
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:HLA-B-CCT5 (FusionGDB2 ID:36621) |
Fusion Gene Summary for HLA-B-CCT5 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: HLA-B-CCT5 | Fusion gene ID: 36621 | Hgene | Tgene | Gene symbol | HLA-B | CCT5 | Gene ID | 3106 | 22948 |
| Gene name | major histocompatibility complex, class I, B | chaperonin containing TCP1 subunit 5 | |
| Synonyms | AS|B-4901|HLAB | CCT-epsilon|CCTE|HEL-S-69|PNAS-102|TCP-1-epsilon | |
| Cytomap | 6p21.33 | 5p15.2 | |
| Type of gene | protein-coding | protein-coding | |
| Description | major histocompatibility complex, class I, BHLA class I antigen HLA-BHLA class I histocompatibility antigen, B alpha chainMHC HLA-B cell surface glycoproteinMHC HLA-B transmembrane glycoproteinMHC class 1 antigenMHC class I antigen HLA-B alpha chain | T-complex protein 1 subunit epsilonchaperonin containing TCP1, subunit 5 (epsilon)epididymis secretory protein Li 69 | |
| Modification date | 20200322 | 20200313 | |
| UniProtAcc | P01889 | P48643 | |
| Ensembl transtripts involved in fusion gene | ENST00000412585, ENST00000359635, ENST00000421349, ENST00000425848, ENST00000435618, ENST00000450871, | ENST00000503026, ENST00000506600, ENST00000515390, ENST00000515676, ENST00000280326, | |
| Fusion gene scores | * DoF score | 21 X 13 X 8=2184 | 58 X 14 X 17=13804 |
| # samples | 24 | 61 | |
| ** MAII score | log2(24/2184*10)=-3.18586654531133 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(61/13804*10)=-4.50013332598527 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: HLA-B [Title/Abstract] AND CCT5 [Title/Abstract] AND fusion [Title/Abstract] | ||
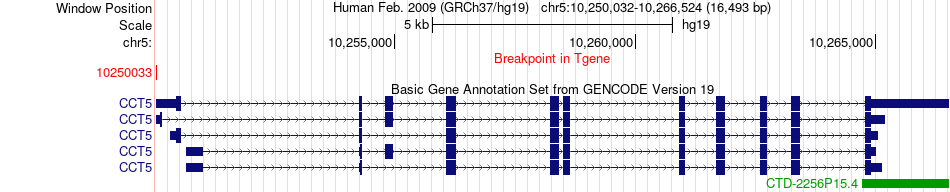
| Most frequent breakpoint | HLA-B(31321649)-CCT5(10250033), # samples:3 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | HLA-B | GO:0002486 | antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway, TAP-independent | 22031944 |
| Hgene | HLA-B | GO:0042270 | protection from natural killer cell mediated cytotoxicity | 2784569|8046333 |
 Fusion gene breakpoints across HLA-B (5'-gene) Fusion gene breakpoints across HLA-B (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
 |
 Fusion gene breakpoints across CCT5 (3'-gene) Fusion gene breakpoints across CCT5 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | CESC | TCGA-LP-A5U3-01A | HLA-B | chr6 | 31321649 | - | CCT5 | chr5 | 10250033 | + |
| ChimerDB4 | KIRC | TCGA-B0-5706-01A | HLA-B | chr6 | 31321649 | - | CCT5 | chr5 | 10250033 | + |
| ChimerDB4 | KIRC | TCGA-CZ-5455-01A | HLA-B | chr6 | 31321649 | - | CCT5 | chr5 | 10250033 | + |
Top |
Fusion Gene ORF analysis for HLA-B-CCT5 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-intron | ENST00000412585 | ENST00000503026 | HLA-B | chr6 | 31321649 | - | CCT5 | chr5 | 10250033 | + |
| 5CDS-intron | ENST00000412585 | ENST00000506600 | HLA-B | chr6 | 31321649 | - | CCT5 | chr5 | 10250033 | + |
| 5CDS-intron | ENST00000412585 | ENST00000515390 | HLA-B | chr6 | 31321649 | - | CCT5 | chr5 | 10250033 | + |
| 5CDS-intron | ENST00000412585 | ENST00000515676 | HLA-B | chr6 | 31321649 | - | CCT5 | chr5 | 10250033 | + |
| In-frame | ENST00000412585 | ENST00000280326 | HLA-B | chr6 | 31321649 | - | CCT5 | chr5 | 10250033 | + |
| intron-3CDS | ENST00000359635 | ENST00000280326 | HLA-B | chr6 | 31321649 | - | CCT5 | chr5 | 10250033 | + |
| intron-3CDS | ENST00000421349 | ENST00000280326 | HLA-B | chr6 | 31321649 | - | CCT5 | chr5 | 10250033 | + |
| intron-3CDS | ENST00000425848 | ENST00000280326 | HLA-B | chr6 | 31321649 | - | CCT5 | chr5 | 10250033 | + |
| intron-3CDS | ENST00000435618 | ENST00000280326 | HLA-B | chr6 | 31321649 | - | CCT5 | chr5 | 10250033 | + |
| intron-3CDS | ENST00000450871 | ENST00000280326 | HLA-B | chr6 | 31321649 | - | CCT5 | chr5 | 10250033 | + |
| intron-intron | ENST00000359635 | ENST00000503026 | HLA-B | chr6 | 31321649 | - | CCT5 | chr5 | 10250033 | + |
| intron-intron | ENST00000359635 | ENST00000506600 | HLA-B | chr6 | 31321649 | - | CCT5 | chr5 | 10250033 | + |
| intron-intron | ENST00000359635 | ENST00000515390 | HLA-B | chr6 | 31321649 | - | CCT5 | chr5 | 10250033 | + |
| intron-intron | ENST00000359635 | ENST00000515676 | HLA-B | chr6 | 31321649 | - | CCT5 | chr5 | 10250033 | + |
| intron-intron | ENST00000421349 | ENST00000503026 | HLA-B | chr6 | 31321649 | - | CCT5 | chr5 | 10250033 | + |
| intron-intron | ENST00000421349 | ENST00000506600 | HLA-B | chr6 | 31321649 | - | CCT5 | chr5 | 10250033 | + |
| intron-intron | ENST00000421349 | ENST00000515390 | HLA-B | chr6 | 31321649 | - | CCT5 | chr5 | 10250033 | + |
| intron-intron | ENST00000421349 | ENST00000515676 | HLA-B | chr6 | 31321649 | - | CCT5 | chr5 | 10250033 | + |
| intron-intron | ENST00000425848 | ENST00000503026 | HLA-B | chr6 | 31321649 | - | CCT5 | chr5 | 10250033 | + |
| intron-intron | ENST00000425848 | ENST00000506600 | HLA-B | chr6 | 31321649 | - | CCT5 | chr5 | 10250033 | + |
| intron-intron | ENST00000425848 | ENST00000515390 | HLA-B | chr6 | 31321649 | - | CCT5 | chr5 | 10250033 | + |
| intron-intron | ENST00000425848 | ENST00000515676 | HLA-B | chr6 | 31321649 | - | CCT5 | chr5 | 10250033 | + |
| intron-intron | ENST00000435618 | ENST00000503026 | HLA-B | chr6 | 31321649 | - | CCT5 | chr5 | 10250033 | + |
| intron-intron | ENST00000435618 | ENST00000506600 | HLA-B | chr6 | 31321649 | - | CCT5 | chr5 | 10250033 | + |
| intron-intron | ENST00000435618 | ENST00000515390 | HLA-B | chr6 | 31321649 | - | CCT5 | chr5 | 10250033 | + |
| intron-intron | ENST00000435618 | ENST00000515676 | HLA-B | chr6 | 31321649 | - | CCT5 | chr5 | 10250033 | + |
| intron-intron | ENST00000450871 | ENST00000503026 | HLA-B | chr6 | 31321649 | - | CCT5 | chr5 | 10250033 | + |
| intron-intron | ENST00000450871 | ENST00000506600 | HLA-B | chr6 | 31321649 | - | CCT5 | chr5 | 10250033 | + |
| intron-intron | ENST00000450871 | ENST00000515390 | HLA-B | chr6 | 31321649 | - | CCT5 | chr5 | 10250033 | + |
| intron-intron | ENST00000450871 | ENST00000515676 | HLA-B | chr6 | 31321649 | - | CCT5 | chr5 | 10250033 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000412585 | HLA-B | chr6 | 31321649 | - | ENST00000280326 | CCT5 | chr5 | 10250033 | + | 5222 | 1547 | 1937 | 3592 | 551 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000412585 | ENST00000280326 | HLA-B | chr6 | 31321649 | - | CCT5 | chr5 | 10250033 | + | 0.00099482 | 0.99900526 |
Top |
Fusion Genomic Features for HLA-B-CCT5 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| HLA-B | chr6 | 31321648 | - | CCT5 | chr5 | 10250032 | + | 0.00183012 | 0.9981699 |
| HLA-B | chr6 | 31321648 | - | CCT5 | chr5 | 10250032 | + | 0.00183012 | 0.9981699 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
 |
Top |
Fusion Protein Features for HLA-B-CCT5 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr6:31321649/chr5:10250033) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| HLA-B | CCT5 |
| FUNCTION: Antigen-presenting major histocompatibility complex class I (MHCI) molecule. In complex with B2M/beta 2 microglobulin displays primarily viral and tumor-derived peptides on antigen-presenting cells for recognition by alpha-beta T cell receptor (TCR) on HLA-B-restricted CD8-positive T cells, guiding antigen-specific T cell immune response to eliminate infected or transformed cells (PubMed:25808313, PubMed:29531227, PubMed:9620674, PubMed:23209413). May also present self-peptides derived from the signal sequence of secreted or membrane proteins, although T cells specific for these peptides are usually inactivated to prevent autoreactivity (PubMed:7743181, PubMed:18991276). Both the peptide and the MHC molecule are recognized by TCR, the peptide is responsible for the fine specificity of antigen recognition and MHC residues account for the MHC restriction of T cells (PubMed:29531227, PubMed:9620674, PubMed:24600035). Typically presents intracellular peptide antigens of 8 to 13 amino acids that arise from cytosolic proteolysis via constitutive proteasome and IFNG-induced immunoproteasome (PubMed:23209413). Can bind different peptides containing allele-specific binding motifs, which are mainly defined by anchor residues at position 2 and 9 (PubMed:25808313, PubMed:29531227). {ECO:0000269|PubMed:18991276, ECO:0000269|PubMed:23209413, ECO:0000269|PubMed:24600035, ECO:0000269|PubMed:25808313, ECO:0000269|PubMed:29531227, ECO:0000269|PubMed:7743181, ECO:0000269|PubMed:9620674}.; FUNCTION: Allele B*07:02: Displays peptides sharing a common signature motif, namely a Pro residue at position 2 and mainly a Leu anchor residue at the C-terminus (PubMed:7743181). Presents a long peptide (APRGPHGGAASGL) derived from the cancer-testis antigen CTAG1A/NY-ESO-1, eliciting a polyclonal CD8-positive T cell response against tumor cells (PubMed:29531227). Presents viral epitopes derived from HIV-1 gag-pol (TPQDLNTML) and Nef (RPQVPLRPM) (PubMed:25808313). Presents an immunodominant epitope derived from SARS-CoV-2 N/nucleoprotein (SPRWYFYYL) (PubMed:32887977). Displays self-peptides including a peptide derived from the signal sequence of HLA-DPB1 (APRTVALTA) (PubMed:7743181). {ECO:0000269|PubMed:25808313, ECO:0000269|PubMed:29531227, ECO:0000269|PubMed:32887977, ECO:0000269|PubMed:7743181}.; FUNCTION: Allele B*08:01: Presents to CD8-positive T cells viral epitopes derived from EBV/HHV-4 EBNA3 (QAKWRLQTL), eliciting cytotoxic T cell response. {ECO:0000269|PubMed:9620674}.; FUNCTION: Allele B*13:02: Presents multiple HIV-1 epitopes derived from gag (RQANFLGKI, GQMREPRGSDI), nef (RQDILDLWI), gag-pol (RQYDQILIE, GQGQWTYQI) and rev (LQLPPLERL), all having in common a Gln residue at position 2 and mainly hydrophobic amino acids Leu, Ile or Val at the C-terminus. Associated with succesful control of HIV-1 infection. {ECO:0000269|PubMed:17251285}.; FUNCTION: Allele B*18:01: Preferentially presents octomeric and nonameric peptides sharing a common motif, namely a Glu at position 2 and Phe or Tyr anchor residues at the C-terminus (PubMed:14978097, PubMed:23749632, PubMed:18991276). Presents an EBV/HHV-4 epitope derived from BZLF1 (SELEIKRY) (PubMed:23749632). May present to CD8-positive T cells an antigenic peptide derived from MAGEA3 (MEVDPIGHLY), triggering an anti-tumor immune response (PubMed:12366779). May display a broad repertoire of self-peptides with a preference for peptides derived from RNA-binding proteins (PubMed:14978097). {ECO:0000269|PubMed:12366779, ECO:0000269|PubMed:14978097, ECO:0000269|PubMed:18991276, ECO:0000269|PubMed:23749632}.; FUNCTION: Allele B*27:05: Presents to CD8-positive T cells immunodominant viral epitopes derived from HCV POLG (ARMILMTHF), HIV-1 gag (KRWIILGLNK), IAV NP (SRYWAIRTR), SARS-CoV-2 N/nucleoprotein (QRNAPRITF), EBV/HHV-4 EBNA4 (HRCQAIRKK) and EBV/HHV-4 EBNA6 (RRIYDLIEL), confering longterm protection against viral infection (PubMed:19139562, PubMed:18385228, PubMed:15113903, PubMed:9620674, PubMed:32887977). Can present self-peptides derived from cytosolic and nuclear proteins. All peptides carry an Arg at position 2 (PubMed:1922338). The peptide-bound form interacts with NK cell inhibitory receptor KIR3DL1 and inhibits NK cell activation in a peptide-specific way, being particularly sensitive to the nature of the amino acid side chain at position 8 of the antigenic peptide (PubMed:8879234, PubMed:15657948). KIR3DL1 fails to recognize HLA-B*27:05 in complex with B2M and EBV/HHV-4 EBNA6 (RRIYDLIEL) peptide, which can lead to increased activation of NK cells during infection (PubMed:15657948). May present an altered repertoire of peptides in the absence of TAP1-TAP2 and TAPBPL (PubMed:9620674). {ECO:0000269|PubMed:15113903, ECO:0000269|PubMed:15657948, ECO:0000269|PubMed:18385228, ECO:0000269|PubMed:19139562, ECO:0000269|PubMed:1922338, ECO:0000269|PubMed:8879234, ECO:0000269|PubMed:9620674}.; FUNCTION: Allele B*40:01: Presents immunodominant viral epitopes derived from EBV/HHV-4 LMP2 (IEDPPFNSL) and SARS-CoV-2 N/nucleoprotein (MEVTPSGTWL), triggering memory CD8-positive T cell response (PubMed:18991276, PubMed:32887977). Displays self-peptides sharing a signature motif, namely a Glu at position 2 and a Leu anchor residue at the C-terminus (PubMed:18991276). {ECO:0000269|PubMed:18991276, ECO:0000269|PubMed:32887977}.; FUNCTION: Allele B*41:01: Displays self-peptides sharing a signature motif, namely a Glu at position 2 and Ala or Pro anchor residues at the C-terminus. {ECO:0000269|PubMed:18991276}.; FUNCTION: Allele B*44:02: Presents immunodominant viral epitopes derived from EBV/HHV-4 EBNA4 (VEITPYKPTW) and EBNA6 (AEGGVGWRHW, EENLLDFVRF), triggering memory CD8-positive T cell response (PubMed:9620674, PubMed:18991276). Displays self-peptides sharing a signature motif, namely a Glu at position 2 and Phe, Tyr or Trp anchor residues at the C-terminus (PubMed:18991276). {ECO:0000269|PubMed:18991276, ECO:0000269|PubMed:9620674}.; FUNCTION: Allele B*45:01: Displays self-peptides sharing a signature motif, namely a Glu at position 2 and Ala or Pro anchor residues at the C-terminus. {ECO:0000269|PubMed:18991276}.; FUNCTION: Allele B*46:01: Preferentially presents nonameric peptides sharing a signature motif, namely Ala and Leu at position 2 and Tyr, Phe, Leu, or Met anchor residues at the C-terminus. The peptide-bound form interacts with KIR2DL3 and inhibits NK cell cytotoxic response in a peptide-specific way. {ECO:0000269|PubMed:28514659}.; FUNCTION: Allele B*47:01: Displays self-peptides sharing a signature motif, namely an Asp at position 2 and Leu or Met anchor residues at the C-terminus. {ECO:0000269|PubMed:18991276}.; FUNCTION: Allele B*49:01: Displays self-peptides sharing a signature motif, namely a Glu at position 2 and Ile or Val anchor residues at the C-terminus. {ECO:0000269|PubMed:18991276}.; FUNCTION: Allele B*50:01: Displays self-peptides sharing a signature motif, namely a Glu at position 2 and Ala or Pro anchor residues at the C-terminus. {ECO:0000269|PubMed:18991276}.; FUNCTION: Allele B*51:01: Presents an octomeric HIV-1 epitope derived from gag-pol (TAFTIPSI) to the public TRAV17/TRBV7-3 TCR clonotype, strongly suppressing HIV-1 replication. {ECO:0000269|PubMed:24600035}.; FUNCTION: Allele B*54:01: Displays peptides sharing a common signature motif, namely a Pro residue at position 2 and Ala anchor residue at the C-terminus. {ECO:0000269|PubMed:7743181}.; FUNCTION: Allele B*55:01: Displays peptides sharing a common signature motif, namely a Pro residue at position 2 and Ala anchor residue at the C-terminus. {ECO:0000269|PubMed:7743181}.; FUNCTION: Allele B*56:01: Displays peptides sharing a common signature motif, namely a Pro residue at position 2 and Ala anchor residue at the C-terminus. {ECO:0000269|PubMed:7743181}.; FUNCTION: Allele B*57:01: The peptide-bound form recognizes KIR3DL1 and inhibits NK cell cytotoxic response. {ECO:0000269|PubMed:22020283, ECO:0000269|PubMed:25480565}.; FUNCTION: Allele B*67:01: Displays peptides sharing a common signature motif, namely a Pro residue at position 2 and Leu anchor residue at the C-terminus. {ECO:0000269|PubMed:7743181}. | FUNCTION: Component of the chaperonin-containing T-complex (TRiC), a molecular chaperone complex that assists the folding of proteins upon ATP hydrolysis (PubMed:25467444). The TRiC complex mediates the folding of WRAP53/TCAB1, thereby regulating telomere maintenance (PubMed:25467444). As part of the TRiC complex may play a role in the assembly of BBSome, a complex involved in ciliogenesis regulating transports vesicles to the cilia (PubMed:20080638). The TRiC complex plays a role in the folding of actin and tubulin (Probable). {ECO:0000269|PubMed:20080638, ECO:0000269|PubMed:25467444, ECO:0000305}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
Top |
Fusion Gene Sequence for HLA-B-CCT5 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >36621_36621_1_HLA-B-CCT5_HLA-B_chr6_31321649_ENST00000412585_CCT5_chr5_10250033_ENST00000280326_length(transcript)=5222nt_BP=1547nt CCGGACTCAGAGTCTCCTCAGACGCCGAGATGCTGGTCATGGCGCCCCGAACCGTCCTCCTGCTGCTCTCGGCGGCCCTGGCCCTGACCG AGACCTGGGCCGGCTCCCACTCCATGAGGTATTTCTACACCTCCGTGTCCCGGCCCGGCCGCGGGGAGCCCCGCTTCATCTCAGTGGGCT ACGTGGACGACACCCAGTTCGTGAGGTTCGACAGCGACGCCGCGAGTCCGAGAGAGGAGCCGCGGGCGCCGTGGATAGAGCAGGAGGGGC CGGAGTATTGGGACCGGAACACACAGATCTACAAGGCCCAGGCACAGACTGACCGAGAGAGCCTGCGGAACCTGCGCGGCTACTACAACC AGAGCGAGGCCGGGTCTCACACCCTCCAGAGCATGTACGGCTGCGACGTGGGGCCGGACGGGCGCCTCCTCCGCGGGCATGACCAGTACG CCTACGACGGCAAGGATTACATCGCCCTGAACGAGGACCTGCGCTCCTGGACCGCCGCGGACACGGCGGCTCAGATCACCCAGCGCAAGT GGGAGGCGGCCCGTGAGGCGGAGCAGCGGAGAGCCTACCTGGAGGGCGAGTGCGTGGAGTGGCTCCGCAGATACCTGGAGAACGGGAAGG ACAAGCTGGAGCGCGCTGACCCCCCAAAGACACACGTGACCCACCACCCCATCTCTGACCATGAGGCCACCCTGAGGTGCTGGGCCCTGG GTTTCTACCCTGCGGAGATCACACTGACCTGGCAGCGGGATGGCGAGGACCAAACTCAGGACACTGAGCTTGTGGAGACCAGACCAGCAG GAGATAGAACCTTCCAGAAGTGGGCAGCTGTGGTGGTGCCTTCTGGAGAAGAGCAGAGATACACATGCCATGTACAGCATGAGGGGCTGC CGAAGCCCCTCACCCTGAGATGGGAGCCGTCTTCCCAGTCCACCGTCCCCATCGTGGGCATTGTTGCTGGCCTGGCTGTCCTAGCAGTTG TGGTCATCGGAGCTGTGGTCGCTGCTGTGATGTGTAGGAGGAAGAGTTCAGGTGGAAAAGGAGGGAGCTACTCTCAGGCTGCGTGCAGCG ACAGTGCCCAGGGCTCTGATGTGTCTCTCACAGCTTGAAAAGCCTGAGACAGCTGTCTTGTGAGGGACTGAGATGCAGGATTTCTTCACG CCTCCCCTTTGTGACTTCAAGAGCCTCTGGCATCTCTTTCTGCAAAGGCACCTGAATGTGTCTGCGTCCCTGTTAGCATAATGTGAGGAG GTGGAGAGACAGCCCACCCTTGTGTCCACTGTGACCCCTGTTCCCATGCTGACCTGTGTTTCCTCCCCAGTCATCTTTCTTGTTCCAGAG AGGTGGGGCTGGATGTCTCCATCTCTGTCTCAACTTTACGTGCACTGAGCTGCAACTTCTTACTTCCCTACTGAAAATAAGAATCTGAAT ATAAATTTGTTTTCTCAAATATTTGCTATGAGAGGTTGATGGATTAATTAAATAAGTCAATTCCTGGAATTTGAGAGAGCAAATAAAGAC CTGAGAACCTTCCAGAAAAAAAAAAAAAAAACCGGAAATGGGTCCTACCATCTTCTCGGAGCCGGAGTGCGAAGAAATAAAGAAATAGTG CTTTAAGTCAATGAATTCCTCCTTGGGACCCACTATCGAGAAACTATCAGTGGTAACGTTTTAAAAAATGACAAATTCAATCTGCTCTTG ACTTGTGTGTCCTAAGATTTCCACTAAGTGTCTTCAAACCTCCCCCTCCCCGGCTTCCTGGATAATAGAAGTTCCCGAAGGCCGCCGATT CCAGAAGATACTGTCTGGCGTGAAATTAGTCTCAGTAGAAACATAAGTCCCGCGCGTCTTGTGCTGCGCGTGCGCAAGCTTTTGGGCCCT CCCGAGAAAGGGAAGTGCATTCTCGCTTCCGTAGCGGTCTCCGCCGGTTGGGGGGAAGTAATTCCGGTTGTTGCACCATGGCGTCCATGG GGACCCTCGCCTTCGATGAATATGGGCGCCCTTTCCTCATCATCAAGGATCAGGACCGCAAGTCCCGTCTTATGGGACTTGAGGCCCTCA AGTCTCATATAATGGCAGCAAAGGCTGTAGCAAATACAATGAGAACATCACTTGGACCAAATGGGCTTGATAAGATGATGGTGGATAAGG ATGGAGATGTGACTGTAACTAATGATGGGGCCACCATCTTAAGCATGATGGATGTTGATCATCAGATTGCCAAGCTGATGGTGGAACTGT CCAAGTCTCAGGATGATGAAATTGGAGATGGAACCACAGGAGTGGTTGTCCTGGCTGGTGCCTTGTTAGAAGAAGCGGAGCAATTGCTAG ACCGAGGCATTCACCCAATCAGAATAGCCGATGGCTATGAGCAGGCTGCTCGTGTTGCTATTGAACACCTGGACAAGATCAGCGATAGCG TCCTTGTTGACATAAAGGACACCGAACCCCTGATTCAGACAGCAAAAACCACGCTGGGCTCCAAAGTGGTCAACAGTTGTCACCGACAGA TGGCTGAGATTGCTGTGAATGCCGTCCTCACTGTAGCAGATATGGAGCGGAGAGACGTTGACTTTGAGCTTATCAAAGTAGAAGGCAAAG TGGGCGGCAGGCTGGAGGACACTAAACTGATTAAGGGCGTGATTGTGGACAAGGATTTCAGTCACCCACAGATGCCAAAAAAAGTGGAAG ATGCGAAGATTGCAATTCTCACATGTCCATTTGAACCACCCAAACCAAAAACAAAGCATAAGCTGGATGTGACCTCTGTCGAAGATTATA AAGCCCTTCAGAAATACGAAAAGGAGAAATTTGAAGAGATGATTCAACAAATTAAAGAGACTGGTGCTAACCTAGCAATTTGTCAGTGGG GCTTTGATGATGAAGCAAATCACTTACTTCTTCAGAACAACTTGCCTGCGGTTCGCTGGGTAGGAGGACCTGAAATTGAGCTGATTGCCA TCGCAACAGGAGGGCGGATCGTCCCCAGGTTCTCAGAGCTCACAGCCGAGAAGCTGGGCTTTGCTGGTCTTGTACAGGAGATCTCATTTG GGACAACTAAGGATAAAATGCTGGTCATCGAGCAGTGTAAGAACTCCAGAGCTGTAACCATTTTTATTAGAGGAGGAAATAAGATGATCA TTGAGGAGGCGAAACGATCCCTTCACGATGCTTTGTGTGTCATCCGGAACCTCATCCGCGATAATCGTGTGGTGTATGGAGGAGGGGCTG CTGAGATATCCTGTGCCCTGGCAGTTAGCCAAGAGGCGGATAAGTGCCCCACCTTAGAACAGTATGCCATGAGAGCGTTTGCCGACGCAC TGGAGGTCATCCCCATGGCCCTCTCTGAAAACAGTGGCATGAATCCCATCCAGACTATGACCGAAGTCCGAGCCAGACAGGTGAAGGAGA TGAACCCTGCTCTTGGCATCGACTGTTTGCACAAGGGGACAAATGATATGAAGCAACAGCATGTCATAGAAACCTTGATTGGCAAAAAGC AACAGATATCTCTTGCAACACAAATGGTTAGAATGATTTTGAAGATTGATGACATTCGTAAGCCTGGAGAATCTGAAGAATGAAGACATT GAGAAAACTATGTAGCAAGATCCACTTCTGTGATTAAGTAAATGGATGTCTCGTGATGCATCTACAGTTATTTATTGTTACATCCTTTTC CAGACACTGTAGATGCTATAATAAAAATAGCTGTTTGGTAACCATAGTTTCACTTGTTCAAAGCTGTGTAATCGTGGGGGTACCATCTCA ACTGCTTTTGTATTCATTGTATTAAAAGAATCTGTTTAAACAACCTTTATCTTCTCTTCGGGTTTAAGAAACGTTTATTGTAACAGTAAT TAAATGCTGCCTTAATTGAAGGGGTTTGGGTGGATTTTTTTTTCTCAAAATAAGCTGTAGGGACTATTTTAACAGCTTAAACAGGAGCTC TCAAGATGCACTTTCATATTGAGAGGAATATGGGCTTGATCCTCTTCCTATCTAAATGGGTGGGCCATTTGATTGTAGAGGGTCCACCAC AGAATTATGGGATGCCTTAAGTGCTGTTACTAGGTTGCTCACAGCCTAACCTGGCGTGTTGTTTAGGGCTGATGGAGACCCATGTGAGCC TTTGCTTTCCTCTGGCCCCGGCCCCACCCTGAACACAGCTCATACACAGAATCAGGACCAGCATGTGCAGAGCTGGCCACCAGCACAGGC TTAGGGCAGTTCAGAACCCACTTGTTTCCCTATCAGAGGGACACAGTGAAGTGGAGGTTAAAGTAAATTACAGGAAATAAGGGAGAAATC TTGCAGTTACCATGTTCAGATAGAGTGACTGAAATTAATTGTACTTACTAAAGTATTAACTAGCTAACAGTGATGGGCCAAGACGCTCCG AGAACTCTACCGGGATTGTCTGTTCTGACAACCCAGTGAGGCAGATACACTTTCTTACTGCTCACATCTTACAGGTGAGTACTCATAATT GGCCAGCATCTCACCACCAGCAAGTAGTAGGGCCAGGTCAATCCCAGGCAGTCTGACCCCAGAGTGGCCCAGCTCATCCCCTACTCTGTT ATTTGCTTGTTAATGATTCTCTAGATTTTCTAAAATAATGTTTCTTAGCATTGTGATGATAAAGCTCATGATGAACTTTATCACTAGTTA TGCCACCTTAACTAGTCAGATTTCCTAGAATTAGGAAATGGTGACTCTTGTCTAAATTTGGTTAAGTGATGAATTTGGGTTACCGTCTCA TGTGAACCTGGAGATTCACCAGTCTTAACTTTTGGGTCATTGTGTTTTCTCTACATTCATGCATTGGATGTTTTGCTAAATAACTCCTGT GGATTTAGGAATGTGTGCTAATAGCAATCTTCCTAATTTTCATGTTTATATGGAACTATGCAGTTGAGTATTGAAAGCTTTAAACTGAGT TTATTTACAAGGACTGAGTCTAGCCTACAGAGAACATACAGCAGCCTTCTTTGGACCACAGTCTTATCCGAGGGGTCTGTGGTTGTATCA GAAGAGCCACTAAACCAATCCCCCTTTCCAAAATTGAACCTCACAGACGTTCCTGTTTTTTGTGATTGAGAAACTGGTCAATGAACAGGA AGACTTAGAGATGTTTCACAAGCCTTTGATTTTTGTTTCATTTATTTGTAATAAACCTCTTCCACGTGATGTCTTTTATTTTTCACGTTC >36621_36621_1_HLA-B-CCT5_HLA-B_chr6_31321649_ENST00000412585_CCT5_chr5_10250033_ENST00000280326_length(amino acids)=551AA_BP= MGGSNSGCCTMASMGTLAFDEYGRPFLIIKDQDRKSRLMGLEALKSHIMAAKAVANTMRTSLGPNGLDKMMVDKDGDVTVTNDGATILSM MDVDHQIAKLMVELSKSQDDEIGDGTTGVVVLAGALLEEAEQLLDRGIHPIRIADGYEQAARVAIEHLDKISDSVLVDIKDTEPLIQTAK TTLGSKVVNSCHRQMAEIAVNAVLTVADMERRDVDFELIKVEGKVGGRLEDTKLIKGVIVDKDFSHPQMPKKVEDAKIAILTCPFEPPKP KTKHKLDVTSVEDYKALQKYEKEKFEEMIQQIKETGANLAICQWGFDDEANHLLLQNNLPAVRWVGGPEIELIAIATGGRIVPRFSELTA EKLGFAGLVQEISFGTTKDKMLVIEQCKNSRAVTIFIRGGNKMIIEEAKRSLHDALCVIRNLIRDNRVVYGGGAAEISCALAVSQEADKC PTLEQYAMRAFADALEVIPMALSENSGMNPIQTMTEVRARQVKEMNPALGIDCLHKGTNDMKQQHVIETLIGKKQQISLATQMVRMILKI -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for HLA-B-CCT5 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for HLA-B-CCT5 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for HLA-B-CCT5 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
