
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:IDE-CAMK1D (FusionGDB2 ID:38230) |
Fusion Gene Summary for IDE-CAMK1D |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: IDE-CAMK1D | Fusion gene ID: 38230 | Hgene | Tgene | Gene symbol | IDE | CAMK1D | Gene ID | 3416 | 57118 |
| Gene name | insulin degrading enzyme | calcium/calmodulin dependent protein kinase ID | |
| Synonyms | INSULYSIN | CKLiK|CaM-K1|CaMKID | |
| Cytomap | 10q23.33 | 10p13 | |
| Type of gene | protein-coding | protein-coding | |
| Description | insulin-degrading enzymeAbeta-degrading proteaseinsulin proteaseinsulinase | calcium/calmodulin-dependent protein kinase type 1DCAMK1D/ANAPC5 fusionCaM kinase IDCamKI-like protein kinasecaM kinase I deltacaM-KI deltacaMKI delta | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | . | Q8IU85 | |
| Ensembl transtripts involved in fusion gene | ENST00000265986, ENST00000371581, ENST00000496903, | ENST00000378845, ENST00000487696, ENST00000378847, | |
| Fusion gene scores | * DoF score | 5 X 5 X 3=75 | 16 X 18 X 6=1728 |
| # samples | 5 | 19 | |
| ** MAII score | log2(5/75*10)=-0.584962500721156 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(19/1728*10)=-3.18503189383252 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: IDE [Title/Abstract] AND CAMK1D [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | IDE(94243012)-CAMK1D(12802947), # samples:3 | ||
| Anticipated loss of major functional domain due to fusion event. | IDE-CAMK1D seems lost the major protein functional domain in Hgene partner, which is a IUPHAR drug target due to the frame-shifted ORF. IDE-CAMK1D seems lost the major protein functional domain in Tgene partner, which is a essential gene due to the frame-shifted ORF. IDE-CAMK1D seems lost the major protein functional domain in Tgene partner, which is a IUPHAR drug target due to the frame-shifted ORF. IDE-CAMK1D seems lost the major protein functional domain in Tgene partner, which is a kinase due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | IDE | GO:0006508 | proteolysis | 18602473|20082125|21185309 |
| Hgene | IDE | GO:0008340 | determination of adult lifespan | 18448515 |
| Hgene | IDE | GO:0010815 | bradykinin catabolic process | 17613531|21185309 |
| Hgene | IDE | GO:0010992 | ubiquitin recycling | 21185309 |
| Hgene | IDE | GO:0032092 | positive regulation of protein binding | 9830016 |
| Hgene | IDE | GO:0042447 | hormone catabolic process | 19321446 |
| Hgene | IDE | GO:0043171 | peptide catabolic process | 20364150 |
| Hgene | IDE | GO:0050435 | amyloid-beta metabolic process | 9830016|17613531|26968463 |
| Hgene | IDE | GO:0051603 | proteolysis involved in cellular protein catabolic process | 9830016 |
| Hgene | IDE | GO:1901142 | insulin metabolic process | 18448515 |
| Hgene | IDE | GO:1901143 | insulin catabolic process | 9231799|9830016|19321446 |
| Tgene | CAMK1D | GO:0032793 | positive regulation of CREB transcription factor activity | 16324104 |
 Fusion gene breakpoints across IDE (5'-gene) Fusion gene breakpoints across IDE (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
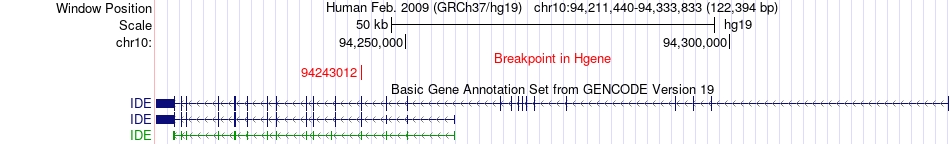 |
 Fusion gene breakpoints across CAMK1D (3'-gene) Fusion gene breakpoints across CAMK1D (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
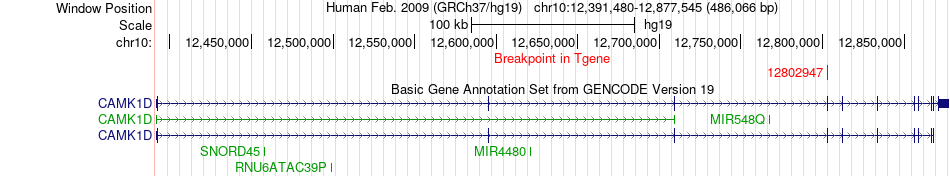 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | PRAD | TCGA-EJ-7314-01A | IDE | chr10 | 94243012 | - | CAMK1D | chr10 | 12802947 | + |
| ChimerDB4 | PRAD | TCGA-EJ-7314 | IDE | chr10 | 94243012 | - | CAMK1D | chr10 | 12802947 | + |
Top |
Fusion Gene ORF analysis for IDE-CAMK1D |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-intron | ENST00000265986 | ENST00000378845 | IDE | chr10 | 94243012 | - | CAMK1D | chr10 | 12802947 | + |
| 5CDS-intron | ENST00000265986 | ENST00000487696 | IDE | chr10 | 94243012 | - | CAMK1D | chr10 | 12802947 | + |
| 5CDS-intron | ENST00000371581 | ENST00000378845 | IDE | chr10 | 94243012 | - | CAMK1D | chr10 | 12802947 | + |
| 5CDS-intron | ENST00000371581 | ENST00000487696 | IDE | chr10 | 94243012 | - | CAMK1D | chr10 | 12802947 | + |
| 5UTR-3CDS | ENST00000496903 | ENST00000378847 | IDE | chr10 | 94243012 | - | CAMK1D | chr10 | 12802947 | + |
| 5UTR-intron | ENST00000496903 | ENST00000378845 | IDE | chr10 | 94243012 | - | CAMK1D | chr10 | 12802947 | + |
| 5UTR-intron | ENST00000496903 | ENST00000487696 | IDE | chr10 | 94243012 | - | CAMK1D | chr10 | 12802947 | + |
| Frame-shift | ENST00000371581 | ENST00000378847 | IDE | chr10 | 94243012 | - | CAMK1D | chr10 | 12802947 | + |
| In-frame | ENST00000265986 | ENST00000378847 | IDE | chr10 | 94243012 | - | CAMK1D | chr10 | 12802947 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000265986 | IDE | chr10 | 94243012 | - | ENST00000378847 | CAMK1D | chr10 | 12802947 | + | 9314 | 1796 | 45 | 2654 | 869 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000265986 | ENST00000378847 | IDE | chr10 | 94243012 | - | CAMK1D | chr10 | 12802947 | + | 0.000448767 | 0.99955124 |
Top |
Fusion Genomic Features for IDE-CAMK1D |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| IDE | chr10 | 94243011 | - | CAMK1D | chr10 | 12802946 | + | 3.89E-07 | 0.99999964 |
| IDE | chr10 | 94243011 | - | CAMK1D | chr10 | 12802946 | + | 3.89E-07 | 0.99999964 |
| IDE | chr10 | 94243011 | - | CAMK1D | chr10 | 12802946 | + | 3.89E-07 | 0.99999964 |
| IDE | chr10 | 94243011 | - | CAMK1D | chr10 | 12802946 | + | 3.89E-07 | 0.99999964 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
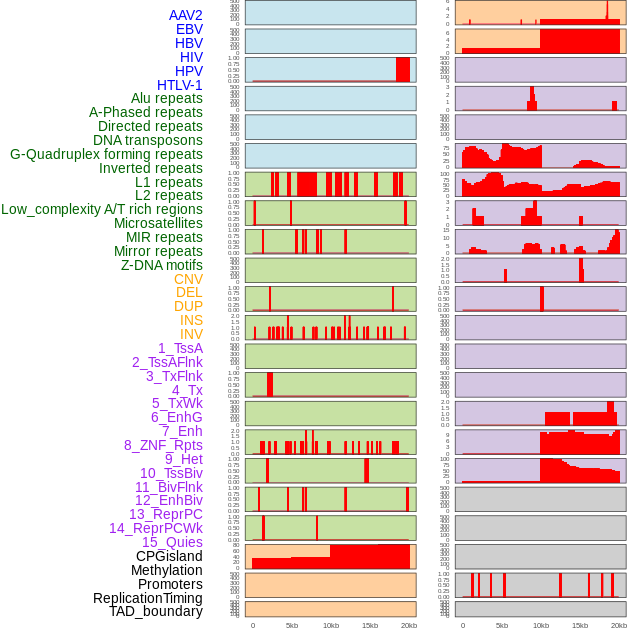 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
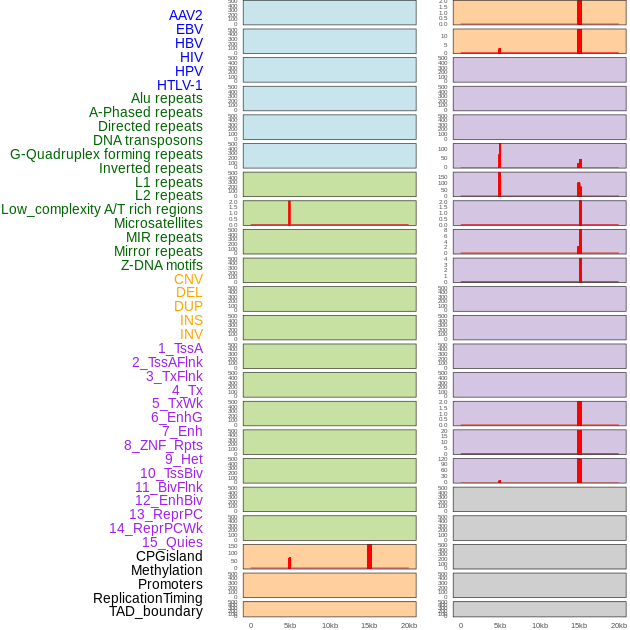 |
Top |
Fusion Protein Features for IDE-CAMK1D |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr10:94243012/chr10:12802947) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| . | CAMK1D |
| FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. | FUNCTION: Calcium/calmodulin-dependent protein kinase that operates in the calcium-triggered CaMKK-CaMK1 signaling cascade and, upon calcium influx, activates CREB-dependent gene transcription, regulates calcium-mediated granulocyte function and respiratory burst and promotes basal dendritic growth of hippocampal neurons. In neutrophil cells, required for cytokine-induced proliferative responses and activation of the respiratory burst. Activates the transcription factor CREB1 in hippocampal neuron nuclei. May play a role in apoptosis of erythroleukemia cells. In vitro, phosphorylates transcription factor CREM isoform Beta. {ECO:0000269|PubMed:11050006, ECO:0000269|PubMed:15840691, ECO:0000269|PubMed:16324104, ECO:0000269|PubMed:17056143}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | IDE | chr10:94243012 | chr10:12802947 | ENST00000265986 | - | 14 | 25 | 336_342 | 579 | 1020.0 | Region | Note=Substrate binding exosite |
| Hgene | IDE | chr10:94243012 | chr10:12802947 | ENST00000265986 | - | 14 | 25 | 359_363 | 579 | 1020.0 | Region | Substrate binding |
| Tgene | CAMK1D | chr10:94243012 | chr10:12802947 | ENST00000378845 | 2 | 10 | 326_364 | 99 | 358.0 | Compositional bias | Note=Ser-rich | |
| Tgene | CAMK1D | chr10:94243012 | chr10:12802947 | ENST00000378847 | 2 | 11 | 326_364 | 99 | 386.0 | Compositional bias | Note=Ser-rich | |
| Tgene | CAMK1D | chr10:94243012 | chr10:12802947 | ENST00000378845 | 2 | 10 | 318_324 | 99 | 358.0 | Motif | Nuclear export signal | |
| Tgene | CAMK1D | chr10:94243012 | chr10:12802947 | ENST00000378847 | 2 | 11 | 318_324 | 99 | 386.0 | Motif | Nuclear export signal | |
| Tgene | CAMK1D | chr10:94243012 | chr10:12802947 | ENST00000378845 | 2 | 10 | 279_319 | 99 | 358.0 | Region | Autoinhibitory domain | |
| Tgene | CAMK1D | chr10:94243012 | chr10:12802947 | ENST00000378845 | 2 | 10 | 299_320 | 99 | 358.0 | Region | Calmodulin-binding | |
| Tgene | CAMK1D | chr10:94243012 | chr10:12802947 | ENST00000378847 | 2 | 11 | 279_319 | 99 | 386.0 | Region | Autoinhibitory domain | |
| Tgene | CAMK1D | chr10:94243012 | chr10:12802947 | ENST00000378847 | 2 | 11 | 299_320 | 99 | 386.0 | Region | Calmodulin-binding |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | IDE | chr10:94243012 | chr10:12802947 | ENST00000265986 | - | 14 | 25 | 853_858 | 579 | 1020.0 | Motif | SlyX motif |
| Hgene | IDE | chr10:94243012 | chr10:12802947 | ENST00000371581 | - | 4 | 15 | 853_858 | 24 | 465.0 | Motif | SlyX motif |
| Hgene | IDE | chr10:94243012 | chr10:12802947 | ENST00000265986 | - | 14 | 25 | 895_901 | 579 | 1020.0 | Nucleotide binding | ATP |
| Hgene | IDE | chr10:94243012 | chr10:12802947 | ENST00000371581 | - | 4 | 15 | 895_901 | 24 | 465.0 | Nucleotide binding | ATP |
| Hgene | IDE | chr10:94243012 | chr10:12802947 | ENST00000371581 | - | 4 | 15 | 336_342 | 24 | 465.0 | Region | Note=Substrate binding exosite |
| Hgene | IDE | chr10:94243012 | chr10:12802947 | ENST00000371581 | - | 4 | 15 | 359_363 | 24 | 465.0 | Region | Substrate binding |
| Tgene | CAMK1D | chr10:94243012 | chr10:12802947 | ENST00000378845 | 2 | 10 | 23_279 | 99 | 358.0 | Domain | Protein kinase | |
| Tgene | CAMK1D | chr10:94243012 | chr10:12802947 | ENST00000378847 | 2 | 11 | 23_279 | 99 | 386.0 | Domain | Protein kinase | |
| Tgene | CAMK1D | chr10:94243012 | chr10:12802947 | ENST00000378845 | 2 | 10 | 29_37 | 99 | 358.0 | Nucleotide binding | ATP | |
| Tgene | CAMK1D | chr10:94243012 | chr10:12802947 | ENST00000378847 | 2 | 11 | 29_37 | 99 | 386.0 | Nucleotide binding | ATP |
Top |
Fusion Gene Sequence for IDE-CAMK1D |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >38230_38230_1_IDE-CAMK1D_IDE_chr10_94243012_ENST00000265986_CAMK1D_chr10_12802947_ENST00000378847_length(transcript)=9314nt_BP=1796nt CCGGCTCGAAGCGCAAGCAGGAAGCGTTTGCGGTGATCCCGGCGACTGCGCTGGCTAATGCGGTACCGGCTAGCGTGGCTTCTGCACCCC GCACTGCCCAGCACCTTCCGCTCAGTCCTCGGCGCCCGCCTGCCGCCTCCGGAGCGCCTGTGTGGTTTCCAAAAAAAGACTTACAGCAAA ATGAATAATCCAGCCATCAAGAGAATAGGAAATCACATTACCAAGTCTCCTGAAGACAAGCGAGAATATCGAGGGCTAGAGCTGGCCAAT GGTATCAAAGTACTTCTTATCAGTGATCCCACCACGGATAAGTCATCAGCAGCACTTGATGTGCACATAGGTTCATTGTCGGATCCTCCA AATATTGCTGGCTTAAGTCATTTTTGTGAACATATGCTTTTTTTGGGAACAAAGAAATACCCTAAAGAAAATGAATACAGCCAGTTTCTC AGTGAGCATGCAGGAAGTTCAAATGCCTTTACTAGTGGAGAGCATACCAATTACTATTTTGATGTTTCTCATGAACACCTAGAAGGTGCC CTAGACAGGTTTGCACAGTTTTTTCTGTGCCCCTTGTTCGATGAAAGTTGCAAAGACAGAGAGGTGAATGCAGTTGATTCAGAACATGAG AAGAATGTGATGAATGATGCCTGGAGACTCTTTCAATTGGAAAAAGCTACAGGGAATCCTAAACACCCCTTCAGTAAATTTGGGACAGGT AACAAATATACTCTGGAGACTAGACCAAACCAAGAAGGCATTGATGTAAGACAAGAGCTACTGAAATTCCATTCTGCTTACTATTCATCC AACTTAATGGCTGTTTGTGTTTTAGGTCGAGAATCTTTAGATGACTTGACTAATCTGGTGGTAAAGTTATTTTCTGAAGTAGAGAACAAA AATGTTCCATTGCCAGAATTTCCTGAACACCCTTTCCAAGAAGAACATCTTAAACAACTTTACAAAATAGTACCCATTAAAGATATTAGG AATCTCTATGTGACATTTCCCATACCTGACCTTCAGAAATACTACAAATCAAATCCTGGTCATTATCTTGGTCATCTCATTGGGCATGAA GGTCCTGGAAGTCTGTTATCAGAACTTAAGTCAAAGGGCTGGGTTAATACTCTTGTTGGTGGGCAGAAGGAAGGAGCCCGAGGTTTTATG TTTTTTATCATTAATGTGGACTTGACCGAGGAAGGATTATTACATGTTGAAGATATAATTTTGCACATGTTTCAATACATTCAGAAGTTA CGTGCAGAAGGACCTCAAGAATGGGTTTTCCAAGAGTGCAAGGACTTGAATGCTGTTGCTTTTAGGTTTAAAGACAAAGAGAGGCCACGG GGCTATACATCTAAGATTGCAGGAATATTGCATTATTATCCCCTAGAAGAGGTGCTCACAGCGGAATATTTACTGGAAGAATTTAGACCT GACTTAATAGAGATGGTTCTCGATAAACTCAGACCAGAAAATGTCCGGGTTGCCATAGTTTCTAAATCTTTTGAAGGAAAAACTGATCGC ACAGAAGAGTGGTATGGAACCCAGTACAAACAAGAAGCTATACCGGATGAAGTCATCAAGAAATGGCAAAATGCTGACCTGAATGGGAAA TTTAAACTTCCTACAAAGAATGAATTTATTCCTACGAATTTTGAGATTTTACCGTTAGAAAAAGAGGCGACACCATACCCTGCTCTTATT AAGGATACAGCTATGAGCAAACTTTGGTTCAAACAAGATGATAAGTTTTTTTTGCCGAAGGCTTGTCTCAACTTTGAATTTTTCAGGGTG TCCGGTGGAGAGCTGTTTGACCGGATAGTGGAGAAGGGGTTTTATACAGAGAAGGATGCCAGCACTCTGATCCGCCAAGTCTTGGACGCC GTGTACTATCTCCACAGAATGGGCATCGTCCACAGAGACCTCAAGCCCGAAAATCTCTTGTACTACAGTCAAGATGAGGAGTCCAAAATA ATGATCAGTGACTTTGGATTGTCAAAAATGGAGGGCAAAGGAGATGTGATGTCCACTGCCTGTGGAACTCCAGGCTATGTCGCTCCTGAA GTCCTCGCCCAGAAACCTTACAGCAAAGCCGTTGACTGCTGGTCCATCGGAGTGATTGCCTACATCTTGCTCTGCGGCTACCCTCCTTTT TATGATGAAAATGACTCCAAGCTCTTTGAGCAGATCCTCAAGGCGGAATATGAGTTTGACTCTCCCTACTGGGATGACATCTCCGACTCT GCAAAAGACTTCATTCGGAACCTGATGGAGAAGGACCCGAATAAAAGATACACGTGTGAGCAGGCAGCTCGGCACCCATGGATCGCTGGT GACACAGCCCTCAACAAAAACATCCACGAGTCCGTCAGCGCCCAGATCCGGAAAAACTTTGCCAAGAGCAAATGGAGACAAGCATTTAAT GCCACGGCCGTCGTCAGACATATGAGAAAACTACACCTCGGCAGCAGCCTGGACAGTTCAAATGCAAGTGTTTCGAGCAGCCTCAGTTTG GCCAGCCAAAAAGACTGTCTGGCACCTTCCACGCTCTGTAGTTTCATTTCTTCTTCGTCGGGGGTCTCAGGAGTTGGAGCCGAGCGGAGA CCCAGGCCCACCACTGTGACGGCAGTGCACTCTGGAAGCAAGTGACTGGCCCTGGAGGTGGGGCCCGGGGTCGGGGCTGGGGAAGGGGAG CCCCAGGGTCGCCAGAGCCGCGAGCCACTCCAGCGAGACCCCACCTTGCATGGTGCCCCTTCCTGCATAGGACTGGAAGACCGAAGTTTT TTTATGGCCATATTTTCTACTGCAATTCTGAAGTGTTCATTTCTCACAAACTGTACTGACTCGAGGGGCGCTGATTTCATAGGATCTGGT GCTGTATATACGAATCTTGCAAAGCTCTAACTGAACGGACCTTCTTATTCCTCTCCCCTAACACCATCGTTTCCACTCTTCTCAGTGTAG GTAACCGTCTATGGTGTGTTTTTTCATTAATGACAAAAAAAAAAAGGTTTCAACTGGATTATTTAAATATTGGTAAATATTGTGCATTAG GGTTTGTTTTTCCTTTTAAGAAGTATGTCCTTTGTATCTCTAAGTTACATGACCTATATCTTTTCCTCTTTAATAGTAGTTTTATGTTAA CCTTTAAGAGATTTGTTTTTCCTCAAAGGAGAATTTAAAGGTATTTTTTAAAATTCTAATAAGAGGATCAGCCGGGTGCAATGACTCATG CCTGTAATCCCAGCACGTTGGGAGGCCAAGTCGGGCGGATCACAAGGTCAGGAGATCAAGGCCATCCTGGCCAACATGGTGAAACCCCAC GTCTACTAAAAATACAAAAAATTAGCCGGGCGTGGTGGCACACACCTGTAGTCCCGGCTACTCGGGAGGCTGAGGCAGGAGAATTGCTTG AACCCGGGAGACGGAGGTTGCAGTGAGCTGAGATCGTGCCACTGCACTCCAGCCTGGGTGACAGAGCAAGACTCTGTCTCAAAAAAAAAA AAAAAAATTCTAACAAGAGGATCATGAATCTGTCTCCAAGGCTAAGAGTGCCCTTCACCTCTACTCCTTCTGTTTCTCTCCCCTTCCCCA GGAGGAGGGGCCCCTCAGTTCTGCGAGGCTCTCTTCTCTGCGGGGGACTTTGGTAGAGAATCTGCACAGAGCCTTTGCTGGCAACCCATC CAGCAGAACCCACACCCACCAGAGACCCTAGAGGCCTCTTGCCATCAGCAACCAGAAATAAGCCTGCTCCCCCGAGACGCAGGGCTCAGC TTCACAGGCCAGACTTGCTGTGAGTGAGATGCCCACACTGCAGAGATGCAGGACCCTCCCTTGCAAAAGGAACCAGCTGCCATGTTGCTG AAAGTGGGCCCACACACAACCCTCATTTCTCACGAGAGCTAACAGATGAAAGCTCAGCTATGCAGTGTTAAAATTCATCTCTTTCTCTGT GGTTTTTAAAAAAATGCATTTCTAATCTTACACTTCACGTGGCCTCCAACACAAAGTTCTTTCTCCACATTTTTACCCTCTTCTCACATG AATGTTTTCCAAGGGAAGGAAGGGAGGGAAGCAGCCTTGGGAAAGGATCCATTTCTGGTGGTAAGGAGAGGGGGCCCTGCCAGGCTAATG GGACTTCAGCCACCACAGAGATGACCAAAGACCACCTCCGCCAGGCCCAGCCAGTGCTTGCAGCCCTCCCATTGGTCAGGGGAGTCTAAA ATCAAAGATACATGGTGGTAGGGTCACCAAACGTCCCATTTGCCCGAGACTGAAGGATTTCTCAGGCTGTGGACATTTAATGCCAAAACC AGGAATATCCGGGGCAAAGCAGGCTGTGTTGTTCGCCTTTGTGGAGCCAGGCAGTGTGTGGCCCAGGGAAAGCTGCGTTTCATGGAGGCT TCCAGGAGGCTGCCCTGTAAAGCCAGGCAGGTGGCTTCAAGTGCCATTCCTGCAAGTGCCCCTGCTTCAGCTCTCAGGCCCATCCAATGC CTCGAGCCCACGGGGGTGTCCCGGCAGGTCTGCGTCATCACGACCCAGTAACGCTGCTGGCTGTGTCCTCATGCCCTCTTCCTCTGGCTG GCCAGTCATGGGAAGTCCTGTTGGTTGAGTCATCTTTGTCACTTCTACACCTTTGAGTATGTACAGGCTGAATGTTGAGTGATGCCCCCC CACCCTCTCAATAAACACACACACACACACACACACACACACACACACACACAATGTTATTAGGCACAGCAGCTCCATCCTGCTCCCTGA GAATGGCAGTGCAGGCACAGAGAAGAGCAGTTGGGAAGACAGGCAGCTGGTCTACCCTGTCCCCGCACACTTCACAGGCACTCCACTGCT TGGCAGGAATTCTGCATGGCATCTGCCACGCATGCTCACAGGGTGAAAAATCAAGCTGTCACTCTGCAAAATCTAAAAACCCTGGAAAGG TCTATAAGATGACATTAGACCCAGTTAAGAAGCAAGTGTCACCTGAGTTGGCCATGCAATGAATGAATATTATAAGCAGTATTGGAGAAC TAGGCGGTTGCTTACTAAGAAGTGTCTTGGAAACATTCGAGTTCATACAGTGCATGACAAAGTAATAGAAGGCTGCAAGCGCTGAAAATG CACCCGAATCTGGTACCTAGGATGAGTACATGAATAGTGATGAATAGCCCTTTGTGCTGTGTGCTATATACAACCATTTGCATTCAGAAA GCAGCATTTGAAGCTGAGTTCCATTACTGGTGATTGAAGTATTTTCAGGAGCAGATACATGTGGTGCATGTTATATGCACTTAGTTTGAA AGTATCCCTCACCCAAAAAGTGCAGAACCTCTGAAAGGTGAGCCGTGGTGCAGGTGCCCCCCTCGCTGTTCGGGTCAGTTCTTGGCCATG CTTGGCCTGTCACACGCCGGGTCTCAACAACATCGTGCGAGGAGACAGTGACTGGTTTCAGCCCAAGAGTCAACTGTTGAGCATGACATC AGGAAGCCTGTACTGTCCTGATCCCACCAGGCTTGGGTTAATTTCCTATTGCTTCAGAGAAAGAGAGGCGCTCTTGTGCTTTTGAGAGAG CTCTCATGTTAAAAAGAGGTTGTCTTTGCACTGCGCTTGGTATGATTTGTTTTCTTATAAAACAGGAAGAGAACCTAGAATGTGCCTTTC GTGAAGGTCTGACAAATGGGGTCTCAGAGTCCTAGGACCCGCACCAAGGTCAGAGTCCACCTGTAGAGGCTGAGCATTTCTACATGCACC CAGGGTGCTGGGCTGAGGCTAAGGGTGGACGCGTAGACCCCATCACAGGCTTCCTTCCTGAGCCCAAGTCCCCCATTTGGTCACTATCCC TTCTCAGAGCAAGTGGATTCTCTTTTCCTCACTTGGAGTATTCAATGTTGAAGTAGTGGCCTGGAGTGGCCTTTCCCTCAGCTGCCTGTC TGGGCCTCCCTGGCTTCCCGGATCTTCATCTCATACGTGAGGCAATAGGAGGAGTTTCTATTCATTGAGACAAACAGTGAGCGGGCACCA ACAGGCCAGCAGCTTGGCCCTTTTCACAGAGGTGGCCTTGGAGTCTGGAGTCTGGGCAGCTGTCTGCCTCTCTACGGCTCTGCTTCCCAG CGGGCACACGGGTCCGGAGCCTCAGAATGAAGTGGTGCAGAGGCGCAGGGCACACCTAGGAGAAGCTCAATGACCACCAAAGCCCTTGAG GAGAGACGGGGTTTGCTGTGTTAGAGAAATGGCAACCCCCAGGACCATGCACGGCTCTGGCAGTGCACAGTGGCTGATTTTCACCAGTTC CTCCTACCCCTGCCTCTGTCGGAGACTAGAAAACATTCTACCCCTTCAGGTCTTCTTTTAAAAGAAAAGAGCTATGGCAAAGCGTGCCAT ACAAAGTTAAAAAAAGGAAGAGTGGCTTTGCGTTCTTGACCATGGCCACAATTCTGGTCACTTCTAGCATCTGACCTTGAGAACGGTGTA GAGTCTCTGAGCCACCGCTGGCCTTTGAGAGAGTCTCACTCTCCAGCTAGCTCTTTGTGTAGTGTCCAGGGCCCCGGCAGTCAGGATAGA GCGAGCAAAAGAAGATGGACAGTGGACAGGGCGCAGTGGCTCACGCCTGTAATCCCAGCGCTTTGGGAGAGCGAGGCGGGCAGATCACAA GGTCAAGAGATCAAGACCATTCTGGCCAACATGGTGAAACCCTGTTTCTACTAAAAATACAAAAATTAGCTGGGCATGGTGGTGCGTGCC TGTAATCCCAGCTACTCGGGAGGCTGAGGCAGGAGAATCGCTTGAACCCGGGAGGCAGAGGTTGCAGTGAGCCGAGATCGTGCCACTGCA CTCCAGCCTGGGTGACAGAGCAAGACTCCGTCTCAAAAAAGATGGCCATGCTGGCCAAGGCTGGGCTCATAGGAAATATCTGTAGTAGGA TGAAAGGAATCTGGCCTAGGGAGCCCCGGACTCGGTTTACCTACCCCGTACTCATCCCACTCATCTGTCTACCCTAGTGTCCATCATGCC CTGGATGAATCGTAGGTGGGAGGGACAGAAGCCGAGAACACCAGGCACACGGGGTGGGGGGAATGGGGGGAGCCCAGGTGATCCCCTTTG GAAAGTGGATTGATCCTCTCCCCTGTGGAATGACAGCTTAACCTGATTAACCCACTGGGGCTTGCTATTTTATGAGCATTTTTAAACAGA AAGGAAAGATGATATCCCAGTGCAGGTAGACTTTATTTTTGAAAGCATCGTGGCCCCTCACCTCTCCCTCTCCCCCAGAACCTTGGCCAG AATTGAGTCTTCGCGTTGGGGAGGCCACGCTGGAACCATTGCTCTAGGGCTGAAGGTGGAAGTAGGGCCACTGCCGCAGCTTCAGCCAGA GCCCTCGCTGCTTCGCAGAATTTAACAAGCACACGACTGGGATTCCTTTGGATATGAGAAAGGAAGGCAAGTGCGGGCAAAGGGAAGCTC CTCCCGTTTTTCATAAACAGAAGTAAGATGTCCTCTTTCCCAGGTTTGCACAATCACTTCTGGCAAATTAATTGAAGGTCCTTGAATTCA GCCAGTGCACGAAACCTTTGCATGTTCTTCTGAAACTGTACTTCTTCCCTATTCTGTCTACCCACACTCTGCGAACTTTGCCTTTTCCTT AAACACATTCCAGACCAAACACTGTTCAGTTTGTTAAAAAAAAAAAAAAAAAAAAAAGATGTATATACCTGTATGTGATCCACCACCCTT TTGAGGGGCACCTGGTCCTCGGTTGGGGCTGCAGGTGTGCCTGACATTTGCAAAAGATATTCCAGTCTTTCGATGTTCAGAATTGAAAAT GTGGAGATAGAAAAGCTCAAACAGACTCCTAAAAGGAGGGTTTCTTTCCTGAAGACTCCAAGAAAATGAGTGTGCAGATGAGTTAGCTTG ACAACCTCAACTCTGGGGAGCAGGCAGGCGGTCTGAGGAAGATAAGGGCCCACGCCCCACTCACCTGGCACCTGTTCGGTGGTCCCGGGG CATCATGGTCTGCAGCTCTGGGCCCCCAGCCCCACTGTGAAGTCAGTCCTGCATGCTGGCCGAGGCCCGGCCACCTCCCGATGCCTCCAG AGTCAGTGGGAAGGAGTCAGCCCTTCCCAGACTTCCCCGCCACCTCCAGCAGGAGCAGCTCAGTTTGTGGCTCTGGGAGCTCCGCTTTTG CAAACCCAAAAAGGCTGTGCATTTGGAAGCCAAACGCTCAGCATGCGGCTGCCGAGTCTGGTTTTGTGGACAAAGCAAACTGTGGAATGG CTTCTCGGTGTCTGTATAAAGGGACAAACGGTTGCATTCACCCTTTGTACTATAACACCGCTTCTGCATTCGCCATATCCGTTTTTTAAC CTTTTTGTCTCCGGGGAACTTCTCATTCGATTATTATGTCTTCTGATGATTTGCCATGTTATTTTAAAGGTGATTTTATTTTCTTATGGA CAGGAAAAAAGAAAATGAGGAGGAGTTGGTTTTACTCTTAGAGATAAGACTTCAGCAGGGGCTCAGAAACCCCAAACTCTAGTTAAATTG CTTCTCGGAGGAAAATCTTAATCTAGACAGATTCAGATTCCCAGGCTCTCCTGGAAGCCCTGCACTTCAGGGCACAGGGGCCCTGGGACG TGGTTGGCAGCCAGGCTGCGAGAGAGCTAAAAAGGAACAAGAACATCAGCTCGGATGTTTGTCTCTCAGCCCTGACACCTGCCACTCTCC TTCCCGGGGCACTTGGCCCTATCAGAGGGACATGAAATGCTGTCCGAATGGTGGCAGAAATGCAGCGACATTGCATGCAGCAGAGTCTTT GCTTCGAAAGATGGGGCGCCATGTTTCAGAGAACATTTATCTTAGTCCCACGTTCAGGTGAGAAGACCTCAAAGGTACCCTCTGTCCAGA ATAAACATTGGAACAATCTCTCAATTGGCTTTTGTCACTACACATAATGTAATCTCCCTTCTTTCAGCTCCCCCAACAGTGAGCATGCTT CCCAAAAAATTACAACATCAAAACCAGGAATGTCCCCGCATCTACTATTACCAGGCTTGCAATTGGTTCAATTTTCCCATAAAAACATGT TTGTCCAAAAGAAGGGAAACTGTGAATTTATTCCATGCATGTAATAAACTCTGCAAGAAGTTTAACCCACATAGGTTTGCGGCACTGCAA AATTGTTCATGGGTCTATGTAGCAAATGATTCTCTGCCTTGACAATCATGTATACATATCAAGATTTCTATCATAATGATAATGAGATTG >38230_38230_1_IDE-CAMK1D_IDE_chr10_94243012_ENST00000265986_CAMK1D_chr10_12802947_ENST00000378847_length(amino acids)=869AA_BP=579 MRWLMRYRLAWLLHPALPSTFRSVLGARLPPPERLCGFQKKTYSKMNNPAIKRIGNHITKSPEDKREYRGLELANGIKVLLISDPTTDKS SAALDVHIGSLSDPPNIAGLSHFCEHMLFLGTKKYPKENEYSQFLSEHAGSSNAFTSGEHTNYYFDVSHEHLEGALDRFAQFFLCPLFDE SCKDREVNAVDSEHEKNVMNDAWRLFQLEKATGNPKHPFSKFGTGNKYTLETRPNQEGIDVRQELLKFHSAYYSSNLMAVCVLGRESLDD LTNLVVKLFSEVENKNVPLPEFPEHPFQEEHLKQLYKIVPIKDIRNLYVTFPIPDLQKYYKSNPGHYLGHLIGHEGPGSLLSELKSKGWV NTLVGGQKEGARGFMFFIINVDLTEEGLLHVEDIILHMFQYIQKLRAEGPQEWVFQECKDLNAVAFRFKDKERPRGYTSKIAGILHYYPL EEVLTAEYLLEEFRPDLIEMVLDKLRPENVRVAIVSKSFEGKTDRTEEWYGTQYKQEAIPDEVIKKWQNADLNGKFKLPTKNEFIPTNFE ILPLEKEATPYPALIKDTAMSKLWFKQDDKFFLPKACLNFEFFRVSGGELFDRIVEKGFYTEKDASTLIRQVLDAVYYLHRMGIVHRDLK PENLLYYSQDEESKIMISDFGLSKMEGKGDVMSTACGTPGYVAPEVLAQKPYSKAVDCWSIGVIAYILLCGYPPFYDENDSKLFEQILKA EYEFDSPYWDDISDSAKDFIRNLMEKDPNKRYTCEQAARHPWIAGDTALNKNIHESVSAQIRKNFAKSKWRQAFNATAVVRHMRKLHLGS -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for IDE-CAMK1D |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for IDE-CAMK1D |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for IDE-CAMK1D |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
