
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:IKZF2-MICU2 (FusionGDB2 ID:39399) |
Fusion Gene Summary for IKZF2-MICU2 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: IKZF2-MICU2 | Fusion gene ID: 39399 | Hgene | Tgene | Gene symbol | IKZF2 | MICU2 | Gene ID | 22807 | 221154 |
| Gene name | IKAROS family zinc finger 2 | mitochondrial calcium uptake 2 | |
| Synonyms | ANF1A2|HELIOS|ZNF1A2|ZNFN1A2 | 1110008L20Rik|EFHA1 | |
| Cytomap | 2q34 | 13q12.11 | |
| Type of gene | protein-coding | protein-coding | |
| Description | zinc finger protein Heliosikaros family zinc finger protein 2zinc finger DNA binding protein Helioszinc finger protein, subfamily 1A, 2 (Helios) | calcium uptake protein 2, mitochondrialEF hand domain family A1EF hand domain family, member A1EF-hand domain-containing family member A1Smhs2 homolog | |
| Modification date | 20200313 | 20200315 | |
| UniProtAcc | Q9UKS7 | Q8IYU8 | |
| Ensembl transtripts involved in fusion gene | ENST00000342002, ENST00000374319, ENST00000413091, ENST00000421754, ENST00000434687, ENST00000451136, ENST00000457361, ENST00000374327, ENST00000442445, | ENST00000479790, ENST00000382374, | |
| Fusion gene scores | * DoF score | 7 X 7 X 5=245 | 13 X 13 X 6=1014 |
| # samples | 7 | 13 | |
| ** MAII score | log2(7/245*10)=-1.8073549220576 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(13/1014*10)=-2.96347412397489 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: IKZF2 [Title/Abstract] AND MICU2 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | IKZF2(213914437)-MICU2(22069457), # samples:2 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Tgene | MICU2 | GO:0006851 | mitochondrial calcium ion transmembrane transport | 24560927 |
| Tgene | MICU2 | GO:0051562 | negative regulation of mitochondrial calcium ion concentration | 24560927 |
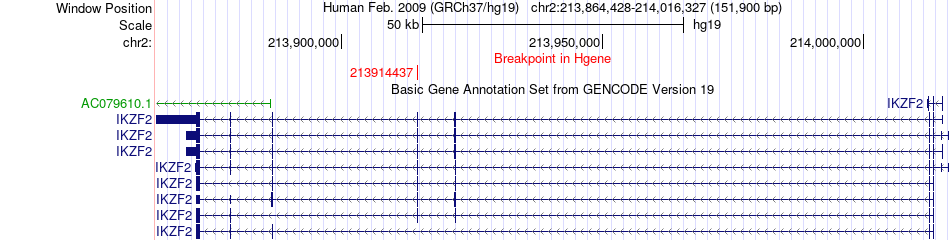
 Fusion gene breakpoints across IKZF2 (5'-gene) Fusion gene breakpoints across IKZF2 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
 |
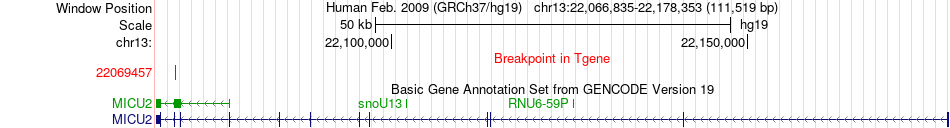
 Fusion gene breakpoints across MICU2 (3'-gene) Fusion gene breakpoints across MICU2 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | PRAD | TCGA-EJ-5512-01A | IKZF2 | chr2 | 213914437 | - | MICU2 | chr13 | 22069457 | - |
| ChimerDB4 | PRAD | TCGA-EJ-5512 | IKZF2 | chr2 | 213914437 | - | MICU2 | chr13 | 22069457 | - |
Top |
Fusion Gene ORF analysis for IKZF2-MICU2 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-5UTR | ENST00000342002 | ENST00000479790 | IKZF2 | chr2 | 213914437 | - | MICU2 | chr13 | 22069457 | - |
| 5CDS-5UTR | ENST00000374319 | ENST00000479790 | IKZF2 | chr2 | 213914437 | - | MICU2 | chr13 | 22069457 | - |
| 5CDS-5UTR | ENST00000413091 | ENST00000479790 | IKZF2 | chr2 | 213914437 | - | MICU2 | chr13 | 22069457 | - |
| 5CDS-5UTR | ENST00000421754 | ENST00000479790 | IKZF2 | chr2 | 213914437 | - | MICU2 | chr13 | 22069457 | - |
| 5CDS-5UTR | ENST00000434687 | ENST00000479790 | IKZF2 | chr2 | 213914437 | - | MICU2 | chr13 | 22069457 | - |
| 5CDS-5UTR | ENST00000451136 | ENST00000479790 | IKZF2 | chr2 | 213914437 | - | MICU2 | chr13 | 22069457 | - |
| 5CDS-5UTR | ENST00000457361 | ENST00000479790 | IKZF2 | chr2 | 213914437 | - | MICU2 | chr13 | 22069457 | - |
| In-frame | ENST00000342002 | ENST00000382374 | IKZF2 | chr2 | 213914437 | - | MICU2 | chr13 | 22069457 | - |
| In-frame | ENST00000374319 | ENST00000382374 | IKZF2 | chr2 | 213914437 | - | MICU2 | chr13 | 22069457 | - |
| In-frame | ENST00000413091 | ENST00000382374 | IKZF2 | chr2 | 213914437 | - | MICU2 | chr13 | 22069457 | - |
| In-frame | ENST00000421754 | ENST00000382374 | IKZF2 | chr2 | 213914437 | - | MICU2 | chr13 | 22069457 | - |
| In-frame | ENST00000434687 | ENST00000382374 | IKZF2 | chr2 | 213914437 | - | MICU2 | chr13 | 22069457 | - |
| In-frame | ENST00000451136 | ENST00000382374 | IKZF2 | chr2 | 213914437 | - | MICU2 | chr13 | 22069457 | - |
| In-frame | ENST00000457361 | ENST00000382374 | IKZF2 | chr2 | 213914437 | - | MICU2 | chr13 | 22069457 | - |
| intron-3CDS | ENST00000374327 | ENST00000382374 | IKZF2 | chr2 | 213914437 | - | MICU2 | chr13 | 22069457 | - |
| intron-3CDS | ENST00000442445 | ENST00000382374 | IKZF2 | chr2 | 213914437 | - | MICU2 | chr13 | 22069457 | - |
| intron-5UTR | ENST00000374327 | ENST00000479790 | IKZF2 | chr2 | 213914437 | - | MICU2 | chr13 | 22069457 | - |
| intron-5UTR | ENST00000442445 | ENST00000479790 | IKZF2 | chr2 | 213914437 | - | MICU2 | chr13 | 22069457 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000457361 | IKZF2 | chr2 | 213914437 | - | ENST00000382374 | MICU2 | chr13 | 22069457 | - | 1555 | 743 | 145 | 1005 | 286 |
| ENST00000342002 | IKZF2 | chr2 | 213914437 | - | ENST00000382374 | MICU2 | chr13 | 22069457 | - | 1555 | 743 | 145 | 1005 | 286 |
| ENST00000434687 | IKZF2 | chr2 | 213914437 | - | ENST00000382374 | MICU2 | chr13 | 22069457 | - | 1696 | 884 | 304 | 1146 | 280 |
| ENST00000374319 | IKZF2 | chr2 | 213914437 | - | ENST00000382374 | MICU2 | chr13 | 22069457 | - | 1618 | 806 | 304 | 1068 | 254 |
| ENST00000451136 | IKZF2 | chr2 | 213914437 | - | ENST00000382374 | MICU2 | chr13 | 22069457 | - | 1308 | 496 | 0 | 758 | 252 |
| ENST00000413091 | IKZF2 | chr2 | 213914437 | - | ENST00000382374 | MICU2 | chr13 | 22069457 | - | 1386 | 574 | 0 | 836 | 278 |
| ENST00000421754 | IKZF2 | chr2 | 213914437 | - | ENST00000382374 | MICU2 | chr13 | 22069457 | - | 1308 | 496 | 0 | 758 | 252 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000457361 | ENST00000382374 | IKZF2 | chr2 | 213914437 | - | MICU2 | chr13 | 22069457 | - | 0.000602682 | 0.9993973 |
| ENST00000342002 | ENST00000382374 | IKZF2 | chr2 | 213914437 | - | MICU2 | chr13 | 22069457 | - | 0.000602682 | 0.9993973 |
| ENST00000434687 | ENST00000382374 | IKZF2 | chr2 | 213914437 | - | MICU2 | chr13 | 22069457 | - | 0.000932356 | 0.9990677 |
| ENST00000374319 | ENST00000382374 | IKZF2 | chr2 | 213914437 | - | MICU2 | chr13 | 22069457 | - | 0.001308477 | 0.9986915 |
| ENST00000451136 | ENST00000382374 | IKZF2 | chr2 | 213914437 | - | MICU2 | chr13 | 22069457 | - | 0.001385558 | 0.99861443 |
| ENST00000413091 | ENST00000382374 | IKZF2 | chr2 | 213914437 | - | MICU2 | chr13 | 22069457 | - | 0.000913586 | 0.9990864 |
| ENST00000421754 | ENST00000382374 | IKZF2 | chr2 | 213914437 | - | MICU2 | chr13 | 22069457 | - | 0.001385558 | 0.99861443 |
Top |
Fusion Genomic Features for IKZF2-MICU2 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
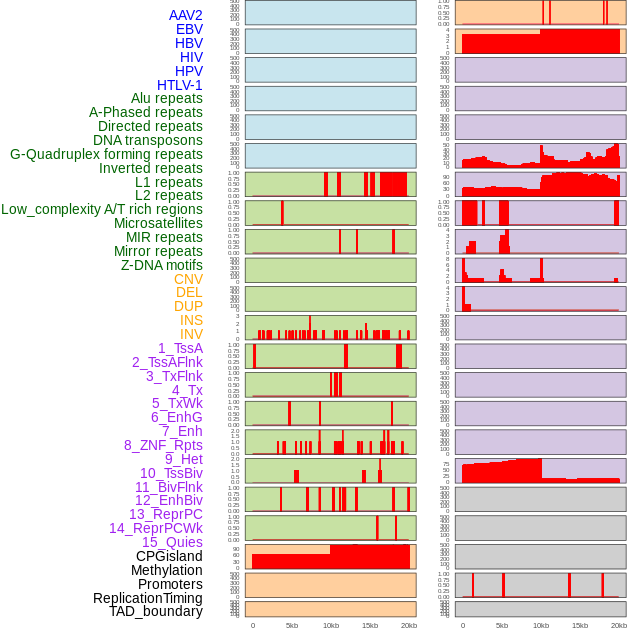
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for IKZF2-MICU2 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr2:213914437/chr13:22069457) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| IKZF2 | MICU2 |
| FUNCTION: Associates with Ikaros at centromeric heterochromatin. | FUNCTION: Key regulator of mitochondrial calcium uniporter (MCU) required to limit calcium uptake by MCU when cytoplasmic calcium is low (PubMed:24503055, PubMed:24560927, PubMed:26903221). MICU1 and MICU2 form a disulfide-linked heterodimer that stimulate and inhibit MCU activity, depending on the concentration of calcium (PubMed:24560927). MICU2 acts as a gatekeeper of MCU that senses calcium level via its EF-hand domains: prevents channel opening at resting calcium, avoiding energy dissipation and cell-death triggering (PubMed:24560927). {ECO:0000269|PubMed:24503055, ECO:0000269|PubMed:24560927, ECO:0000269|PubMed:26387864, ECO:0000269|PubMed:26903221}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | IKZF2 | chr2:213914437 | chr13:22069457 | ENST00000374319 | - | 6 | 9 | 112_134 | 165 | 501.0 | Zinc finger | C2H2-type 1 |
| Hgene | IKZF2 | chr2:213914437 | chr13:22069457 | ENST00000374319 | - | 6 | 9 | 140_162 | 165 | 501.0 | Zinc finger | C2H2-type 2 |
| Hgene | IKZF2 | chr2:213914437 | chr13:22069457 | ENST00000434687 | - | 6 | 9 | 112_134 | 191 | 527.0 | Zinc finger | C2H2-type 1 |
| Hgene | IKZF2 | chr2:213914437 | chr13:22069457 | ENST00000434687 | - | 6 | 9 | 140_162 | 191 | 527.0 | Zinc finger | C2H2-type 2 |
| Hgene | IKZF2 | chr2:213914437 | chr13:22069457 | ENST00000434687 | - | 6 | 9 | 168_190 | 191 | 527.0 | Zinc finger | C2H2-type 3 |
| Hgene | IKZF2 | chr2:213914437 | chr13:22069457 | ENST00000457361 | - | 5 | 8 | 112_134 | 191 | 527.0 | Zinc finger | C2H2-type 1 |
| Hgene | IKZF2 | chr2:213914437 | chr13:22069457 | ENST00000457361 | - | 5 | 8 | 140_162 | 191 | 527.0 | Zinc finger | C2H2-type 2 |
| Hgene | IKZF2 | chr2:213914437 | chr13:22069457 | ENST00000457361 | - | 5 | 8 | 168_190 | 191 | 527.0 | Zinc finger | C2H2-type 3 |
| Tgene | MICU2 | chr2:213914437 | chr13:22069457 | ENST00000382374 | 9 | 12 | 362_397 | 347 | 435.0 | Domain | EF-hand 4 |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | IKZF2 | chr2:213914437 | chr13:22069457 | ENST00000374319 | - | 6 | 9 | 168_190 | 165 | 501.0 | Zinc finger | C2H2-type 3 |
| Hgene | IKZF2 | chr2:213914437 | chr13:22069457 | ENST00000374319 | - | 6 | 9 | 196_219 | 165 | 501.0 | Zinc finger | C2H2-type 4 |
| Hgene | IKZF2 | chr2:213914437 | chr13:22069457 | ENST00000374319 | - | 6 | 9 | 471_493 | 165 | 501.0 | Zinc finger | C2H2-type 5 |
| Hgene | IKZF2 | chr2:213914437 | chr13:22069457 | ENST00000374319 | - | 6 | 9 | 499_523 | 165 | 501.0 | Zinc finger | C2H2-type 6 |
| Hgene | IKZF2 | chr2:213914437 | chr13:22069457 | ENST00000434687 | - | 6 | 9 | 196_219 | 191 | 527.0 | Zinc finger | C2H2-type 4 |
| Hgene | IKZF2 | chr2:213914437 | chr13:22069457 | ENST00000434687 | - | 6 | 9 | 471_493 | 191 | 527.0 | Zinc finger | C2H2-type 5 |
| Hgene | IKZF2 | chr2:213914437 | chr13:22069457 | ENST00000434687 | - | 6 | 9 | 499_523 | 191 | 527.0 | Zinc finger | C2H2-type 6 |
| Hgene | IKZF2 | chr2:213914437 | chr13:22069457 | ENST00000457361 | - | 5 | 8 | 196_219 | 191 | 527.0 | Zinc finger | C2H2-type 4 |
| Hgene | IKZF2 | chr2:213914437 | chr13:22069457 | ENST00000457361 | - | 5 | 8 | 471_493 | 191 | 527.0 | Zinc finger | C2H2-type 5 |
| Hgene | IKZF2 | chr2:213914437 | chr13:22069457 | ENST00000457361 | - | 5 | 8 | 499_523 | 191 | 527.0 | Zinc finger | C2H2-type 6 |
| Tgene | MICU2 | chr2:213914437 | chr13:22069457 | ENST00000382374 | 9 | 12 | 2_50 | 347 | 435.0 | Compositional bias | Note=Ala-rich | |
| Tgene | MICU2 | chr2:213914437 | chr13:22069457 | ENST00000382374 | 9 | 12 | 172_207 | 347 | 435.0 | Domain | EF-hand 1 | |
| Tgene | MICU2 | chr2:213914437 | chr13:22069457 | ENST00000382374 | 9 | 12 | 227_262 | 347 | 435.0 | Domain | EF-hand 2 | |
| Tgene | MICU2 | chr2:213914437 | chr13:22069457 | ENST00000382374 | 9 | 12 | 293_328 | 347 | 435.0 | Domain | EF-hand 3 |
Top |
Fusion Gene Sequence for IKZF2-MICU2 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >39399_39399_1_IKZF2-MICU2_IKZF2_chr2_213914437_ENST00000342002_MICU2_chr13_22069457_ENST00000382374_length(transcript)=1555nt_BP=743nt GGGCTTGGCTTGGAGGGGGCAAGGGAGGGAAAGAGAGAAGGGGGAAACACAAAAAACTTCTTTCTTTCTCCCTCCGTTTATCTTCAGCCC GACATTGTCACCTCCTCTTTGAGGGGTTAGAAGAAGCTGAGATCTCCCGACAGAGCTGGAAATGCATTGCACTTTGACTATGGAAACAGA GGCTATTGATGGCTATATAACGTGTGACAATGAGCTTTCACCCGAAAGGGAGCACTCCAATATGGCAATTGACCTCACCTCAAGCACACC CAATGGACAGCATGCCTCACCAAGTCACATGACAAGCACAAATTCAGTAAAGCTAGAAATGCAGAGTGATGAAGAGTGTGACAGGAAACC CCTGAGCCGTGAAGATGAGATCAGGGGCCATGATGAGGGTAGCAGCCTAGAAGAACCCCTAATTGAGAGCAGCGAGGTGGCTGACAACAG GAAAGTCCAGGAGCTTCAAGGCGAGGGAGGAATCCGGCTTCCGAATGGTAAACTGAAATGTGACGTCTGTGGCATGGTTTGCATTGGGCC CAATGTGCTTATGGTACATAAAAGGAGTCACACTGGTGAACGCCCCTTCCACTGTAACCAGTGTGGAGCTTCTTTTACTCAGAAGGGCAA CCTTCTGAGACACATAAAGTTACACTCTGGAGAGAAGCCGTTCAAATGTCCTTTCTGTAGCTACGCCTGTAGAAGAAGGGACGCCCTCAC AGGACACCTCAGGACCCATTCTGCGGAGTTTAAGAGAGCTGTGAAAGTAGCAACAGGACAAGAACTCTCAAACAATATTTTGGACACTGT CTTTAAGATCTTTGATTTGGATGGTGATGAATGTCTTAGTCATGAAGAGTTTCTTGGGGTGTTAAAAAACAGAATGCATCGAGGTTTATG GGTACCACAACATCAGAGTATACAAGAATACTGGAAGTGTGTGAAGAAAGAAAGCATTAAAGGAGTAAAAGAAGTCTGGAAACAAGCTGG AAAAGGTCTTTTTTAATAAAAGATATAATAGTATGGCAATTATATTGTTCCAAATGTCAAAATTTGTGATTTTTTAGAAGTACTTGCTAT TTATCTTCTTAAGTCTTCATTGATATTCTGTGTGAAATAAGCATGTCTTGTACTTGCTTTCTGATTCATAATTTTATTAAAGAACTTAGT AGAAAGAAAAGTAAGTATAAAAATAGATATTGGATTCTGTCAGAAGGCCTAGATTTGAAATAATGTTTTGTACTTCGGTAAGATGGAAAA CTTAGTGATTCACTGATTTCTTAGACACTCTAATATGATATGCTTTCTGGAAGGATAAAACAAATACATATGGGAAAAAGTACTTGAGAC CAAGGCCAGCATCAATTCCAGACATCTTCATGTTCCTAATAGGCTAAATGAAGTTAAAAACTTATTTCAGATTTTTCTCATCTGTACCTT ATATCTCATAAATTTATTGCATATTTTATGTCAGTAGCTTAGCTGTTTATTGTCTTTAAAATAACATGTAAACTTCAATGTTCTATCTGG >39399_39399_1_IKZF2-MICU2_IKZF2_chr2_213914437_ENST00000342002_MICU2_chr13_22069457_ENST00000382374_length(amino acids)=286AA_BP=199 MEMHCTLTMETEAIDGYITCDNELSPEREHSNMAIDLTSSTPNGQHASPSHMTSTNSVKLEMQSDEECDRKPLSREDEIRGHDEGSSLEE PLIESSEVADNRKVQELQGEGGIRLPNGKLKCDVCGMVCIGPNVLMVHKRSHTGERPFHCNQCGASFTQKGNLLRHIKLHSGEKPFKCPF CSYACRRRDALTGHLRTHSAEFKRAVKVATGQELSNNILDTVFKIFDLDGDECLSHEEFLGVLKNRMHRGLWVPQHQSIQEYWKCVKKES -------------------------------------------------------------- >39399_39399_2_IKZF2-MICU2_IKZF2_chr2_213914437_ENST00000374319_MICU2_chr13_22069457_ENST00000382374_length(transcript)=1618nt_BP=806nt GCTCCTCGCTGAAGATGGAGGAAGTAAAAACAGGATTACCCTTAGCTACAGATCCACTGCCTTAGTTTCCACCACCAACTGCAGTGCACA AACACACGTTAGGCACAGGAAAGAAAGAAAGACAGAGGACACATTAACAGTAAACACAAACAAAAGGGTGATGGGATTATTTTACTGCAT GCACTGCTGAGCCCGACATTGTCACCTCCTCTTTGAGGGGTTAGAAGAAGCTGAGATCTCCCGACAGAGCTGGAAATGGTGATGAATCTT TTTTAATCAAAGGACAATTTCTTTTCATTGCACTTTGACTATGGAAACAGAGGCTATTGATGGCTATATAACGTGTGACAATGAGCTTTC ACCCGAAAGGGAGCACTCCAATATGGCAATTGACCTCACCTCAAGCACACCCAATGGACAGCATGCCTCACCAAGTCACATGACAAGCAC AAATTCAGTAAAGCTAGAAATGCAGAGTGATGAAGAGTGTGACAGGAAACCCCTGAGCCGTGAAGATGAGATCAGGGGCCATGATGAGGG TAGCAGCCTAGAAGAACCCCTAATTGAGAGCAGCGAGGTGGCTGACAACAGGAAAGTCCAGGAGCTTCAAGGCGAGGGAGGAATCCGGCT TCCGAATGGTGAACGCCCCTTCCACTGTAACCAGTGTGGAGCTTCTTTTACTCAGAAGGGCAACCTTCTGAGACACATAAAGTTACACTC TGGAGAGAAGCCGTTCAAATGTCCTTTCTGTAGCTACGCCTGTAGAAGAAGGGACGCCCTCACAGGACACCTCAGGACCCATTCTGCGGA GTTTAAGAGAGCTGTGAAAGTAGCAACAGGACAAGAACTCTCAAACAATATTTTGGACACTGTCTTTAAGATCTTTGATTTGGATGGTGA TGAATGTCTTAGTCATGAAGAGTTTCTTGGGGTGTTAAAAAACAGAATGCATCGAGGTTTATGGGTACCACAACATCAGAGTATACAAGA ATACTGGAAGTGTGTGAAGAAAGAAAGCATTAAAGGAGTAAAAGAAGTCTGGAAACAAGCTGGAAAAGGTCTTTTTTAATAAAAGATATA ATAGTATGGCAATTATATTGTTCCAAATGTCAAAATTTGTGATTTTTTAGAAGTACTTGCTATTTATCTTCTTAAGTCTTCATTGATATT CTGTGTGAAATAAGCATGTCTTGTACTTGCTTTCTGATTCATAATTTTATTAAAGAACTTAGTAGAAAGAAAAGTAAGTATAAAAATAGA TATTGGATTCTGTCAGAAGGCCTAGATTTGAAATAATGTTTTGTACTTCGGTAAGATGGAAAACTTAGTGATTCACTGATTTCTTAGACA CTCTAATATGATATGCTTTCTGGAAGGATAAAACAAATACATATGGGAAAAAGTACTTGAGACCAAGGCCAGCATCAATTCCAGACATCT TCATGTTCCTAATAGGCTAAATGAAGTTAAAAACTTATTTCAGATTTTTCTCATCTGTACCTTATATCTCATAAATTTATTGCATATTTT >39399_39399_2_IKZF2-MICU2_IKZF2_chr2_213914437_ENST00000374319_MICU2_chr13_22069457_ENST00000382374_length(amino acids)=254AA_BP=167 MTMETEAIDGYITCDNELSPEREHSNMAIDLTSSTPNGQHASPSHMTSTNSVKLEMQSDEECDRKPLSREDEIRGHDEGSSLEEPLIESS EVADNRKVQELQGEGGIRLPNGERPFHCNQCGASFTQKGNLLRHIKLHSGEKPFKCPFCSYACRRRDALTGHLRTHSAEFKRAVKVATGQ -------------------------------------------------------------- >39399_39399_3_IKZF2-MICU2_IKZF2_chr2_213914437_ENST00000413091_MICU2_chr13_22069457_ENST00000382374_length(transcript)=1386nt_BP=574nt ATGGAAACAGAGGCTATTGATGGCTATATAACGTGTGACAATGAGCTTTCACCCGAAAGGGAGCACTCCAATATGGCAATTGACCTCACC TCAAGCACACCCAATGGACAGCATGCCTCACCAAGTCACATGACAAGCACAAATTCAGTAAAGCTAGAAATGCAGAGTGATGAAGAGTGT GACAGGAAACCCCTGAGCCGTGAAGATGAGATCAGGGGCCATGATGAGGGTAGCAGCCTAGAAGAACCCCTAATTGAGAGCAGCGAGGTG GCTGACAACAGGAAAGTCCAGGAGCTTCAAGGCGAGGGAGGAATCCGGCTTCCGAATGGTAAACTGAAATGTGACGTCTGTGGCATGGTT TGCATTGGGCCCAATGTGCTTATGGTACATAAAAGGAGTCACACTGGTGAACGCCCCTTCCACTGTAACCAGTGTGGAGCTTCTTTTACT CAGAAGGGCAACCTTCTGAGACACATAAAGTTACACTCTGGAGAGAAGCCGTTCAAATGTCCTTTCTGTAGCTACGCCTGTAGAAGAAGG GACGCCCTCACAGGACACCTCAGGACCCATTCTGCGGAGTTTAAGAGAGCTGTGAAAGTAGCAACAGGACAAGAACTCTCAAACAATATT TTGGACACTGTCTTTAAGATCTTTGATTTGGATGGTGATGAATGTCTTAGTCATGAAGAGTTTCTTGGGGTGTTAAAAAACAGAATGCAT CGAGGTTTATGGGTACCACAACATCAGAGTATACAAGAATACTGGAAGTGTGTGAAGAAAGAAAGCATTAAAGGAGTAAAAGAAGTCTGG AAACAAGCTGGAAAAGGTCTTTTTTAATAAAAGATATAATAGTATGGCAATTATATTGTTCCAAATGTCAAAATTTGTGATTTTTTAGAA GTACTTGCTATTTATCTTCTTAAGTCTTCATTGATATTCTGTGTGAAATAAGCATGTCTTGTACTTGCTTTCTGATTCATAATTTTATTA AAGAACTTAGTAGAAAGAAAAGTAAGTATAAAAATAGATATTGGATTCTGTCAGAAGGCCTAGATTTGAAATAATGTTTTGTACTTCGGT AAGATGGAAAACTTAGTGATTCACTGATTTCTTAGACACTCTAATATGATATGCTTTCTGGAAGGATAAAACAAATACATATGGGAAAAA GTACTTGAGACCAAGGCCAGCATCAATTCCAGACATCTTCATGTTCCTAATAGGCTAAATGAAGTTAAAAACTTATTTCAGATTTTTCTC ATCTGTACCTTATATCTCATAAATTTATTGCATATTTTATGTCAGTAGCTTAGCTGTTTATTGTCTTTAAAATAACATGTAAACTTCAAT >39399_39399_3_IKZF2-MICU2_IKZF2_chr2_213914437_ENST00000413091_MICU2_chr13_22069457_ENST00000382374_length(amino acids)=278AA_BP=191 METEAIDGYITCDNELSPEREHSNMAIDLTSSTPNGQHASPSHMTSTNSVKLEMQSDEECDRKPLSREDEIRGHDEGSSLEEPLIESSEV ADNRKVQELQGEGGIRLPNGKLKCDVCGMVCIGPNVLMVHKRSHTGERPFHCNQCGASFTQKGNLLRHIKLHSGEKPFKCPFCSYACRRR DALTGHLRTHSAEFKRAVKVATGQELSNNILDTVFKIFDLDGDECLSHEEFLGVLKNRMHRGLWVPQHQSIQEYWKCVKKESIKGVKEVW -------------------------------------------------------------- >39399_39399_4_IKZF2-MICU2_IKZF2_chr2_213914437_ENST00000421754_MICU2_chr13_22069457_ENST00000382374_length(transcript)=1308nt_BP=496nt ATGGAAACAGAGGCTATTGATGGCTATATAACGTGTGACAATGAGCTTTCACCCGAAAGGGAGCACTCCAATATGGCAATTGACCTCACC TCAAGCACACCCAATGGACAGCATGCCTCACCAAGTCACATGACAAGCACAAATTCAGTAAAGCTAGAAATGCAGAGTGATGAAGAGTGT GACAGGAAACCCCTGAGCCGTGAAGATGAGATCAGGGGCCATGATGAGGGTAGCAGCCTAGAAGAACCCCTAATTGAGAGCAGCGAGGTG GCTGACAACAGGAAAGTCCAGGAGCTTCAAGGCGAGGGAGGAATCCGGCTTCCGAATGGTGAACGCCCCTTCCACTGTAACCAGTGTGGA GCTTCTTTTACTCAGAAGGGCAACCTTCTGAGACACATAAAGTTACACTCTGGAGAGAAGCCGTTCAAATGTCCTTTCTGTAGCTACGCC TGTAGAAGAAGGGACGCCCTCACAGGACACCTCAGGACCCATTCTGCGGAGTTTAAGAGAGCTGTGAAAGTAGCAACAGGACAAGAACTC TCAAACAATATTTTGGACACTGTCTTTAAGATCTTTGATTTGGATGGTGATGAATGTCTTAGTCATGAAGAGTTTCTTGGGGTGTTAAAA AACAGAATGCATCGAGGTTTATGGGTACCACAACATCAGAGTATACAAGAATACTGGAAGTGTGTGAAGAAAGAAAGCATTAAAGGAGTA AAAGAAGTCTGGAAACAAGCTGGAAAAGGTCTTTTTTAATAAAAGATATAATAGTATGGCAATTATATTGTTCCAAATGTCAAAATTTGT GATTTTTTAGAAGTACTTGCTATTTATCTTCTTAAGTCTTCATTGATATTCTGTGTGAAATAAGCATGTCTTGTACTTGCTTTCTGATTC ATAATTTTATTAAAGAACTTAGTAGAAAGAAAAGTAAGTATAAAAATAGATATTGGATTCTGTCAGAAGGCCTAGATTTGAAATAATGTT TTGTACTTCGGTAAGATGGAAAACTTAGTGATTCACTGATTTCTTAGACACTCTAATATGATATGCTTTCTGGAAGGATAAAACAAATAC ATATGGGAAAAAGTACTTGAGACCAAGGCCAGCATCAATTCCAGACATCTTCATGTTCCTAATAGGCTAAATGAAGTTAAAAACTTATTT CAGATTTTTCTCATCTGTACCTTATATCTCATAAATTTATTGCATATTTTATGTCAGTAGCTTAGCTGTTTATTGTCTTTAAAATAACAT >39399_39399_4_IKZF2-MICU2_IKZF2_chr2_213914437_ENST00000421754_MICU2_chr13_22069457_ENST00000382374_length(amino acids)=252AA_BP=165 METEAIDGYITCDNELSPEREHSNMAIDLTSSTPNGQHASPSHMTSTNSVKLEMQSDEECDRKPLSREDEIRGHDEGSSLEEPLIESSEV ADNRKVQELQGEGGIRLPNGERPFHCNQCGASFTQKGNLLRHIKLHSGEKPFKCPFCSYACRRRDALTGHLRTHSAEFKRAVKVATGQEL -------------------------------------------------------------- >39399_39399_5_IKZF2-MICU2_IKZF2_chr2_213914437_ENST00000434687_MICU2_chr13_22069457_ENST00000382374_length(transcript)=1696nt_BP=884nt GCTCCTCGCTGAAGATGGAGGAAGTAAAAACAGGATTACCCTTAGCTACAGATCCACTGCCTTAGTTTCCACCACCAACTGCAGTGCACA AACACACGTTAGGCACAGGAAAGAAAGAAAGACAGAGGACACATTAACAGTAAACACAAACAAAAGGGTGATGGGATTATTTTACTGCAT GCACTGCTGAGCCCGACATTGTCACCTCCTCTTTGAGGGGTTAGAAGAAGCTGAGATCTCCCGACAGAGCTGGAAATGGTGATGAATCTT TTTTAATCAAAGGACAATTTCTTTTCATTGCACTTTGACTATGGAAACAGAGGCTATTGATGGCTATATAACGTGTGACAATGAGCTTTC ACCCGAAAGGGAGCACTCCAATATGGCAATTGACCTCACCTCAAGCACACCCAATGGACAGCATGCCTCACCAAGTCACATGACAAGCAC AAATTCAGTAAAGCTAGAAATGCAGAGTGATGAAGAGTGTGACAGGAAACCCCTGAGCCGTGAAGATGAGATCAGGGGCCATGATGAGGG TAGCAGCCTAGAAGAACCCCTAATTGAGAGCAGCGAGGTGGCTGACAACAGGAAAGTCCAGGAGCTTCAAGGCGAGGGAGGAATCCGGCT TCCGAATGGTAAACTGAAATGTGACGTCTGTGGCATGGTTTGCATTGGGCCCAATGTGCTTATGGTACATAAAAGGAGTCACACTGGTGA ACGCCCCTTCCACTGTAACCAGTGTGGAGCTTCTTTTACTCAGAAGGGCAACCTTCTGAGACACATAAAGTTACACTCTGGAGAGAAGCC GTTCAAATGTCCTTTCTGTAGCTACGCCTGTAGAAGAAGGGACGCCCTCACAGGACACCTCAGGACCCATTCTGCGGAGTTTAAGAGAGC TGTGAAAGTAGCAACAGGACAAGAACTCTCAAACAATATTTTGGACACTGTCTTTAAGATCTTTGATTTGGATGGTGATGAATGTCTTAG TCATGAAGAGTTTCTTGGGGTGTTAAAAAACAGAATGCATCGAGGTTTATGGGTACCACAACATCAGAGTATACAAGAATACTGGAAGTG TGTGAAGAAAGAAAGCATTAAAGGAGTAAAAGAAGTCTGGAAACAAGCTGGAAAAGGTCTTTTTTAATAAAAGATATAATAGTATGGCAA TTATATTGTTCCAAATGTCAAAATTTGTGATTTTTTAGAAGTACTTGCTATTTATCTTCTTAAGTCTTCATTGATATTCTGTGTGAAATA AGCATGTCTTGTACTTGCTTTCTGATTCATAATTTTATTAAAGAACTTAGTAGAAAGAAAAGTAAGTATAAAAATAGATATTGGATTCTG TCAGAAGGCCTAGATTTGAAATAATGTTTTGTACTTCGGTAAGATGGAAAACTTAGTGATTCACTGATTTCTTAGACACTCTAATATGAT ATGCTTTCTGGAAGGATAAAACAAATACATATGGGAAAAAGTACTTGAGACCAAGGCCAGCATCAATTCCAGACATCTTCATGTTCCTAA TAGGCTAAATGAAGTTAAAAACTTATTTCAGATTTTTCTCATCTGTACCTTATATCTCATAAATTTATTGCATATTTTATGTCAGTAGCT >39399_39399_5_IKZF2-MICU2_IKZF2_chr2_213914437_ENST00000434687_MICU2_chr13_22069457_ENST00000382374_length(amino acids)=280AA_BP=193 MTMETEAIDGYITCDNELSPEREHSNMAIDLTSSTPNGQHASPSHMTSTNSVKLEMQSDEECDRKPLSREDEIRGHDEGSSLEEPLIESS EVADNRKVQELQGEGGIRLPNGKLKCDVCGMVCIGPNVLMVHKRSHTGERPFHCNQCGASFTQKGNLLRHIKLHSGEKPFKCPFCSYACR RRDALTGHLRTHSAEFKRAVKVATGQELSNNILDTVFKIFDLDGDECLSHEEFLGVLKNRMHRGLWVPQHQSIQEYWKCVKKESIKGVKE -------------------------------------------------------------- >39399_39399_6_IKZF2-MICU2_IKZF2_chr2_213914437_ENST00000451136_MICU2_chr13_22069457_ENST00000382374_length(transcript)=1308nt_BP=496nt ATGGAAACAGAGGCTATTGATGGCTATATAACGTGTGACAATGAGCTTTCACCCGAAAGGGAGCACTCCAATATGGCAATTGACCTCACC TCAAGCACACCCAATGGACAGCATGCCTCACCAAGTCACATGACAAGCACAAATTCAGTAAAGCTAGAAATGCAGAGTGATGAAGAGTGT GACAGGAAACCCCTGAGCCGTGAAGATGAGATCAGGGGCCATGATGAGGGTAGCAGCCTAGAAGAACCCCTAATTGAGAGCAGCGAGGTG GCTGACAACAGGAAAGTCCAGGAGCTTCAAGGCGAGGGAGGAATCCGGCTTCCGAATGGTGAACGCCCCTTCCACTGTAACCAGTGTGGA GCTTCTTTTACTCAGAAGGGCAACCTTCTGAGACACATAAAGTTACACTCTGGAGAGAAGCCGTTCAAATGTCCTTTCTGTAGCTACGCC TGTAGAAGAAGGGACGCCCTCACAGGACACCTCAGGACCCATTCTGCGGAGTTTAAGAGAGCTGTGAAAGTAGCAACAGGACAAGAACTC TCAAACAATATTTTGGACACTGTCTTTAAGATCTTTGATTTGGATGGTGATGAATGTCTTAGTCATGAAGAGTTTCTTGGGGTGTTAAAA AACAGAATGCATCGAGGTTTATGGGTACCACAACATCAGAGTATACAAGAATACTGGAAGTGTGTGAAGAAAGAAAGCATTAAAGGAGTA AAAGAAGTCTGGAAACAAGCTGGAAAAGGTCTTTTTTAATAAAAGATATAATAGTATGGCAATTATATTGTTCCAAATGTCAAAATTTGT GATTTTTTAGAAGTACTTGCTATTTATCTTCTTAAGTCTTCATTGATATTCTGTGTGAAATAAGCATGTCTTGTACTTGCTTTCTGATTC ATAATTTTATTAAAGAACTTAGTAGAAAGAAAAGTAAGTATAAAAATAGATATTGGATTCTGTCAGAAGGCCTAGATTTGAAATAATGTT TTGTACTTCGGTAAGATGGAAAACTTAGTGATTCACTGATTTCTTAGACACTCTAATATGATATGCTTTCTGGAAGGATAAAACAAATAC ATATGGGAAAAAGTACTTGAGACCAAGGCCAGCATCAATTCCAGACATCTTCATGTTCCTAATAGGCTAAATGAAGTTAAAAACTTATTT CAGATTTTTCTCATCTGTACCTTATATCTCATAAATTTATTGCATATTTTATGTCAGTAGCTTAGCTGTTTATTGTCTTTAAAATAACAT >39399_39399_6_IKZF2-MICU2_IKZF2_chr2_213914437_ENST00000451136_MICU2_chr13_22069457_ENST00000382374_length(amino acids)=252AA_BP=165 METEAIDGYITCDNELSPEREHSNMAIDLTSSTPNGQHASPSHMTSTNSVKLEMQSDEECDRKPLSREDEIRGHDEGSSLEEPLIESSEV ADNRKVQELQGEGGIRLPNGERPFHCNQCGASFTQKGNLLRHIKLHSGEKPFKCPFCSYACRRRDALTGHLRTHSAEFKRAVKVATGQEL -------------------------------------------------------------- >39399_39399_7_IKZF2-MICU2_IKZF2_chr2_213914437_ENST00000457361_MICU2_chr13_22069457_ENST00000382374_length(transcript)=1555nt_BP=743nt GGGCTTGGCTTGGAGGGGGCAAGGGAGGGAAAGAGAGAAGGGGGAAACACAAAAAACTTCTTTCTTTCTCCCTCCGTTTATCTTCAGCCC GACATTGTCACCTCCTCTTTGAGGGGTTAGAAGAAGCTGAGATCTCCCGACAGAGCTGGAAATGCATTGCACTTTGACTATGGAAACAGA GGCTATTGATGGCTATATAACGTGTGACAATGAGCTTTCACCCGAAAGGGAGCACTCCAATATGGCAATTGACCTCACCTCAAGCACACC CAATGGACAGCATGCCTCACCAAGTCACATGACAAGCACAAATTCAGTAAAGCTAGAAATGCAGAGTGATGAAGAGTGTGACAGGAAACC CCTGAGCCGTGAAGATGAGATCAGGGGCCATGATGAGGGTAGCAGCCTAGAAGAACCCCTAATTGAGAGCAGCGAGGTGGCTGACAACAG GAAAGTCCAGGAGCTTCAAGGCGAGGGAGGAATCCGGCTTCCGAATGGTAAACTGAAATGTGACGTCTGTGGCATGGTTTGCATTGGGCC CAATGTGCTTATGGTACATAAAAGGAGTCACACTGGTGAACGCCCCTTCCACTGTAACCAGTGTGGAGCTTCTTTTACTCAGAAGGGCAA CCTTCTGAGACACATAAAGTTACACTCTGGAGAGAAGCCGTTCAAATGTCCTTTCTGTAGCTACGCCTGTAGAAGAAGGGACGCCCTCAC AGGACACCTCAGGACCCATTCTGCGGAGTTTAAGAGAGCTGTGAAAGTAGCAACAGGACAAGAACTCTCAAACAATATTTTGGACACTGT CTTTAAGATCTTTGATTTGGATGGTGATGAATGTCTTAGTCATGAAGAGTTTCTTGGGGTGTTAAAAAACAGAATGCATCGAGGTTTATG GGTACCACAACATCAGAGTATACAAGAATACTGGAAGTGTGTGAAGAAAGAAAGCATTAAAGGAGTAAAAGAAGTCTGGAAACAAGCTGG AAAAGGTCTTTTTTAATAAAAGATATAATAGTATGGCAATTATATTGTTCCAAATGTCAAAATTTGTGATTTTTTAGAAGTACTTGCTAT TTATCTTCTTAAGTCTTCATTGATATTCTGTGTGAAATAAGCATGTCTTGTACTTGCTTTCTGATTCATAATTTTATTAAAGAACTTAGT AGAAAGAAAAGTAAGTATAAAAATAGATATTGGATTCTGTCAGAAGGCCTAGATTTGAAATAATGTTTTGTACTTCGGTAAGATGGAAAA CTTAGTGATTCACTGATTTCTTAGACACTCTAATATGATATGCTTTCTGGAAGGATAAAACAAATACATATGGGAAAAAGTACTTGAGAC CAAGGCCAGCATCAATTCCAGACATCTTCATGTTCCTAATAGGCTAAATGAAGTTAAAAACTTATTTCAGATTTTTCTCATCTGTACCTT ATATCTCATAAATTTATTGCATATTTTATGTCAGTAGCTTAGCTGTTTATTGTCTTTAAAATAACATGTAAACTTCAATGTTCTATCTGG >39399_39399_7_IKZF2-MICU2_IKZF2_chr2_213914437_ENST00000457361_MICU2_chr13_22069457_ENST00000382374_length(amino acids)=286AA_BP=199 MEMHCTLTMETEAIDGYITCDNELSPEREHSNMAIDLTSSTPNGQHASPSHMTSTNSVKLEMQSDEECDRKPLSREDEIRGHDEGSSLEE PLIESSEVADNRKVQELQGEGGIRLPNGKLKCDVCGMVCIGPNVLMVHKRSHTGERPFHCNQCGASFTQKGNLLRHIKLHSGEKPFKCPF CSYACRRRDALTGHLRTHSAEFKRAVKVATGQELSNNILDTVFKIFDLDGDECLSHEEFLGVLKNRMHRGLWVPQHQSIQEYWKCVKKES -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for IKZF2-MICU2 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for IKZF2-MICU2 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for IKZF2-MICU2 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
