
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:HNRNPM-EEF2K (FusionGDB2 ID:37274) |
Fusion Gene Summary for HNRNPM-EEF2K |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: HNRNPM-EEF2K | Fusion gene ID: 37274 | Hgene | Tgene | Gene symbol | HNRNPM | EEF2K | Gene ID | 4670 | 29904 |
| Gene name | heterogeneous nuclear ribonucleoprotein M | eukaryotic elongation factor 2 kinase | |
| Synonyms | CEAR|HNRNPM4|HNRPM|HNRPM4|HTGR1|NAGR1|hnRNP M | CaMKIII|HSU93850|eEF-2K | |
| Cytomap | 19p13.2 | 16p12.2 | |
| Type of gene | protein-coding | protein-coding | |
| Description | heterogeneous nuclear ribonucleoprotein MCEA receptorN-acetylglucosamine receptor 1heterogenous nuclear ribonucleoprotein M4hnRNA-binding protein M4 | eukaryotic elongation factor 2 kinasealternative protein EEF2Kcalcium/calmodulin-dependent eukaryotic elongation factor-2 kinasecalmodulin-dependent protein kinase IIIeEF-2 kinaseelongation factor-2 kinaseeukaroytic elongation factor 2 kinase | |
| Modification date | 20200322 | 20200313 | |
| UniProtAcc | P52272 | Q96G04 | |
| Ensembl transtripts involved in fusion gene | ENST00000325495, ENST00000348943, ENST00000602219, | ENST00000561791, ENST00000263026, | |
| Fusion gene scores | * DoF score | 16 X 12 X 10=1920 | 3 X 3 X 3=27 |
| # samples | 19 | 3 | |
| ** MAII score | log2(19/1920*10)=-3.33703498727757 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(3/27*10)=0.15200309344505 effective Gene in Pan-Cancer Fusion Genes (eGinPCFGs). DoF>8 and MAII>0 | |
| Context | PubMed: HNRNPM [Title/Abstract] AND EEF2K [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | HNRNPM(8532468)-EEF2K(22284947), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Tgene | EEF2K | GO:0046777 | protein autophosphorylation | 9144159|23184662 |
 Fusion gene breakpoints across HNRNPM (5'-gene) Fusion gene breakpoints across HNRNPM (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
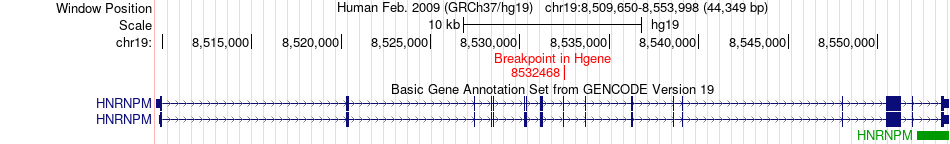 |
 Fusion gene breakpoints across EEF2K (3'-gene) Fusion gene breakpoints across EEF2K (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
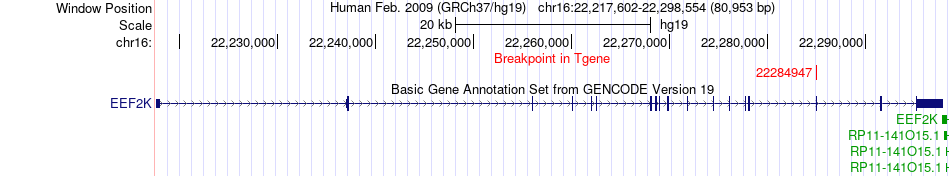 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | STAD | TCGA-CD-8531-01A | HNRNPM | chr19 | 8532468 | + | EEF2K | chr16 | 22284947 | + |
Top |
Fusion Gene ORF analysis for HNRNPM-EEF2K |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-intron | ENST00000325495 | ENST00000561791 | HNRNPM | chr19 | 8532468 | + | EEF2K | chr16 | 22284947 | + |
| 5CDS-intron | ENST00000348943 | ENST00000561791 | HNRNPM | chr19 | 8532468 | + | EEF2K | chr16 | 22284947 | + |
| In-frame | ENST00000325495 | ENST00000263026 | HNRNPM | chr19 | 8532468 | + | EEF2K | chr16 | 22284947 | + |
| In-frame | ENST00000348943 | ENST00000263026 | HNRNPM | chr19 | 8532468 | + | EEF2K | chr16 | 22284947 | + |
| intron-3CDS | ENST00000602219 | ENST00000263026 | HNRNPM | chr19 | 8532468 | + | EEF2K | chr16 | 22284947 | + |
| intron-intron | ENST00000602219 | ENST00000561791 | HNRNPM | chr19 | 8532468 | + | EEF2K | chr16 | 22284947 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000348943 | HNRNPM | chr19 | 8532468 | + | ENST00000263026 | EEF2K | chr16 | 22284947 | + | 4000 | 949 | 151 | 1362 | 403 |
| ENST00000325495 | HNRNPM | chr19 | 8532468 | + | ENST00000263026 | EEF2K | chr16 | 22284947 | + | 3926 | 875 | 41 | 1288 | 415 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000348943 | ENST00000263026 | HNRNPM | chr19 | 8532468 | + | EEF2K | chr16 | 22284947 | + | 0.000306509 | 0.9996935 |
| ENST00000325495 | ENST00000263026 | HNRNPM | chr19 | 8532468 | + | EEF2K | chr16 | 22284947 | + | 0.000357985 | 0.999642 |
Top |
Fusion Genomic Features for HNRNPM-EEF2K |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| HNRNPM | chr19 | 8532468 | + | EEF2K | chr16 | 22284946 | + | 1.69E-09 | 1 |
| HNRNPM | chr19 | 8532468 | + | EEF2K | chr16 | 22284946 | + | 1.69E-09 | 1 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
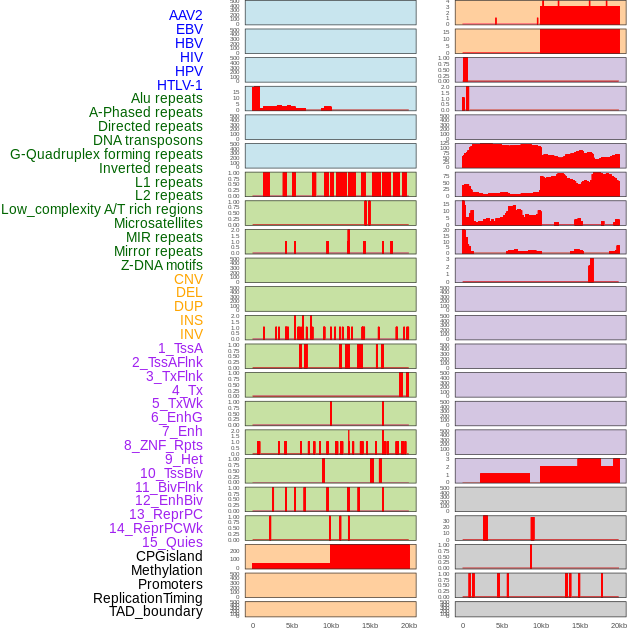 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
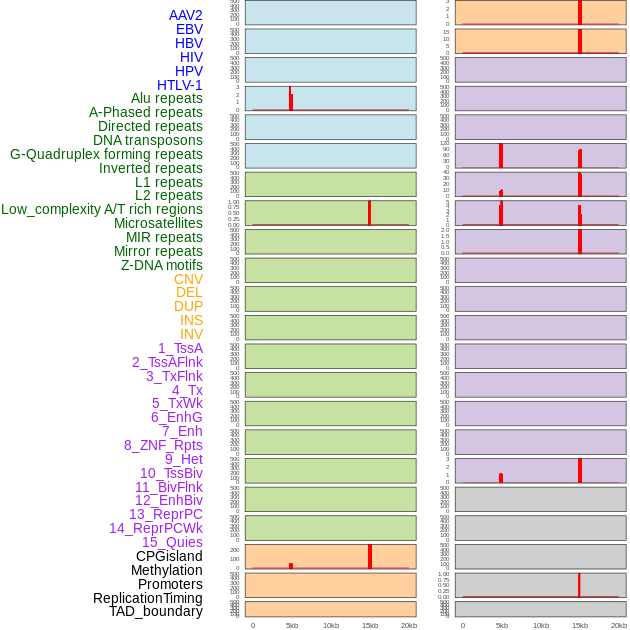 |
Top |
Fusion Protein Features for HNRNPM-EEF2K |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr19:8532468/chr16:22284947) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| HNRNPM | EEF2K |
| FUNCTION: Pre-mRNA binding protein in vivo, binds avidly to poly(G) and poly(U) RNA homopolymers in vitro. Involved in splicing. Acts as a receptor for carcinoembryonic antigen in Kupffer cells, may initiate a series of signaling events leading to tyrosine phosphorylation of proteins and induction of IL-1 alpha, IL-6, IL-10 and tumor necrosis factor alpha cytokines. | FUNCTION: Catalyzes the trimethylation of eukaryotic elongation factor 2 (EEF2) on 'Lys-525'. {ECO:0000269|PubMed:25231979}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | HNRNPM | chr19:8532468 | chr16:22284947 | ENST00000325495 | + | 8 | 16 | 204_281 | 278 | 731.0 | Domain | RRM 2 |
| Hgene | HNRNPM | chr19:8532468 | chr16:22284947 | ENST00000325495 | + | 8 | 16 | 71_149 | 278 | 731.0 | Domain | RRM 1 |
| Hgene | HNRNPM | chr19:8532468 | chr16:22284947 | ENST00000348943 | + | 9 | 17 | 71_149 | 239 | 692.0 | Domain | RRM 1 |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | HNRNPM | chr19:8532468 | chr16:22284947 | ENST00000325495 | + | 8 | 16 | 390_396 | 278 | 731.0 | Compositional bias | Note=Poly-Gly |
| Hgene | HNRNPM | chr19:8532468 | chr16:22284947 | ENST00000325495 | + | 8 | 16 | 612_616 | 278 | 731.0 | Compositional bias | Note=Poly-Gly |
| Hgene | HNRNPM | chr19:8532468 | chr16:22284947 | ENST00000348943 | + | 9 | 17 | 390_396 | 239 | 692.0 | Compositional bias | Note=Poly-Gly |
| Hgene | HNRNPM | chr19:8532468 | chr16:22284947 | ENST00000348943 | + | 9 | 17 | 612_616 | 239 | 692.0 | Compositional bias | Note=Poly-Gly |
| Hgene | HNRNPM | chr19:8532468 | chr16:22284947 | ENST00000325495 | + | 8 | 16 | 653_729 | 278 | 731.0 | Domain | RRM 3 |
| Hgene | HNRNPM | chr19:8532468 | chr16:22284947 | ENST00000348943 | + | 9 | 17 | 204_281 | 239 | 692.0 | Domain | RRM 2 |
| Hgene | HNRNPM | chr19:8532468 | chr16:22284947 | ENST00000348943 | + | 9 | 17 | 653_729 | 239 | 692.0 | Domain | RRM 3 |
| Hgene | HNRNPM | chr19:8532468 | chr16:22284947 | ENST00000325495 | + | 8 | 16 | 400_608 | 278 | 731.0 | Region | Note=27 X 6 AA repeats of [GEVSTPAN]-[ILMV]-[DE]-[RH]-[MLVI]-[GAV] |
| Hgene | HNRNPM | chr19:8532468 | chr16:22284947 | ENST00000348943 | + | 9 | 17 | 400_608 | 239 | 692.0 | Region | Note=27 X 6 AA repeats of [GEVSTPAN]-[ILMV]-[DE]-[RH]-[MLVI]-[GAV] |
| Hgene | HNRNPM | chr19:8532468 | chr16:22284947 | ENST00000325495 | + | 8 | 16 | 400_405 | 278 | 731.0 | Repeat | Note=1 |
| Hgene | HNRNPM | chr19:8532468 | chr16:22284947 | ENST00000325495 | + | 8 | 16 | 407_412 | 278 | 731.0 | Repeat | Note=2 |
| Hgene | HNRNPM | chr19:8532468 | chr16:22284947 | ENST00000325495 | + | 8 | 16 | 415_420 | 278 | 731.0 | Repeat | Note=3 |
| Hgene | HNRNPM | chr19:8532468 | chr16:22284947 | ENST00000325495 | + | 8 | 16 | 426_431 | 278 | 731.0 | Repeat | Note=4 |
| Hgene | HNRNPM | chr19:8532468 | chr16:22284947 | ENST00000325495 | + | 8 | 16 | 433_438 | 278 | 731.0 | Repeat | Note=5 |
| Hgene | HNRNPM | chr19:8532468 | chr16:22284947 | ENST00000325495 | + | 8 | 16 | 440_445 | 278 | 731.0 | Repeat | Note=6 |
| Hgene | HNRNPM | chr19:8532468 | chr16:22284947 | ENST00000325495 | + | 8 | 16 | 446_451 | 278 | 731.0 | Repeat | Note=7 |
| Hgene | HNRNPM | chr19:8532468 | chr16:22284947 | ENST00000325495 | + | 8 | 16 | 453_458 | 278 | 731.0 | Repeat | Note=8 |
| Hgene | HNRNPM | chr19:8532468 | chr16:22284947 | ENST00000325495 | + | 8 | 16 | 461_466 | 278 | 731.0 | Repeat | Note=9 |
| Hgene | HNRNPM | chr19:8532468 | chr16:22284947 | ENST00000325495 | + | 8 | 16 | 468_473 | 278 | 731.0 | Repeat | Note=10 |
| Hgene | HNRNPM | chr19:8532468 | chr16:22284947 | ENST00000325495 | + | 8 | 16 | 475_480 | 278 | 731.0 | Repeat | Note=11 |
| Hgene | HNRNPM | chr19:8532468 | chr16:22284947 | ENST00000325495 | + | 8 | 16 | 482_487 | 278 | 731.0 | Repeat | Note=12 |
| Hgene | HNRNPM | chr19:8532468 | chr16:22284947 | ENST00000325495 | + | 8 | 16 | 493_498 | 278 | 731.0 | Repeat | Note=13 |
| Hgene | HNRNPM | chr19:8532468 | chr16:22284947 | ENST00000325495 | + | 8 | 16 | 500_505 | 278 | 731.0 | Repeat | Note=14 |
| Hgene | HNRNPM | chr19:8532468 | chr16:22284947 | ENST00000325495 | + | 8 | 16 | 507_512 | 278 | 731.0 | Repeat | Note=15 |
| Hgene | HNRNPM | chr19:8532468 | chr16:22284947 | ENST00000325495 | + | 8 | 16 | 514_519 | 278 | 731.0 | Repeat | Note=16 |
| Hgene | HNRNPM | chr19:8532468 | chr16:22284947 | ENST00000325495 | + | 8 | 16 | 521_526 | 278 | 731.0 | Repeat | Note=17 |
| Hgene | HNRNPM | chr19:8532468 | chr16:22284947 | ENST00000325495 | + | 8 | 16 | 528_533 | 278 | 731.0 | Repeat | Note=18 |
| Hgene | HNRNPM | chr19:8532468 | chr16:22284947 | ENST00000325495 | + | 8 | 16 | 540_545 | 278 | 731.0 | Repeat | Note=19 |
| Hgene | HNRNPM | chr19:8532468 | chr16:22284947 | ENST00000325495 | + | 8 | 16 | 547_552 | 278 | 731.0 | Repeat | Note=20 |
| Hgene | HNRNPM | chr19:8532468 | chr16:22284947 | ENST00000325495 | + | 8 | 16 | 554_559 | 278 | 731.0 | Repeat | Note=21 |
| Hgene | HNRNPM | chr19:8532468 | chr16:22284947 | ENST00000325495 | + | 8 | 16 | 562_566 | 278 | 731.0 | Repeat | Note=22 |
| Hgene | HNRNPM | chr19:8532468 | chr16:22284947 | ENST00000325495 | + | 8 | 16 | 567_572 | 278 | 731.0 | Repeat | Note=23 |
| Hgene | HNRNPM | chr19:8532468 | chr16:22284947 | ENST00000325495 | + | 8 | 16 | 575_579 | 278 | 731.0 | Repeat | Note=24 |
| Hgene | HNRNPM | chr19:8532468 | chr16:22284947 | ENST00000325495 | + | 8 | 16 | 580_585 | 278 | 731.0 | Repeat | Note=25 |
| Hgene | HNRNPM | chr19:8532468 | chr16:22284947 | ENST00000325495 | + | 8 | 16 | 588_593 | 278 | 731.0 | Repeat | Note=26 |
| Hgene | HNRNPM | chr19:8532468 | chr16:22284947 | ENST00000325495 | + | 8 | 16 | 603_608 | 278 | 731.0 | Repeat | Note=27 |
| Hgene | HNRNPM | chr19:8532468 | chr16:22284947 | ENST00000348943 | + | 9 | 17 | 400_405 | 239 | 692.0 | Repeat | Note=1 |
| Hgene | HNRNPM | chr19:8532468 | chr16:22284947 | ENST00000348943 | + | 9 | 17 | 407_412 | 239 | 692.0 | Repeat | Note=2 |
| Hgene | HNRNPM | chr19:8532468 | chr16:22284947 | ENST00000348943 | + | 9 | 17 | 415_420 | 239 | 692.0 | Repeat | Note=3 |
| Hgene | HNRNPM | chr19:8532468 | chr16:22284947 | ENST00000348943 | + | 9 | 17 | 426_431 | 239 | 692.0 | Repeat | Note=4 |
| Hgene | HNRNPM | chr19:8532468 | chr16:22284947 | ENST00000348943 | + | 9 | 17 | 433_438 | 239 | 692.0 | Repeat | Note=5 |
| Hgene | HNRNPM | chr19:8532468 | chr16:22284947 | ENST00000348943 | + | 9 | 17 | 440_445 | 239 | 692.0 | Repeat | Note=6 |
| Hgene | HNRNPM | chr19:8532468 | chr16:22284947 | ENST00000348943 | + | 9 | 17 | 446_451 | 239 | 692.0 | Repeat | Note=7 |
| Hgene | HNRNPM | chr19:8532468 | chr16:22284947 | ENST00000348943 | + | 9 | 17 | 453_458 | 239 | 692.0 | Repeat | Note=8 |
| Hgene | HNRNPM | chr19:8532468 | chr16:22284947 | ENST00000348943 | + | 9 | 17 | 461_466 | 239 | 692.0 | Repeat | Note=9 |
| Hgene | HNRNPM | chr19:8532468 | chr16:22284947 | ENST00000348943 | + | 9 | 17 | 468_473 | 239 | 692.0 | Repeat | Note=10 |
| Hgene | HNRNPM | chr19:8532468 | chr16:22284947 | ENST00000348943 | + | 9 | 17 | 475_480 | 239 | 692.0 | Repeat | Note=11 |
| Hgene | HNRNPM | chr19:8532468 | chr16:22284947 | ENST00000348943 | + | 9 | 17 | 482_487 | 239 | 692.0 | Repeat | Note=12 |
| Hgene | HNRNPM | chr19:8532468 | chr16:22284947 | ENST00000348943 | + | 9 | 17 | 493_498 | 239 | 692.0 | Repeat | Note=13 |
| Hgene | HNRNPM | chr19:8532468 | chr16:22284947 | ENST00000348943 | + | 9 | 17 | 500_505 | 239 | 692.0 | Repeat | Note=14 |
| Hgene | HNRNPM | chr19:8532468 | chr16:22284947 | ENST00000348943 | + | 9 | 17 | 507_512 | 239 | 692.0 | Repeat | Note=15 |
| Hgene | HNRNPM | chr19:8532468 | chr16:22284947 | ENST00000348943 | + | 9 | 17 | 514_519 | 239 | 692.0 | Repeat | Note=16 |
| Hgene | HNRNPM | chr19:8532468 | chr16:22284947 | ENST00000348943 | + | 9 | 17 | 521_526 | 239 | 692.0 | Repeat | Note=17 |
| Hgene | HNRNPM | chr19:8532468 | chr16:22284947 | ENST00000348943 | + | 9 | 17 | 528_533 | 239 | 692.0 | Repeat | Note=18 |
| Hgene | HNRNPM | chr19:8532468 | chr16:22284947 | ENST00000348943 | + | 9 | 17 | 540_545 | 239 | 692.0 | Repeat | Note=19 |
| Hgene | HNRNPM | chr19:8532468 | chr16:22284947 | ENST00000348943 | + | 9 | 17 | 547_552 | 239 | 692.0 | Repeat | Note=20 |
| Hgene | HNRNPM | chr19:8532468 | chr16:22284947 | ENST00000348943 | + | 9 | 17 | 554_559 | 239 | 692.0 | Repeat | Note=21 |
| Hgene | HNRNPM | chr19:8532468 | chr16:22284947 | ENST00000348943 | + | 9 | 17 | 562_566 | 239 | 692.0 | Repeat | Note=22 |
| Hgene | HNRNPM | chr19:8532468 | chr16:22284947 | ENST00000348943 | + | 9 | 17 | 567_572 | 239 | 692.0 | Repeat | Note=23 |
| Hgene | HNRNPM | chr19:8532468 | chr16:22284947 | ENST00000348943 | + | 9 | 17 | 575_579 | 239 | 692.0 | Repeat | Note=24 |
| Hgene | HNRNPM | chr19:8532468 | chr16:22284947 | ENST00000348943 | + | 9 | 17 | 580_585 | 239 | 692.0 | Repeat | Note=25 |
| Hgene | HNRNPM | chr19:8532468 | chr16:22284947 | ENST00000348943 | + | 9 | 17 | 588_593 | 239 | 692.0 | Repeat | Note=26 |
| Hgene | HNRNPM | chr19:8532468 | chr16:22284947 | ENST00000348943 | + | 9 | 17 | 603_608 | 239 | 692.0 | Repeat | Note=27 |
| Tgene | EEF2K | chr19:8532468 | chr16:22284947 | ENST00000263026 | 14 | 18 | 116_326 | 588 | 726.0 | Domain | Alpha-type protein kinase | |
| Tgene | EEF2K | chr19:8532468 | chr16:22284947 | ENST00000263026 | 14 | 18 | 296_302 | 588 | 726.0 | Nucleotide binding | ATP | |
| Tgene | EEF2K | chr19:8532468 | chr16:22284947 | ENST00000263026 | 14 | 18 | 81_94 | 588 | 726.0 | Region | Calmodulin-binding |
Top |
Fusion Gene Sequence for HNRNPM-EEF2K |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >37274_37274_1_HNRNPM-EEF2K_HNRNPM_chr19_8532468_ENST00000325495_EEF2K_chr16_22284947_ENST00000263026_length(transcript)=3926nt_BP=875nt GTGCAGCCCGTTCGCTCACACAAAGCCCAGACGCGGAGAAAATGGCGGCAGGGGTCGAAGCGGCGGCGGAGGTGGCGGCGACGGAGATCA AAATGGAGGAAGAGAGCGGCGCGCCCGGCGTGCCGAGCGGCAACGGGGCTCCGGGCCCTAAGGGTGAAGGAGAACGACCTGCTCAGAATG AGAAGAGGAAGGAGAAAAACATAAAAAGAGGAGGCAATCGCTTTGAGCCATATGCCAATCCAACTAAAAGATACAGAGCCTTCATTACAA ACATACCTTTTGATGTGAAATGGCAGTCACTTAAAGACCTGGTTAAAGAAAAAGTTGGTGAGGTAACATACGTGGAGCTCTTAATGGACG CTGAAGGAAAGTCAAGGGGATGTGCTGTTGTTGAATTCAAGATGGAAGAGAGCATGAAAAAAGCTGCGGAAGTCCTAAACAAGCATAGTC TGAGCGGAAGACCACTGAAAGTCAAAGAAGATCCTGATGGTGAACATGCCAGGAGAGCAATGCAAAAGGTGATGGCTACGACTGGTGGGA TGGGTATGGGACCAGGTGGCCCAGGAATGATTACTATCCCACCCAGTATCCTAAATAATCCCAACATCCCAAATGAGATTATCCATGCAT TACAGGCTGGAAGACTTGGAAGCACAGTATTTGTAGCAAATCTGGATTATAAAGTTGGCTGGAAGAAACTGAAGGAAGTATTTAGTATGG CTGGTGTGGTGGTCCGAGCAGACATTCTTGAAGATAAAGATGGAAAAAGTCGTGGAATAGGCACTGTTACTTTTGAACAGTCCATTGAAG CTGTGCAAGCTATATCTATGTTCAATGGCCAGCTGCTATTTGATAGACCAATGCACGTCAAGATGGAGACAGAAGAGAACAAAACCAAAG GATTTGATTACTTACTAAAGGCCGCTGAAGCTGGCGACAGGCAGTCCATGATCCTAGTGGCGCGAGCTTTTGACTCTGGCCAGAACCTCA GCCCGGACAGGTGCCAAGACTGGCTAGAGGCCCTGCACTGGTACAACACTGCCCTGGAGATGACGGACTGTGATGAGGGCGGTGAGTACG ACGGAATGCAGGACGAGCCCCGGTACATGATGCTGGCCAGGGAGGCCGAGATGCTGTTCACAGGAGGCTACGGGCTGGAGAAGGACCCGC AGAGATCAGGGGACTTGTATACCCAGGCAGCAGAGGCAGCGATGGAAGCCATGAAGGGCCGACTGGCCAACCAGTACTACCAAAAGGCTG AAGAGGCCTGGGCCCAGATGGAGGAGTAACCAGGAAAATCACTGCCGGCTAGTCCCAAGCAAACGGGCTAGGAGGAAAGATTAAAAAAAC AACAACAACAACTTATTTAGTTTGGGGAGGGGAAGCATTTTTAAGTGTGTTGTAAAATCAAATTTTATATTTCATTTTTTGACTCTTGAA AAATGTCTTTGCTCCTTGGCAGCTACCAGCAGAGACTCTATAGCTGTCTCTTAGGGCAGTATTTTGGGGAAGTGGGGCTTGAAGAAGCAG CCTAATGAACCAACATACCGTTTTGTGTGTGGTTTTTTTTGTTTGTTTGTTTGTTTGTTTTGAGACAGAGTCTTGCTCTGTCACCCAGGC TGGAGTGCAGTGACATGATCTTAGCTCACTGCAACCTCCGCCTCCTGGGTTCAAGTGATTCTCCTGCCTCAGCCTCCCAAGTAGCTGGGA TTACTGGTGCACACCACCACACTCAGCTAATTTTTGCATTTTTAGTAGAGATGGGGTTTCACCATGTTGGCCAGGCTGGTCTCGAACTCC TAACCTCAGGTGATCCACCTGCCTCAGCCTCCCAAAGTGCTGGGATTACAGGTGTGAGCCACCATGCCTGCCCATTTTGTGGTTCTATTT TCATTTTTATTTCTTTTTTTTTTTTTGTCACGAGATATAAGAAAGTGCTTTTTGCCTTGAATGGACAATTTTAGGGCTGTGCTCACTAGT CTTTTCAGGCTGGACTGAAATGTCGGGCCCATGGAGCCCTGTGTTTTGTGCATCGGGATGAGAAATGAAGCACTTCACGCTGGCTTTCCT AAGTCACGGGGCGTGTATTGCCGTGGCTTAGTGCAAAGCATTCTTTCTCAGAGCATTTAGAGGCATGCGTGGCATTTTTTCAGTGGGTGT GAGATTGCACAATACCCAGGCTCCCTTCTACTGTGGGGAAGGGCCTGCATGTTGGCTGTTTTTTAAACTTCTAGTTCAATTTCCTTCCAT AATGCTACTGATTTTCTGGCATACAGCCGAATTCCATCTTTTAAGCATGCTTTCTACGGTGGGCTTTTCAAAACAGGTTTGAGTTTTGTA TGCACACGTTTACTACCTCTAACTCCTACATCAGCTAGTGTGGAAGAGGGTGCACCTCAAAGCTTTTACACGTAAGGACAGCGGCTTGGA ATGTGAGAGCCTTTTCTCCAAGCAGACCCACACTCTGCATCTCAGTGGCAGCTCCCACAACGTGACTGCAATGTCTCTTATACAGTATTC CTTGGTGTTTTCTTAGTGTCTGGATGTTCTTACGTGAAATCTGCTCCCCAGCCCTGGTCCTTGGCATTTTCTGCTTGAAGCTGGGCTGAT TTTCTTGTAATTTACAGCAGGACGCTTTCAGCAGCAGTCTCTTGGGATTTTATCTAAGATGTTTGAGGATGAGAGGGCAAGAACTATAAA CACTCATTAATTCTAGTAGTCTCCCCATGGCCAGACAATGGCGATTGTTATTTAATGAGCTTTTCCTTTCAATGGAATTCAGCTCTCACA TTAGTATGATTTCATTTGATGTTTCAAATAGCAAAGATGCTAGGTGCGGTGGCTCCCGCCTGTAATCTCAGCACTTTGAGGAGGGCCAAG GTGGGAGGATTGCTTGAGCTCAGGACTTCAAGACCAGCCTGGGTAAACATGGCGAGACCCTGTCTCTACCAAAACAACAAAAAAAAGACA GACTGCTTTGATCAACCCTAAATGCAAAAGCAGCCTATTTTTCTTTGTTTAAAAGTCAAAACATAAAAAAGCAGAGTATAACATACAAAC CATTCTTAACTATTCATTAAAATGGGTCCTTCAACACCTTAGTGGGGTTTGTTGTTGTTGCTTATGCAGAGAGATTATTTTCTTTTTATT ATTTTATAATTTTTGAAATAGAGATGGGGTCTCACTGTGTTGCCCAGGCTGGTCTCGAACTCCTGGACTTAAGTGAGCCTCCCGCCTCAG TCTCCCAAAGCGCTGGGATTACAGGCAGGAGCCACTGAGCCCAGCCAAGACTTCAGTGTTGACTGCTTTGGAGGCACAAACCCATGCAAG CGTTAGTTCCAAAGTTCAGTGTGTACCCTTAAATGAACAATGAAGCAGGTAAAATTACCCTTGAAAAAAATCCCTTGGACCACCCATAAA TGACAGTGACTTTTTCAATATGGACTCATCATAGCCAGTTTTCCTTTTGAAGTTGGAACTGATCACCCTTTTGTCATCTGTACCAGATCA GTAGTTGGCTTGTGTTACATTTTGTGTGTGTGTGTGCGTGTTTTAAACCAGTGCATATAAATTGTATGTTAAATGTAAGTAACTTTAAGT TGACTTATCTCTTCACAGTAATCAAGCCTCACGTAATTCATGCTTTTTAAATTCAGCCAGCCCCCCCTCTCTGAAATTTTATTATGTAAA TAATTTGTGTTCCCTGATCACTCGTTTAAGTTCTTAGTTGTATGTCATCTCTTCTCTAGCAGGAATTGGCAAACTTTTTTGTAAAGGGGT AGAAAGTGAAGATTTTAGGCTTTGCAGGCCATATAGCCTCTGCTGCAAATGCTCAGCCCTGCTGTTGTAATGTAAAAGCTGCCACAGACA >37274_37274_1_HNRNPM-EEF2K_HNRNPM_chr19_8532468_ENST00000325495_EEF2K_chr16_22284947_ENST00000263026_length(amino acids)=415AA_BP=278 MAAGVEAAAEVAATEIKMEEESGAPGVPSGNGAPGPKGEGERPAQNEKRKEKNIKRGGNRFEPYANPTKRYRAFITNIPFDVKWQSLKDL VKEKVGEVTYVELLMDAEGKSRGCAVVEFKMEESMKKAAEVLNKHSLSGRPLKVKEDPDGEHARRAMQKVMATTGGMGMGPGGPGMITIP PSILNNPNIPNEIIHALQAGRLGSTVFVANLDYKVGWKKLKEVFSMAGVVVRADILEDKDGKSRGIGTVTFEQSIEAVQAISMFNGQLLF DRPMHVKMETEENKTKGFDYLLKAAEAGDRQSMILVARAFDSGQNLSPDRCQDWLEALHWYNTALEMTDCDEGGEYDGMQDEPRYMMLAR -------------------------------------------------------------- >37274_37274_2_HNRNPM-EEF2K_HNRNPM_chr19_8532468_ENST00000348943_EEF2K_chr16_22284947_ENST00000263026_length(transcript)=4000nt_BP=949nt CGCGCCGGGAACAGCCAGTCGGTGCCTAACGCGAGTGTATCTCGAGAGAGAAGCGATCAACAGCTGCCGGTCTGCGCCTGCGCGCGGCGG GGGCGTGGCCCGGGGCGAGTGGGGCCAAGGAGGCAGCCGGGAGCGGCGGGCGCAGGTGTTACTGGTTGCGTCGGGTCACGTGGGCGCGCA GGCGCAGCGCGGTGCAGCCCGTTCGCTCACACAAAGCCCAGACGCGGAGAAAATGGCGGCAGGGGTCGAAGCGGCGGCGGAGGTGGCGGC GACGGAGATCAAAATGGAGGAAGAGAGCGGCGCGCCCGGCGTGCCGAGCGGCAACGGGGCTCCGGGCCCTAAGGGTGAAGGAGAACGACC TGCTCAGAATGAGAAGAGGAAGGAGAAAAACATAAAAAGAGGAGGCAATCGCTTTGAGCCATATGCCAATCCAACTAAAAGATACAGAGC CTTCATTACAAACATACCTTTTGATGTGAAATGGCAGTCACTTAAAGACCTGGTTAAAGAAAAAGTTGGTGAGGTAACATACGTGGAGCT CTTAATGGACGCTGAAGGAAAGTCAAGGGGATGTGCTGTTGTTGAATTCAAGATGGAAGAGAGCATGAAAAAAGCTGCGGAAGTCCTAAA CAAGCATAGTCTGAGCGGAAGACCACTGAAAGTCAAAGAAGATCCTGATGGTGAACATGCCAGGAGAGCAATGCAAAAGGCTGGAAGACT TGGAAGCACAGTATTTGTAGCAAATCTGGATTATAAAGTTGGCTGGAAGAAACTGAAGGAAGTATTTAGTATGGCTGGTGTGGTGGTCCG AGCAGACATTCTTGAAGATAAAGATGGAAAAAGTCGTGGAATAGGCACTGTTACTTTTGAACAGTCCATTGAAGCTGTGCAAGCTATATC TATGTTCAATGGCCAGCTGCTATTTGATAGACCAATGCACGTCAAGATGGAGACAGAAGAGAACAAAACCAAAGGATTTGATTACTTACT AAAGGCCGCTGAAGCTGGCGACAGGCAGTCCATGATCCTAGTGGCGCGAGCTTTTGACTCTGGCCAGAACCTCAGCCCGGACAGGTGCCA AGACTGGCTAGAGGCCCTGCACTGGTACAACACTGCCCTGGAGATGACGGACTGTGATGAGGGCGGTGAGTACGACGGAATGCAGGACGA GCCCCGGTACATGATGCTGGCCAGGGAGGCCGAGATGCTGTTCACAGGAGGCTACGGGCTGGAGAAGGACCCGCAGAGATCAGGGGACTT GTATACCCAGGCAGCAGAGGCAGCGATGGAAGCCATGAAGGGCCGACTGGCCAACCAGTACTACCAAAAGGCTGAAGAGGCCTGGGCCCA GATGGAGGAGTAACCAGGAAAATCACTGCCGGCTAGTCCCAAGCAAACGGGCTAGGAGGAAAGATTAAAAAAACAACAACAACAACTTAT TTAGTTTGGGGAGGGGAAGCATTTTTAAGTGTGTTGTAAAATCAAATTTTATATTTCATTTTTTGACTCTTGAAAAATGTCTTTGCTCCT TGGCAGCTACCAGCAGAGACTCTATAGCTGTCTCTTAGGGCAGTATTTTGGGGAAGTGGGGCTTGAAGAAGCAGCCTAATGAACCAACAT ACCGTTTTGTGTGTGGTTTTTTTTGTTTGTTTGTTTGTTTGTTTTGAGACAGAGTCTTGCTCTGTCACCCAGGCTGGAGTGCAGTGACAT GATCTTAGCTCACTGCAACCTCCGCCTCCTGGGTTCAAGTGATTCTCCTGCCTCAGCCTCCCAAGTAGCTGGGATTACTGGTGCACACCA CCACACTCAGCTAATTTTTGCATTTTTAGTAGAGATGGGGTTTCACCATGTTGGCCAGGCTGGTCTCGAACTCCTAACCTCAGGTGATCC ACCTGCCTCAGCCTCCCAAAGTGCTGGGATTACAGGTGTGAGCCACCATGCCTGCCCATTTTGTGGTTCTATTTTCATTTTTATTTCTTT TTTTTTTTTTGTCACGAGATATAAGAAAGTGCTTTTTGCCTTGAATGGACAATTTTAGGGCTGTGCTCACTAGTCTTTTCAGGCTGGACT GAAATGTCGGGCCCATGGAGCCCTGTGTTTTGTGCATCGGGATGAGAAATGAAGCACTTCACGCTGGCTTTCCTAAGTCACGGGGCGTGT ATTGCCGTGGCTTAGTGCAAAGCATTCTTTCTCAGAGCATTTAGAGGCATGCGTGGCATTTTTTCAGTGGGTGTGAGATTGCACAATACC CAGGCTCCCTTCTACTGTGGGGAAGGGCCTGCATGTTGGCTGTTTTTTAAACTTCTAGTTCAATTTCCTTCCATAATGCTACTGATTTTC TGGCATACAGCCGAATTCCATCTTTTAAGCATGCTTTCTACGGTGGGCTTTTCAAAACAGGTTTGAGTTTTGTATGCACACGTTTACTAC CTCTAACTCCTACATCAGCTAGTGTGGAAGAGGGTGCACCTCAAAGCTTTTACACGTAAGGACAGCGGCTTGGAATGTGAGAGCCTTTTC TCCAAGCAGACCCACACTCTGCATCTCAGTGGCAGCTCCCACAACGTGACTGCAATGTCTCTTATACAGTATTCCTTGGTGTTTTCTTAG TGTCTGGATGTTCTTACGTGAAATCTGCTCCCCAGCCCTGGTCCTTGGCATTTTCTGCTTGAAGCTGGGCTGATTTTCTTGTAATTTACA GCAGGACGCTTTCAGCAGCAGTCTCTTGGGATTTTATCTAAGATGTTTGAGGATGAGAGGGCAAGAACTATAAACACTCATTAATTCTAG TAGTCTCCCCATGGCCAGACAATGGCGATTGTTATTTAATGAGCTTTTCCTTTCAATGGAATTCAGCTCTCACATTAGTATGATTTCATT TGATGTTTCAAATAGCAAAGATGCTAGGTGCGGTGGCTCCCGCCTGTAATCTCAGCACTTTGAGGAGGGCCAAGGTGGGAGGATTGCTTG AGCTCAGGACTTCAAGACCAGCCTGGGTAAACATGGCGAGACCCTGTCTCTACCAAAACAACAAAAAAAAGACAGACTGCTTTGATCAAC CCTAAATGCAAAAGCAGCCTATTTTTCTTTGTTTAAAAGTCAAAACATAAAAAAGCAGAGTATAACATACAAACCATTCTTAACTATTCA TTAAAATGGGTCCTTCAACACCTTAGTGGGGTTTGTTGTTGTTGCTTATGCAGAGAGATTATTTTCTTTTTATTATTTTATAATTTTTGA AATAGAGATGGGGTCTCACTGTGTTGCCCAGGCTGGTCTCGAACTCCTGGACTTAAGTGAGCCTCCCGCCTCAGTCTCCCAAAGCGCTGG GATTACAGGCAGGAGCCACTGAGCCCAGCCAAGACTTCAGTGTTGACTGCTTTGGAGGCACAAACCCATGCAAGCGTTAGTTCCAAAGTT CAGTGTGTACCCTTAAATGAACAATGAAGCAGGTAAAATTACCCTTGAAAAAAATCCCTTGGACCACCCATAAATGACAGTGACTTTTTC AATATGGACTCATCATAGCCAGTTTTCCTTTTGAAGTTGGAACTGATCACCCTTTTGTCATCTGTACCAGATCAGTAGTTGGCTTGTGTT ACATTTTGTGTGTGTGTGTGCGTGTTTTAAACCAGTGCATATAAATTGTATGTTAAATGTAAGTAACTTTAAGTTGACTTATCTCTTCAC AGTAATCAAGCCTCACGTAATTCATGCTTTTTAAATTCAGCCAGCCCCCCCTCTCTGAAATTTTATTATGTAAATAATTTGTGTTCCCTG ATCACTCGTTTAAGTTCTTAGTTGTATGTCATCTCTTCTCTAGCAGGAATTGGCAAACTTTTTTGTAAAGGGGTAGAAAGTGAAGATTTT AGGCTTTGCAGGCCATATAGCCTCTGCTGCAAATGCTCAGCCCTGCTGTTGTAATGTAAAAGCTGCCACAGACACTACATGAACACGAAT >37274_37274_2_HNRNPM-EEF2K_HNRNPM_chr19_8532468_ENST00000348943_EEF2K_chr16_22284947_ENST00000263026_length(amino acids)=403AA_BP=266 MVASGHVGAQAQRGAARSLTQSPDAEKMAAGVEAAAEVAATEIKMEEESGAPGVPSGNGAPGPKGEGERPAQNEKRKEKNIKRGGNRFEP YANPTKRYRAFITNIPFDVKWQSLKDLVKEKVGEVTYVELLMDAEGKSRGCAVVEFKMEESMKKAAEVLNKHSLSGRPLKVKEDPDGEHA RRAMQKAGRLGSTVFVANLDYKVGWKKLKEVFSMAGVVVRADILEDKDGKSRGIGTVTFEQSIEAVQAISMFNGQLLFDRPMHVKMETEE NKTKGFDYLLKAAEAGDRQSMILVARAFDSGQNLSPDRCQDWLEALHWYNTALEMTDCDEGGEYDGMQDEPRYMMLAREAEMLFTGGYGL -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for HNRNPM-EEF2K |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for HNRNPM-EEF2K |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for HNRNPM-EEF2K |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
