
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:IFT57-MLXIP (FusionGDB2 ID:38438) |
Fusion Gene Summary for IFT57-MLXIP |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: IFT57-MLXIP | Fusion gene ID: 38438 | Hgene | Tgene | Gene symbol | IFT57 | MLXIP | Gene ID | 55081 | 22877 |
| Gene name | intraflagellar transport 57 | MLX interacting protein | |
| Synonyms | ESRRBL1|HIPPI|MHS4R2|OFD18 | MIR|MONDOA|bHLHe36 | |
| Cytomap | 3q13.12-q13.13 | 12q24.31 | |
| Type of gene | protein-coding | protein-coding | |
| Description | intraflagellar transport protein 57 homologHIP1 protein interactorHIP1-interacting proteindermal papilla-derived protein 8estrogen-related receptor beta like 1estrogen-related receptor beta-like protein 1huntingtin interacting protein-1 interacting | MLX-interacting proteinMlx interactorclass E basic helix-loop-helix protein 36transcriptional activator MondoA | |
| Modification date | 20200313 | 20200329 | |
| UniProtAcc | Q9NWB7 | Q9NP71 | |
| Ensembl transtripts involved in fusion gene | ENST00000264538, ENST00000468021, | ENST00000377037, ENST00000535996, ENST00000538698, ENST00000319080, | |
| Fusion gene scores | * DoF score | 5 X 6 X 2=60 | 11 X 11 X 6=726 |
| # samples | 6 | 12 | |
| ** MAII score | log2(6/60*10)=0 | log2(12/726*10)=-2.59693514238723 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: IFT57 [Title/Abstract] AND MLXIP [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | IFT57(107886639)-MLXIP(122613683), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | IFT57 | GO:0006915 | apoptotic process | 11788820 |
| Hgene | IFT57 | GO:0006919 | activation of cysteine-type endopeptidase activity involved in apoptotic process | 11788820 |
| Hgene | IFT57 | GO:0042981 | regulation of apoptotic process | 11788820 |
| Tgene | MLXIP | GO:0045944 | positive regulation of transcription by RNA polymerase II | 11073985 |
 Fusion gene breakpoints across IFT57 (5'-gene) Fusion gene breakpoints across IFT57 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
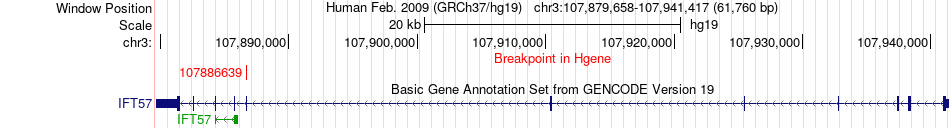 |
 Fusion gene breakpoints across MLXIP (3'-gene) Fusion gene breakpoints across MLXIP (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
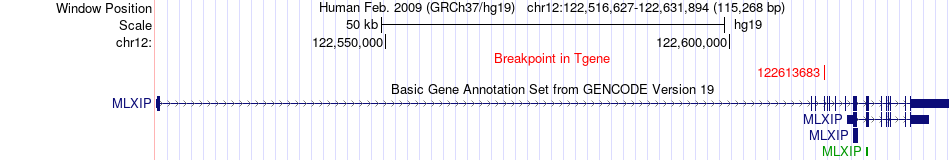 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | OV | TCGA-59-A5PD | IFT57 | chr3 | 107886639 | - | MLXIP | chr12 | 122613683 | + |
Top |
Fusion Gene ORF analysis for IFT57-MLXIP |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-intron | ENST00000264538 | ENST00000377037 | IFT57 | chr3 | 107886639 | - | MLXIP | chr12 | 122613683 | + |
| 5CDS-intron | ENST00000264538 | ENST00000535996 | IFT57 | chr3 | 107886639 | - | MLXIP | chr12 | 122613683 | + |
| 5CDS-intron | ENST00000264538 | ENST00000538698 | IFT57 | chr3 | 107886639 | - | MLXIP | chr12 | 122613683 | + |
| In-frame | ENST00000264538 | ENST00000319080 | IFT57 | chr3 | 107886639 | - | MLXIP | chr12 | 122613683 | + |
| intron-3CDS | ENST00000468021 | ENST00000319080 | IFT57 | chr3 | 107886639 | - | MLXIP | chr12 | 122613683 | + |
| intron-intron | ENST00000468021 | ENST00000377037 | IFT57 | chr3 | 107886639 | - | MLXIP | chr12 | 122613683 | + |
| intron-intron | ENST00000468021 | ENST00000535996 | IFT57 | chr3 | 107886639 | - | MLXIP | chr12 | 122613683 | + |
| intron-intron | ENST00000468021 | ENST00000538698 | IFT57 | chr3 | 107886639 | - | MLXIP | chr12 | 122613683 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000264538 | IFT57 | chr3 | 107886639 | - | ENST00000319080 | MLXIP | chr12 | 122613683 | + | 8786 | 1097 | 227 | 3250 | 1007 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000264538 | ENST00000319080 | IFT57 | chr3 | 107886639 | - | MLXIP | chr12 | 122613683 | + | 0.000771772 | 0.99922824 |
Top |
Fusion Genomic Features for IFT57-MLXIP |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| IFT57 | chr3 | 107886639 | - | MLXIP | chr12 | 122613683 | + | 0.48196718 | 0.5180328 |
| IFT57 | chr3 | 107886639 | - | MLXIP | chr12 | 122613683 | + | 0.48196718 | 0.5180328 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
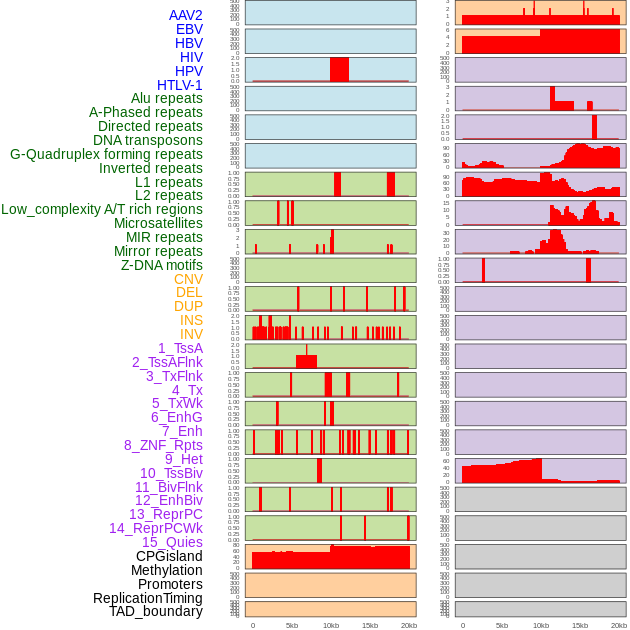 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
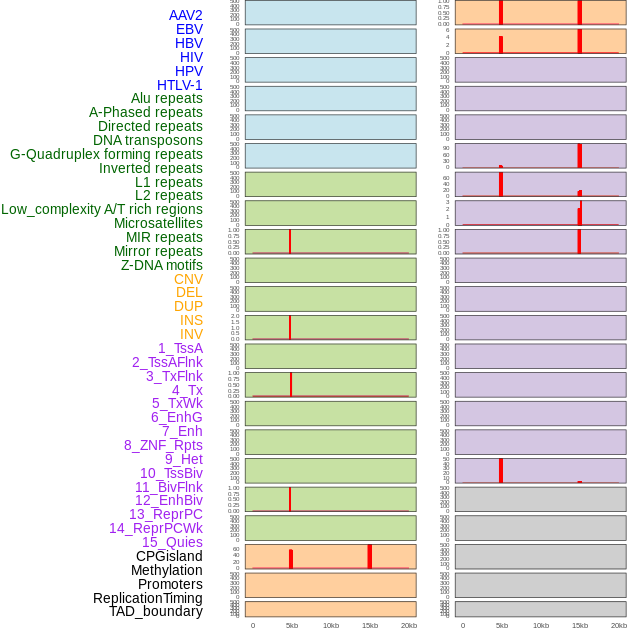 |
Top |
Fusion Protein Features for IFT57-MLXIP |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr3:107886639/chr12:122613683) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| IFT57 | MLXIP |
| FUNCTION: Required for the formation of cilia. Plays an indirect role in sonic hedgehog signaling, cilia being required for all activity of the hedgehog pathway (By similarity). Has pro-apoptotic function via its interaction with HIP1, leading to recruit caspase-8 (CASP8) and trigger apoptosis. Has the ability to bind DNA sequence motif 5'-AAAGACATG-3' present in the promoter of caspase genes such as CASP1, CASP8 and CASP10, suggesting that it may act as a transcription regulator; however the relevance of such function remains unclear. {ECO:0000250, ECO:0000269|PubMed:11788820, ECO:0000269|PubMed:17107665, ECO:0000269|PubMed:17623017}. | FUNCTION: Transcriptional repressor. Binds to the canonical and non-canonical E box sequences 5'-CACGTG-3' (By similarity). {ECO:0000250}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | MLXIP | chr3:107886639 | chr12:122613683 | ENST00000319080 | 2 | 17 | 719_769 | 202 | 920.0 | Domain | bHLH | |
| Tgene | MLXIP | chr3:107886639 | chr12:122613683 | ENST00000538698 | 0 | 9 | 719_769 | 0 | 527.0 | Domain | bHLH | |
| Tgene | MLXIP | chr3:107886639 | chr12:122613683 | ENST00000319080 | 2 | 17 | 322_445 | 202 | 920.0 | Region | Transactivation domain | |
| Tgene | MLXIP | chr3:107886639 | chr12:122613683 | ENST00000319080 | 2 | 17 | 769_790 | 202 | 920.0 | Region | Note=Leucine-zipper | |
| Tgene | MLXIP | chr3:107886639 | chr12:122613683 | ENST00000538698 | 0 | 9 | 322_445 | 0 | 527.0 | Region | Transactivation domain | |
| Tgene | MLXIP | chr3:107886639 | chr12:122613683 | ENST00000538698 | 0 | 9 | 73_327 | 0 | 527.0 | Region | Required for cytoplasmic localization | |
| Tgene | MLXIP | chr3:107886639 | chr12:122613683 | ENST00000538698 | 0 | 9 | 769_790 | 0 | 527.0 | Region | Note=Leucine-zipper |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | IFT57 | chr3:107886639 | chr12:122613683 | ENST00000264538 | - | 7 | 11 | 304_369 | 283 | 430.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | IFT57 | chr3:107886639 | chr12:122613683 | ENST00000264538 | - | 7 | 11 | 335_426 | 283 | 430.0 | Region | Note=pDED |
| Tgene | MLXIP | chr3:107886639 | chr12:122613683 | ENST00000319080 | 2 | 17 | 73_327 | 202 | 920.0 | Region | Required for cytoplasmic localization |
Top |
Fusion Gene Sequence for IFT57-MLXIP |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >38438_38438_1_IFT57-MLXIP_IFT57_chr3_107886639_ENST00000264538_MLXIP_chr12_122613683_ENST00000319080_length(transcript)=8786nt_BP=1097nt ATGAGGGGCGGGGCTGCTTCCCGACCACCATTTCCACAGAGGCAGGGCAAGAAGAGGGCCACAGGTGACGTCATTCAGGAAAGCCGCCCT GATTGGAGGGAAGGAGGCGGAGTCTAGTTTCCCTGGTGACGGATTGTCCGGTGGTCGCCTGGTAACCGGTCGTGGCTGTACTGGCGGCGG CAGGGCTGGCGGCTTAGGCCGCAGAGGTCTGTGGGCCTGAGCCCACGCTGGACTCTGTCCGTTCTGCGATGACTGCTGCTCTGGCCGTCG TCACGACGTCGGGTTTGGAAGATGGGGTGCCTAGGTCCCGTGGCGAAGGGACCGGGGAAGTGGTCTTGGAGCGGGGGCCCGGCGCGGCCT ACCACATGTTCGTGGTGATGGAGGACTTGGTGGAGAAGCTGAAGCTGCTCCGCTACGAGGAGGAGTTCCTCCGGAAGAGCAACCTGAAGG CCCCGTCCAGACACTATTTTGCACTGCCTACCAACCCTGGCGAACAGTTCTACATGTTTTGTACTCTTGCTGCTTGGTTGATTAATAAAG CGGGACGTCCCTTTGAGCAGCCTCAAGAATATGATGACCCTAATGCAACAATATCTAACATACTATCCGAGCTTCGGTCATTTGGAAGAA CTGCAGATTTTCCTCCTTCAAAATTAAAGTCAGGTTATGGAGAACATGTATGCTATGTTCTTGATTGCTTCGCTGAAGAAGCATTGAAAT ATATTGGTTTCACCTGGAAAAGGCCAATATACCCAGTAGAAGAATTAGAAGAAGAAAGCGTTGCAGAAGATGATGCAGAATTAACATTAA ATAAAGTGGATGAAGAATTTGTGGAAGAAGAGACAGATAATGAAGAAAACTTTATTGATCTCAACGTTTTAAAGGCCCAGACATATCACT TGGATATGAACGAGACTGCCAAACAAGAAGATATTTTGGAATCCACAACAGATGCTGCAGAATGGAGCCTAGAAGTGGAACGTGTACTAC CGCAACTGAAAGTCACGATTAGGACTGACAATAAGGATTGGAGAATCCATGTTGACCAAATGCACCAGCACAGAAGTGGAATTGAATCTG CTCTAAAGGAGACCAAGGCCATCACCACGGAAGGGAAGTACTGGAAGAGCCGCATCGAGATTGTGATCCGGGAGTATCACAAGTGGAGAA CCTACTTCAAGAAAAGGCTACAGCAGCACAAGGATGAGGACCTCTCCAGCCTGGTCCAGGACGATGACATGCTGTATTGGCACAAGCACG GGGATGGATGGAAGACCCCCGTCCCCATGGAGGAGGATCCCCTGCTGGACACAGACATGCTCATGTCGGAATTCAGCGACACCCTCTTCT CCACACTTTCTTCACACCAGCCGGTGGCCTGGCCCAATCCCCGGGAAATAGCACATCTGGGAAATGCAGACATGATCCAGCCGGGACTGA TTCCTTTGCAGCCTAACCTGGACTTCATGGACACCTTTGAGCCTTTCCAGGACCTCTTCTCTTCTAGCCGCTCCATTTTTGGCTCCATGC TACCTGCATCTGCCTCAGCACCTGTACCAGATCCCAACAACCCACCTGCACAGGAGAGCATCCTGCCGACCACAGCCCTCCCCACTGTGA GCCTTCCTGACAGCCTCATCGCGCCCCCTACCGCCCCATCCCTGGCTCACATGGATGAGCAGGGCTGTGAACACACCTCCCGGACTGAGG ACCCGTTTATCCAGCCCACGGACTTCGGTCCCTCAGAGCCGCCACTGAGTGTCCCGCAGCCCTTCCTCCCTGTCTTCACCATGCCCCTGC TGTCTCCCAGCCCCGCCCCACCGCCCATCTCCCCCGTGTTACCATTAGTTCCTCCTCCTGCCACTGCCCTGAACCCCCCGGCTCCACCCA CCTTCCATCAGCCACAGAAGTTTGCTGGAGTCAACAAAGCGCCGTCTGTCATCACCCACACGGCCTCTGCCACCCTCACCCACGATGCCC CCGCCACCACCTTTAGCCAGAGTCAGGGCCTTGTGATCACCACCCATCACCCTGCCCCGTCAGCGGCCCCTTGTGGGCTGGCACTGTCTC CTGTCACCCGGCCTCCCCAGCCACGGTTAACTTTTGTGCACCCCAAACCTGTATCCTTGACTGGGGGCAGGCCTAAGCAGCCCCACAAAA TAGTGCCTGCTCCCAAACCAGAGCCCGTGTCCTTGGTGTTGAAGAATGCCCGTATCGCCCCAGCTGCCTTTTCAGGCCAACCACAAGCGG TGATCATGACGTCAGGGCCTCTGAAGAGAGAAGGGATGTTGGCCTCCACCGTGTCCCAGTCCAACGTGGTCATTGCGCCTGCTGCCATCG CCAGGGCTCCTGGGGTCCCGGAGTTCCACAGCAGCATCCTGGTGACAGATCTCGGCCATGGCACGAGCAGCCCGCCTGCCCCCGTCTCCC GGCTCTTCCCAAGCACAGCGCAAGACCCCCTGGGGAAGGGCGAGCAGGTCCCGCTGCATGGGGGCAGCCCCCAGGTCACTGTCACAGGGC CCAGTCGGGACTGCCCAAACTCAGGGCAGGCCTCTCCGTGTGCATCGGAGCAGAGCCCCAGTCCTCAATCTCCCCAGAACAACTGCTCAG GGAAATCCGACCCCAAAAATGTGGCTGCACTAAAGAACCGGCAGATGAAGCACATCTCAGCTGAGCAGAAAAGGCGCTTCAACATCAAGA TGTGCTTCGACATGCTCAACAGCCTCATCTCCAACAATTCCAAGCTGACCAGTCACGCCATCACACTGCAGAAGACTGTGGAGTACATCA CCAAGCTGCAGCAGGAGAGAGGCCAGATGCAGGAGGAGGCCCGGCGGCTGCGGGAGGAGATCGAGGAGCTCAATGCCACCATCATCTCCT GCCAGCAGCTGCTCCCTGCCACGGGAGTCCCCGTTACCCGGCGCCAGTTTGATCACATGAAAGACATGTTTGACGAATACGTGAAAACCC GGACCTTGCAGAATTGGAAGTTCTGGATTTTCAGCATCATCATCAAGCCGCTGTTTGAGTCGTTCAAGGGCATGGTGTCCACCAGCAGCC TGGAGGAGCTGCACCGGACGGCGCTCTCCTGGCTGGACCAGCACTGCTCCCTGCCCATCCTCAGGCCGATGGTATTGAGCACGCTGCGGC AGCTGAGCACCTCCACCTCCATCCTCACAGACCCGGCACAGCTGCCAGAGCAGGCGTCCAAGGCTGTCACCAGGATTGGCAAGAGATTGG GAGAGTCCTAGCTGCTTAGCTGGCATGTGGCCGCATGAGATGCCAGGAGACCCTTCCCTGCCCATGGAGAGTAGGCTGCGCCCCCCAGCC CTTCCTGACGCTCAGCCTCGGGGCCTCTCTCCAACTCTGCCGGCCCACCGTGGCATCGGGAGGCCATGCTCAGGTCTGAAGCAGGTTTGG GGCCTGCTGACAGCAATAGCCCGCCTTTGGGAACCCCTTGCTGTGAACTCTCTCACTCAGTGACCTCAGTCACCAACCTCCTCTGCCCTC GGGGCAGCCCACACAAAAGGGAAGTGCTGGCCGTGCTGGTCCTGCCCTGCTGGTGGCCTGCCGGGCCTGGCGCCGGTGAGCGGAATCGAT GGGATGAGGGTGACAGGGCCTGCTCCTGTCCTGAGGCCCAGCCTTGTCCCTCCTGCCACGTCCTGTCCACATGCATGCCTCTGCCTGATG CCCTGCTCCACTCTCTGGTCTGCCCGTGGGGCAGTTGGAAGGCGTCTTTCTTTCTCCCCTCAACTCTGACAGCACCCAGCCCTTGTGGAT GGACTTGGGCTTCTATTCAGGCTTATGCATGGCAGGCTGCCAGGGGGAAGTGCCTTCTTCAGAGGTCCTCCAGGACACATGTGTGCAGAA ACGGTGGATGTGGAACACACAGGACCAGAATGGAAGCGTGTGATGCACGGTGGCTGCTCTGGCTGAGAGGCCCTGCTGGGCATGTTTCAT CTGTCCCCTTTTAGCTCCACCTGACATTGCAGGATCCATGGGGACTCAGCCCAGGGCCTTCTCGGATGTCACCTCACCGCTGTGGCCCTT CTGCCGTTCTTCTCCACTTGGCTCCAGCTGCAGCTGTTGACAGATCAAGCATGTCCTGTGGGAGCTTAGAACCCTGAAGTTCTAGTGTCT GAAAGATCAGACTCCACGTCCTGCTGTCAGCCTTGTCATCTTGTCTGATGTCTTTCAGCTGGGAGCCCCAAACCAGGACAGTTCTCGGAC CAAAGATGCCCCCACACTCAAAAGTCTGTCCCGTCTTGTGTTTGGAGAAGGAAACAATGTTGGCAGGCAGCACTCTGTGGTGGTCAGCCC TCAGAGCTGTTTCTAGGCATCTCTCAGATCAGACAGCAAAGAATCTACCCAGATCTGGGCTGGGTGGAGGTGTGGCTGGGCTGGGGGCCA TTCTGAGCCTGCAGTGAGAGTTTGGCCCAGCCTCAGTCCTTGCTCTTCTCTGGCTACCTCTGCAGGGAGCTGCAGGGGCAAGCACTCTCT CCAGCACTCAGGAAGCCCGGCCGAGGGTACCTCCTCGTGGAAAGAATGCACTTTAAAGCTCTGCTGAGGAGTTCGGAGCCCAGGCTTTCA GGCGACCTCTGCCCTCCCTGCCTCTCCTCACCCTCCCTCTCTTCCTGCAGGGCCTGGGAAGGGCTTTGAGGGAGCCTGGGAGCCATGTGA AGAGGGGCACGCCTGGGCTGTCCCACAGTTTAGATCCAGTTGGAGGTTCTCCCTGGCTCCTGCAGGCCTGCGGGGATCTCTCCCCACTTC AGGCCTCCGGCCAGCTGCCTGCCCTCTTGTCTGTGCTTCAGCCCTGCACAAAAGCAGCTTGGTGACACCACTCAGCCACCCAGAGTACGT GTTTACAGGCTTTCCAGATCACCTTCCTGTGGGGTGAACGTAATGAGGCGGGGCTGGTCCTTGGAATTTCCCCTGGAAAATGGTAACAGA CTCCATCCTTGACCCGGGGATGAGCATGAAGGCATTGTCCCAAAGGCAGAGGCCACCGTGGTAGGAATTCCACCAAGGCCAGAAGGGAAA AAGGAAGAACCCACCGTGTCTGGCTGTGCGGGCCCTGGGGAGGGTCGTGAGTGCAGCCCCTCTCTACTTCTGTGCCTTTGTAAAACGTGT AGATAACCGCAGTGGTTGGCTGAGCCAAGAACTCTCCTAAATCAGTGGCTTTCTCCCCACCCCTTGCTGGGGAGTCATTTTTAAAAAAAT CTGTGGGATATAAAATTGGCCTCCTGCTGCTTCAGCCTACCTCTCCCTCTGCTGACTTAATGTCGTGATTCTGTTTCTTCAGATATTTAA GGCTGTTAGGTTGTGTGAGCCTTGAAGTGTGTGTGTGTGTCCCAGCGACTGTCCACTGTCCAGGAGATGCATGTCTTTGTATTGGAGATA TTTCTGTAACTCATTCTCTTGGTGCTCACGATTGCCATGGCCATAGGGCCACAGTGCCGTATCTGCTGCAGACATGATTGTTTCTTGTTC TAGAGGTTTTCTTGTTTTCGAATCTTGCCTGATGAATCCAGCCAGACCAAGGGGCCTAGATTTGACCTCTGTCCTGGGCTCCTGGGCCAG GTGCAGGAACATCTGAGGCCACTCTGCTGGCCACCTCCAGTGGGTGCTGACCACAGGATGGGCTTTGTTTACACTCATTTTCACCCTGAT TCTTGCCCCCACTTTCATAAAAGAAACTTCAAAATGCTGACGCTTTGGAGAGTAAGAAAATCAATCTTGGCTGGGCACGGTGGCTCCTGC CTGTGATCCTAGCACTTTGGGAGGCTGAAGCTGAAGGATCACTTGAGCTCAGGAGTTGGAGACCAACCCTGGCAACATAACAAGACCCTG TCTCTACAAAAAAAAAAAAAAAAAAAAAAAATTTAGCCGTGTGTAGCAGTGAGTGCCTGTGGTCCCAGCTACTTGGGCCTGAGGCTGGAG GATTGCTTGAGCCTAGAAGTTGCAGTGAGCTATGATCATGCCACACTGTACTCCAGCCTGGATGACAGAGTGAAACCCTGCCTCTAAATA AAAGAAAATAGAGGCAGACTGTGGTGGCTCACGCCTGTAATCCCAGCACTTTGGTAGGCCAAGGCGGGTGGATCACCTGCAGTCAGGAGT TCACAACCAGCCTGACCAACATGGTGAAACCCTGTCTATACTAAAAATACAAAACATTAGCCACGTGTGGTGGTACACGCCTGTAATCCC AGCTACTCGGGAGGCTGAGTCAGGAGAATCACTTGAACCTGGGAGGCGGAGGTTGCAGTGAGCTGAGATTGTACCACTGCACTCCAGCCT GGGTGACAGAGCAAAACCCTATCTCAAAAAAAAAAAGAAGAAAAAAATAGAAAATCCAAAAAGAAAAACCAGAAGGCCTGCTGGCGTATA AATCCCTGGGCGGGTTTCTGATAAATTGCCCTGTGCTGTCAGCCCCTGCACTGCACTGCTGCCTTCCATGGTGGGTGGGAGACCCCAGAG CGGGGCAGGCGCCACTGAGGGCTTTTCTTGGAGTCGGCCAGCAGGCCACGCAGCATCCGGCACCCTGGGCGGGCTGGCTAGCTGCCTTCT TAGGACATCTCTACTTTGAAGGATTTTACCGCAGGAAGCAATAGCAGCGCTGGCCATTGGTGCTGATGACAGCATTGGGTCTGGTTGAGG GGAAGGGGCTGGAAAGCAGCAGACCCCCCCAGTGCTGCGCATGGTCCCGGAGCTGTCAGCCAGGGGAGTGGGGTCAGCCGTCAGCCAGCC CTCCCATTTCCCGCCCATGGGCCCTGACCACACTCCCTTTTCTAGAAGTCAATCCTAAGGTTTCTCTGCTCTGGCTAAGAGGATGTAAAT TTGGATTCTTAGAGGGCATGGCACCCCCAGTCCCTGCCCAGATAAAGTAGCACAGTGGCAGGCAGCACCTCTGTCTGTTGCTGACGTTGG GGGGCTTACACACCCACCTCATCTCCGTGCACAGCCATGACTGGCCCTGCCGGCAGCTGGGGTGCAGGTAAGGGTCTCTCTCATAGAGGG GAGCTGCAGCTGAGAACTGGCGAGGCCCCTTCCTCCAAGGCCCTAGCTGGCCCCCGGGTGAACCTGAGGTGGCAGGTTCAGGTTTTCAAG ATGGTGAGGTCTCGCTGTCTGCTGGACAGTACGTTAGGCTCTCAGAACTCATGGGTGTGGAGCTGGGCCTGTCCCGGGCCAGTGGACCCC TGTGTGTGGGGGATTTGGGGTGCTGTGGGCCTGGTTATGCACTGGCAGATGGACCTTGCTTTGGTCCAGCTCTTTTCCTTACCCTGGCTC TGACGTGGGAAGGCTTGGAGGGCCCGTCTCATCACCCCCGTTCGCCCTCAGCTGTCCCTTTCCCTTGTCGCCTGGCCGCTGCCTCGCCCG CCTGAGGCCTCCTAGCAGGCAGCCTGGGTGTGAGTTGAGCCTCTCTCTTTTCCCTCTGGTGGGAAAGTGGCCTTTCCCTCAACACCTGCT CCCCGGCCCCAGAGGAACCCACCTGTTTTGGAGCTCAGCTTGGCCCAGCGTTTCCTTGGGGAAGGGAAAGGAGGGCTGGACAGCACTGAT CCGGGCAGGCAGCGTGTGCAGCAGTGGCCAGCCAGAGTGCCAAAGATGCACGGGGATGTGGTGTGTGGCTCCGGGCCCTCGACATCTCTG CTTTGGGGGATTTTTACCTTGTCTGCACACTTGTCAGGGGAGAGGGGACAGCAAGGTGGGAGGTTGAAGAGCTTTGAGGCTCAGCAGCAT GTTTGTGGCATTCGGTGGACACCATGGCCTTGGGCGGCTGGACAGGTTTTTGTGATGTGAGGGACACGCATGGGGCACATGGTAAGCTTG GCAAGGGCTCCAGGAACGCTGACGAAGGGTTTTAGGACCCCCACCCCCATGCCTGTACCAGGGCTGGCCTCCAGAGCGGGTGAGGACAGA GCAGCTGTGGGCTTTTCATTCTGAGGTCTTGGCCCCCCTGGCCACCGCAAGGGACTCTTTGCTTGTCAGGGCTTGCAAAAACCAACCTTC GAGAAAGAAAAGGGAACTCTTCACGTTGAATGTTGACTTTGTGTGTATGCGTGTGTGTGTGTGTGTGTGCACGCGCGCGTGTGCGTGTTG ACTTCATGGAATTTTGTTTTGTGAAATTCCCCTCCAATCGTGTCAGAATTTACCTCCATGCCCCAGTCACACTGTTGGTTCTGCGCTCTG AACCTGGGTGTAGCTCATTTGAAGGACTCTCTTCTGCGTTTCCTAACAGTTATTTGGTGGTCTCAAGAGTTGAGGTTGTGGAGGGTTGGG AGAAACTGAAGTTCTATACATTTCCATAGAGTTTACATCCTGCAGTTAAAAGGCAGGGAGGGCTCAGCCCGGGCCCCACAGCTCCAGGCC ATCCCCTACGGGCTGCCCACAGTGCCCCCTTTTCTCTAGCCGAATCTTTTTCGAACAGCCCGGGAAAGGAAAACGGATTCACTTGCTGAT TTTGTTCACGGCGGAAGCACCATGTTCCGTTCCTTTTTCAGGTTCAGTTTGTTGTGTAAATGGCGGTTTTTTCTGGTGTGAGCTTTGGTG ATGGTGGCAGGGCTCCTTTGAAGAGATGGTTCCACCTCGTGGTCTGAAGAACAAACCAGAGAAGAGTCTGGTTTGGCCAGAGGCCCCCTC CGGTCCACGTCACCCTGAGTACACCCCTCTGATTGCTCTGCTGTCAAGAAGCACGTTTCCACCAGCTGTATTCAACACTACAATGCATTT >38438_38438_1_IFT57-MLXIP_IFT57_chr3_107886639_ENST00000264538_MLXIP_chr12_122613683_ENST00000319080_length(amino acids)=1007AA_BP=290 MDSVRSAMTAALAVVTTSGLEDGVPRSRGEGTGEVVLERGPGAAYHMFVVMEDLVEKLKLLRYEEEFLRKSNLKAPSRHYFALPTNPGEQ FYMFCTLAAWLINKAGRPFEQPQEYDDPNATISNILSELRSFGRTADFPPSKLKSGYGEHVCYVLDCFAEEALKYIGFTWKRPIYPVEEL EEESVAEDDAELTLNKVDEEFVEEETDNEENFIDLNVLKAQTYHLDMNETAKQEDILESTTDAAEWSLEVERVLPQLKVTIRTDNKDWRI HVDQMHQHRSGIESALKETKAITTEGKYWKSRIEIVIREYHKWRTYFKKRLQQHKDEDLSSLVQDDDMLYWHKHGDGWKTPVPMEEDPLL DTDMLMSEFSDTLFSTLSSHQPVAWPNPREIAHLGNADMIQPGLIPLQPNLDFMDTFEPFQDLFSSSRSIFGSMLPASASAPVPDPNNPP AQESILPTTALPTVSLPDSLIAPPTAPSLAHMDEQGCEHTSRTEDPFIQPTDFGPSEPPLSVPQPFLPVFTMPLLSPSPAPPPISPVLPL VPPPATALNPPAPPTFHQPQKFAGVNKAPSVITHTASATLTHDAPATTFSQSQGLVITTHHPAPSAAPCGLALSPVTRPPQPRLTFVHPK PVSLTGGRPKQPHKIVPAPKPEPVSLVLKNARIAPAAFSGQPQAVIMTSGPLKREGMLASTVSQSNVVIAPAAIARAPGVPEFHSSILVT DLGHGTSSPPAPVSRLFPSTAQDPLGKGEQVPLHGGSPQVTVTGPSRDCPNSGQASPCASEQSPSPQSPQNNCSGKSDPKNVAALKNRQM KHISAEQKRRFNIKMCFDMLNSLISNNSKLTSHAITLQKTVEYITKLQQERGQMQEEARRLREEIEELNATIISCQQLLPATGVPVTRRQ FDHMKDMFDEYVKTRTLQNWKFWIFSIIIKPLFESFKGMVSTSSLEELHRTALSWLDQHCSLPILRPMVLSTLRQLSTSTSILTDPAQLP -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for IFT57-MLXIP |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for IFT57-MLXIP |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for IFT57-MLXIP |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
