCAACAAGGACTTCAACTAACCAACTAAAGAACTGTTCCCCAGAGCATTGTTCCTGAGAAGGAAAAGAGTCCAAACACC TACCCACACCTGCTTTGTGCCAAGAATCCACAGTTGGATTGCAAGGACAGTGTATGTTGTCCTTTTGGAAAAATGAGAGTGAGCCCAAAT GAAGAACAAGCAAAGGCGTTCAAGACCCAGCTGTTGAGAGTAGAAAAGCAGAAGAAAGGACCCGAGGTCAGCAAGTGCCCTCCCCACAAT GGGGCAGATCTGCCAGCGAGAATCGGCTACAGCAGCTGAAAAACCAAAACTTCATCTACTTGCTGAAAGTGAGCATTGTTGATGCTCTTG AGAGCATCAGCCAGGACATTAATGTGCACCACTGTGGTGGCGTGGAAAGATGGCAAAAAGAGCCCTGCATGATTTTATTGACAAGTATCT CTATGCCATGCGGCTCTCCGATGAAACTCTCATAGATATCATGACTCGCTTCAGGAAGGAGATGAAGAATGGCCTCTCCCGGGATTTTAA TCCAACAGCCACAGTCAAGATGTTGCCAACATTCGTAAGGTCCATTCCTGATGGCTCTGAAAAGGGAGATTTCATTGCCCTGGATCTTGG TGGGTCTTCCTTTCGAATTCTGCGGGTGCAAGTGAATCATGAGAAAAACCAGAATGTTCACATGGAGTCCGAGGTTTATGACACCCCAGA GAACATCGTGCACGGCAGTGGAAGCCAGCTTTTTGATCATGTTGCTGAGTGCCTGGGAGATTTCATGGAGAAAAGGAAGATCAAGGACAA GAAGTTACCTGTGGGATTCACGTTTTCTTTTCCTTGCCAACAATCCAAAATAGATGAGGCCATCCTGATCACCTGGACAAAGCGATTTAA AGCGAGCGGAGTGGAAGGAGCAGATGTGGTCAAACTGCTTAACAAAGCCATCAAAAAGCGAGGGGACTATGATGCCAACATCGTAGCTGT GGTGAATGACACAGTGGGCACCATGATGACCTGTGGCTATGACGACCAGCACTGTGAAGTCGGCCTGATCATCGGCACTGGCACCAATGC TTGCTACATGGAGGAACTGAGGCACATTGATCTGGTGGAAGGAGACGAGGGGAGGATGTGTATCAATACAGAATGGGGAGCCTTTGGAGA CGATGGATCATTAGAAGACATCCGGACAGAGTTTGACAGGGAGATAGACCGGGGATCCCTCAACCCTGGAAAACAGCTGTTTGAGAAGAT GGTCAGTGGCATGTACTTGGGAGAGCTGGTTCGACTGATCCTAGTCAAGATGGCCAAGGAGGGCCTCTTATTTGAAGGGCGGATCACCCC GGAGCTGCTCACCCGAGGGAAGTTTAACACCAGTGATGTGTCAGCCATCGAAAAGAATAAGGAAGGCCTCCACAATGCCAAAGAAATCCT GACCCGCCTGGGAGTGGAGCCGTCCGATGATGACTGTGTCTCAGTCCAGCACGTTTGCACCATTGTCTCATTTCGCTCAGCCAACTTGGT GGCTGCCACACTGGGCGCCATCTTGAACCGCCTGCGTGATAACAAGGGCACACCCAGGCTGCGGACCACGGTTGGTGTCGACGGATCTCT TTACAAGACGCACCCACAGTATTCCCGGCGTTTCCACAAGACTCTAAGGCGCTTGGTGCCAGACTCCGATGTGCGCTTCCTCCTCTCGGA GAGTGGCAGCGGCAAGGGGGCTGCCATGGTGACGGCGGTGGCCTACCGCTTGGCCGAGCAGCACCGGCAGATAGAGGAGACCCTGGCTCA TTTCCACCTCACCAAGGACATGCTGCTGGAGGTGAAGAAGAGGATGCGGGCCGAGATGGAGCTGGGGCTGAGGAAGCAGACGCACAACAA TGCCGTGGTTAAGATGCTGCCCTCCTTCGTCCGGAGAACTCCCGACGGGACCGAGAATGGTGACTTCTTGGCCCTGGATCTTGGAGGAAC CAATTTCCGTGTGCTGCTGGTGAAAATCCGTAGTGGGAAAAAGAGAACGGTGGAAATGCACAACAAGATCTACGCCATTCCTATTGAAAT CATGCAGGGCACTGGGGAAGAGCTGTTTGATCACATTGTCTCCTGCATCTCTGACTTCTTGGACTACATGGGGATCAAAGGCCCCAGGAT GCCTCTGGGCTTCACGTTCTCATTTCCCTGCCAGCAGACGAGTCTGGACGCGGGAATCTTGATCACGTGGACAAAGGGTTTTAAGGCAAC AGACTGCGTGGGCCACGATGTAGTCACCTTACTAAGGGATGCGATAAAAAGGAGAGAGGGAACAATGGCAAAAGCCTTTGATGAAGCATT TTCCTCTGAGACAACAGCGGCAATACCTGAAGACCATGAAGCATCTGCTGAGTCCTTTTGTTCCATCTTCTTTCTTCCTGATCTTGATCG >36585_36585_3_HK1-UTP23_HK1_chr10_71146174_ENST00000360289_UTP23_chr8_117861127_ENST00000520733_length(amino acids)=688AA_BP=639 MWWRGKMAKRALHDFIDKYLYAMRLSDETLIDIMTRFRKEMKNGLSRDFNPTATVKMLPTFVRSIPDGSEKGDFIALDLGGSSFRILRVQ VNHEKNQNVHMESEVYDTPENIVHGSGSQLFDHVAECLGDFMEKRKIKDKKLPVGFTFSFPCQQSKIDEAILITWTKRFKASGVEGADVV KLLNKAIKKRGDYDANIVAVVNDTVGTMMTCGYDDQHCEVGLIIGTGTNACYMEELRHIDLVEGDEGRMCINTEWGAFGDDGSLEDIRTE FDREIDRGSLNPGKQLFEKMVSGMYLGELVRLILVKMAKEGLLFEGRITPELLTRGKFNTSDVSAIEKNKEGLHNAKEILTRLGVEPSDD DCVSVQHVCTIVSFRSANLVAATLGAILNRLRDNKGTPRLRTTVGVDGSLYKTHPQYSRRFHKTLRRLVPDSDVRFLLSESGSGKGAAMV TAVAYRLAEQHRQIEETLAHFHLTKDMLLEVKKRMRAEMELGLRKQTHNNAVVKMLPSFVRRTPDGTENGDFLALDLGGTNFRVLLVKIR SGKKRTVEMHNKIYAIPIEIMQGTGEELFDHIVSCISDFLDYMGIKGPRMPLGFTFSFPCQQTSLDAGILITWTKGFKATDCVGHDVVTL -------------------------------------------------------------- >36585_36585_4_HK1-UTP23_HK1_chr10_71146174_ENST00000404387_UTP23_chr8_117861127_ENST00000520733_length(transcript)=2139nt_BP=1947nt ATGGGGCAGATCTGCCAGCGAGAATCGGCTACAGCAGCTGAAAAACCAAAACTTCATCTACTTGCTGAAAGTGAGATTGACAAGTATCTC TATGCCATGCGGCTCTCCGATGAAACTCTCATAGATATCATGACTCGCTTCAGGAAGGAGATGAAGAATGGCCTCTCCCGGGATTTTAAT CCAACAGCCACAGTCAAGATGTTGCCAACATTCGTAAGGTCCATTCCTGATGGCTCTGAAAAGGGAGATTTCATTGCCCTGGATCTTGGT GGGTCTTCCTTTCGAATTCTGCGGGTGCAAGTGAATCATGAGAAAAACCAGAATGTTCACATGGAGTCCGAGGTTTATGACACCCCAGAG AACATCGTGCACGGCAGTGGAAGCCAGCTTTTTGATCATGTTGCTGAGTGCCTGGGAGATTTCATGGAGAAAAGGAAGATCAAGGACAAG AAGTTACCTGTGGGATTCACGTTTTCTTTTCCTTGCCAACAATCCAAAATAGATGAGGCCATCCTGATCACCTGGACAAAGCGATTTAAA GCGAGCGGAGTGGAAGGAGCAGATGTGGTCAAACTGCTTAACAAAGCCATCAAAAAGCGAGGGGACTATGATGCCAACATCGTAGCTGTG GTGAATGACACAGTGGGCACCATGATGACCTGTGGCTATGACGACCAGCACTGTGAAGTCGGCCTGATCATCGGCACTGGCACCAATGCT TGCTACATGGAGGAACTGAGGCACATTGATCTGGTGGAAGGAGACGAGGGGAGGATGTGTATCAATACAGAATGGGGAGCCTTTGGAGAC GATGGATCATTAGAAGACATCCGGACAGAGTTTGACAGGGAGATAGACCGGGGATCCCTCAACCCTGGAAAACAGCTGTTTGAGAAGATG GTCAGTGGCATGTACTTGGGAGAGCTGGTTCGACTGATCCTAGTCAAGATGGCCAAGGAGGGCCTCTTATTTGAAGGGCGGATCACCCCG GAGCTGCTCACCCGAGGGAAGTTTAACACCAGTGATGTGTCAGCCATCGAAAAGAATAAGGAAGGCCTCCACAATGCCAAAGAAATCCTG ACCCGCCTGGGAGTGGAGCCGTCCGATGATGACTGTGTCTCAGTCCAGCACGTTTGCACCATTGTCTCATTTCGCTCAGCCAACTTGGTG GCTGCCACACTGGGCGCCATCTTGAACCGCCTGCGTGATAACAAGGGCACACCCAGGCTGCGGACCACGGTTGGTGTCGACGGATCTCTT TACAAGACGCACCCACAGTATTCCCGGCGTTTCCACAAGACTCTAAGGCGCTTGGTGCCAGACTCCGATGTGCGCTTCCTCCTCTCGGAG AGTGGCAGCGGCAAGGGGGCTGCCATGGTGACGGCGGTGGCCTACCGCTTGGCCGAGCAGCACCGGCAGATAGAGGAGACCCTGGCTCAT TTCCACCTCACCAAGGACATGCTGCTGGAGGTGAAGAAGAGGATGCGGGCCGAGATGGAGCTGGGGCTGAGGAAGCAGACGCACAACAAT GCCGTGGTTAAGATGCTGCCCTCCTTCGTCCGGAGAACTCCCGACGGGACCGAGAATGGTGACTTCTTGGCCCTGGATCTTGGAGGAACC AATTTCCGTGTGCTGCTGGTGAAAATCCGTAGTGGGAAAAAGAGAACGGTGGAAATGCACAACAAGATCTACGCCATTCCTATTGAAATC ATGCAGGGCACTGGGGAAGAGCTGTTTGATCACATTGTCTCCTGCATCTCTGACTTCTTGGACTACATGGGGATCAAAGGCCCCAGGATG CCTCTGGGCTTCACGTTCTCATTTCCCTGCCAGCAGACGAGTCTGGACGCGGGAATCTTGATCACGTGGACAAAGGGTTTTAAGGCAACA GACTGCGTGGGCCACGATGTAGTCACCTTACTAAGGGATGCGATAAAAAGGAGAGAGGGAACAATGGCAAAAGCCTTTGATGAAGCATTT TCCTCTGAGACAACAGCGGCAATACCTGAAGACCATGAAGCATCTGCTGAGTCCTTTTGTTCCATCTTCTTTCTTCCTGATCTTGATCGC >36585_36585_4_HK1-UTP23_HK1_chr10_71146174_ENST00000404387_UTP23_chr8_117861127_ENST00000520733_length(amino acids)=698AA_BP=649 MGQICQRESATAAEKPKLHLLAESEIDKYLYAMRLSDETLIDIMTRFRKEMKNGLSRDFNPTATVKMLPTFVRSIPDGSEKGDFIALDLG GSSFRILRVQVNHEKNQNVHMESEVYDTPENIVHGSGSQLFDHVAECLGDFMEKRKIKDKKLPVGFTFSFPCQQSKIDEAILITWTKRFK ASGVEGADVVKLLNKAIKKRGDYDANIVAVVNDTVGTMMTCGYDDQHCEVGLIIGTGTNACYMEELRHIDLVEGDEGRMCINTEWGAFGD DGSLEDIRTEFDREIDRGSLNPGKQLFEKMVSGMYLGELVRLILVKMAKEGLLFEGRITPELLTRGKFNTSDVSAIEKNKEGLHNAKEIL TRLGVEPSDDDCVSVQHVCTIVSFRSANLVAATLGAILNRLRDNKGTPRLRTTVGVDGSLYKTHPQYSRRFHKTLRRLVPDSDVRFLLSE SGSGKGAAMVTAVAYRLAEQHRQIEETLAHFHLTKDMLLEVKKRMRAEMELGLRKQTHNNAVVKMLPSFVRRTPDGTENGDFLALDLGGT NFRVLLVKIRSGKKRTVEMHNKIYAIPIEIMQGTGEELFDHIVSCISDFLDYMGIKGPRMPLGFTFSFPCQQTSLDAGILITWTKGFKAT -------------------------------------------------------------- >36585_36585_5_HK1-UTP23_HK1_chr10_71146174_ENST00000448642_UTP23_chr8_117861127_ENST00000520733_length(transcript)=2621nt_BP=2429nt ATCTATCTTGCTGTGTTTGGACAGGCCAGCCCCTGAAACATCTTGGGCAATGGAGGGTTAACTTCTCAAAGTTTAATAGGCAAGACCAGC AACCATGCAACAAGAAGATACTCTATAAATGATGATTCAATGAATGCCAAGCCCTGTTCTATGCACTGAAGACCAAAAGAAATAAAAGAC ATCATTCCTGCTCTGACTTCAACTAACCAACTAAAGAACTGTTCCCCAGAGCATTGTTCCTGAGAAGGAAAAGAGTCCAAACACCTACCC ACACCTGCTTTGTGCCAAGAATCCACAGTTGGATTGCAAGGACAGTGCGTTCAAGACCCAGCTGTTGAGAGTAGAAAAGCAGAAGAAAGG ACCCGAGGTCAGCAAGTGCCCTCCCCACAATGGGGCAGATCTGCCAGCGAGAATCGAGACAGGGTCTTGCTATGTTGCCCAGGCTGTTCT TAAATCCCTGGCGTCAAGTGATCCTCCTGCCTCAGCCTCCTAAAGTGCTGGGATTACAGGCTACAGCAGCTGAAAAACCAAAACTTCATC TACTTGCTGAAAGTGAGATTGACAAGTATCTCTATGCCATGCGGCTCTCCGATGAAACTCTCATAGATATCATGACTCGCTTCAGGAAGG AGATGAAGAATGGCCTCTCCCGGGATTTTAATCCAACAGCCACAGTCAAGATGTTGCCAACATTCGTAAGGTCCATTCCTGATGGCTCTG AAAAGGGAGATTTCATTGCCCTGGATCTTGGTGGGTCTTCCTTTCGAATTCTGCGGGTGCAAGTGAATCATGAGAAAAACCAGAATGTTC ACATGGAGTCCGAGGTTTATGACACCCCAGAGAACATCGTGCACGGCAGTGGAAGCCAGCTTTTTGATCATGTTGCTGAGTGCCTGGGAG ATTTCATGGAGAAAAGGAAGATCAAGGACAAGAAGTTACCTGTGGGATTCACGTTTTCTTTTCCTTGCCAACAATCCAAAATAGATGAGG CCATCCTGATCACCTGGACAAAGCGATTTAAAGCGAGCGGAGTGGAAGGAGCAGATGTGGTCAAACTGCTTAACAAAGCCATCAAAAAGC GAGGGGACTATGATGCCAACATCGTAGCTGTGGTGAATGACACAGTGGGCACCATGATGACCTGTGGCTATGACGACCAGCACTGTGAAG TCGGCCTGATCATCGGCACTGGCACCAATGCTTGCTACATGGAGGAACTGAGGCACATTGATCTGGTGGAAGGAGACGAGGGGAGGATGT GTATCAATACAGAATGGGGAGCCTTTGGAGACGATGGATCATTAGAAGACATCCGGACAGAGTTTGACAGGGAGATAGACCGGGGATCCC TCAACCCTGGAAAACAGCTGTTTGAGAAGATGGTCAGTGGCATGTACTTGGGAGAGCTGGTTCGACTGATCCTAGTCAAGATGGCCAAGG AGGGCCTCTTATTTGAAGGGCGGATCACCCCGGAGCTGCTCACCCGAGGGAAGTTTAACACCAGTGATGTGTCAGCCATCGAAAAGAATA AGGAAGGCCTCCACAATGCCAAAGAAATCCTGACCCGCCTGGGAGTGGAGCCGTCCGATGATGACTGTGTCTCAGTCCAGCACGTTTGCA CCATTGTCTCATTTCGCTCAGCCAACTTGGTGGCTGCCACACTGGGCGCCATCTTGAACCGCCTGCGTGATAACAAGGGCACACCCAGGC TGCGGACCACGGTTGGTGTCGACGGATCTCTTTACAAGACGCACCCACAGTATTCCCGGCGTTTCCACAAGACTCTAAGGCGCTTGGTGC CAGACTCCGATGTGCGCTTCCTCCTCTCGGAGAGTGGCAGCGGCAAGGGGGCTGCCATGGTGACGGCGGTGGCCTACCGCTTGGCCGAGC AGCACCGGCAGATAGAGGAGACCCTGGCTCATTTCCACCTCACCAAGGACATGCTGCTGGAGGTGAAGAAGAGGATGCGGGCCGAGATGG AGCTGGGGCTGAGGAAGCAGACGCACAACAATGCCGTGGTTAAGATGCTGCCCTCCTTCGTCCGGAGAACTCCCGACGGGACCGAGAATG GTGACTTCTTGGCCCTGGATCTTGGAGGAACCAATTTCCGTGTGCTGCTGGTGAAAATCCGTAGTGGGAAAAAGAGAACGGTGGAAATGC ACAACAAGATCTACGCCATTCCTATTGAAATCATGCAGGGCACTGGGGAAGAGCTGTTTGATCACATTGTCTCCTGCATCTCTGACTTCT TGGACTACATGGGGATCAAAGGCCCCAGGATGCCTCTGGGCTTCACGTTCTCATTTCCCTGCCAGCAGACGAGTCTGGACGCGGGAATCT TGATCACGTGGACAAAGGGTTTTAAGGCAACAGACTGCGTGGGCCACGATGTAGTCACCTTACTAAGGGATGCGATAAAAAGGAGAGAGG GAACAATGGCAAAAGCCTTTGATGAAGCATTTTCCTCTGAGACAACAGCGGCAATACCTGAAGACCATGAAGCATCTGCTGAGTCCTTTT GTTCCATCTTCTTTCTTCCTGATCTTGATCGCCCCCTGATGCATCTTCATCCTGAATAAAAATGACCCCCAAAAAGCTGACAAAACAAGT >36585_36585_5_HK1-UTP23_HK1_chr10_71146174_ENST00000448642_UTP23_chr8_117861127_ENST00000520733_length(amino acids)=729AA_BP=680 MGQICQRESRQGLAMLPRLFLNPWRQVILLPQPPKVLGLQATAAEKPKLHLLAESEIDKYLYAMRLSDETLIDIMTRFRKEMKNGLSRDF NPTATVKMLPTFVRSIPDGSEKGDFIALDLGGSSFRILRVQVNHEKNQNVHMESEVYDTPENIVHGSGSQLFDHVAECLGDFMEKRKIKD KKLPVGFTFSFPCQQSKIDEAILITWTKRFKASGVEGADVVKLLNKAIKKRGDYDANIVAVVNDTVGTMMTCGYDDQHCEVGLIIGTGTN ACYMEELRHIDLVEGDEGRMCINTEWGAFGDDGSLEDIRTEFDREIDRGSLNPGKQLFEKMVSGMYLGELVRLILVKMAKEGLLFEGRIT PELLTRGKFNTSDVSAIEKNKEGLHNAKEILTRLGVEPSDDDCVSVQHVCTIVSFRSANLVAATLGAILNRLRDNKGTPRLRTTVGVDGS LYKTHPQYSRRFHKTLRRLVPDSDVRFLLSESGSGKGAAMVTAVAYRLAEQHRQIEETLAHFHLTKDMLLEVKKRMRAEMELGLRKQTHN NAVVKMLPSFVRRTPDGTENGDFLALDLGGTNFRVLLVKIRSGKKRTVEMHNKIYAIPIEIMQGTGEELFDHIVSCISDFLDYMGIKGPR MPLGFTFSFPCQQTSLDAGILITWTKGFKATDCVGHDVVTLLRDAIKRREGTMAKAFDEAFSSETTAAIPEDHEASAESFCSIFFLPDLD -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for HK1-UTP23 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for HK1-UTP23 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for HK1-UTP23 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
