
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:IMPAD1-TCF12 (FusionGDB2 ID:39694) |
Fusion Gene Summary for IMPAD1-TCF12 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: IMPAD1-TCF12 | Fusion gene ID: 39694 | Hgene | Tgene | Gene symbol | IMPAD1 | TCF12 | Gene ID | 54928 | 6938 |
| Gene name | 3'(2'), 5'-bisphosphate nucleotidase 2 | transcription factor 12 | |
| Synonyms | GPAPP|IMP 3|IMP-3|IMPA3|IMPAD1 | CRS3|HEB|HTF4|HsT17266|TCF-12|bHLHb20|p64 | |
| Cytomap | 8q12.1 | 15q21.3 | |
| Type of gene | protein-coding | protein-coding | |
| Description | Golgi-resident adenosine 3',5'-bisphosphate 3'-phosphataseGolgi 3-prime phosphoadenosine 5-prime phosphate 3-prime phosphataseIMPase 3golgi-resident PAP phosphatasegolgi-resident nucleotide phosphataseinositol monophosphatase domain containing 1inos | transcription factor 12DNA-binding protein HTF4E-box-binding proteinclass B basic helix-loop-helix protein 20helix-loop-helix transcription factor 4transcription factor HTF-4 | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | . | . | |
| Ensembl transtripts involved in fusion gene | ENST00000262644, | ENST00000333725, ENST00000343827, ENST00000452095, ENST00000537840, ENST00000543579, ENST00000557843, ENST00000559703, ENST00000559710, ENST00000560764, ENST00000267811, ENST00000438423, | |
| Fusion gene scores | * DoF score | 4 X 4 X 4=64 | 21 X 22 X 8=3696 |
| # samples | 4 | 26 | |
| ** MAII score | log2(4/64*10)=-0.678071905112638 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(26/3696*10)=-3.8293812283876 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: IMPAD1 [Title/Abstract] AND TCF12 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | IMPAD1(57890608)-TCF12(57355947), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Tgene | TCF12 | GO:0045944 | positive regulation of transcription by RNA polymerase II | 11802795 |
 Fusion gene breakpoints across IMPAD1 (5'-gene) Fusion gene breakpoints across IMPAD1 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
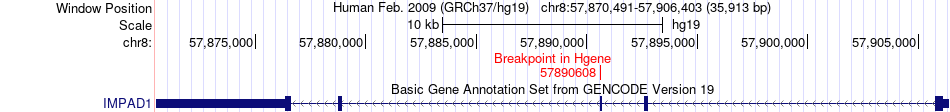 |
 Fusion gene breakpoints across TCF12 (3'-gene) Fusion gene breakpoints across TCF12 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
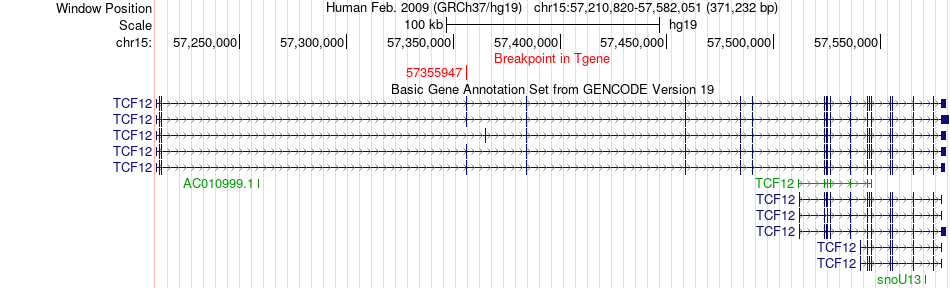 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | ESCA | TCGA-L5-A43J | IMPAD1 | chr8 | 57890608 | - | TCF12 | chr15 | 57355947 | + |
Top |
Fusion Gene ORF analysis for IMPAD1-TCF12 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-intron | ENST00000262644 | ENST00000333725 | IMPAD1 | chr8 | 57890608 | - | TCF12 | chr15 | 57355947 | + |
| 5CDS-intron | ENST00000262644 | ENST00000343827 | IMPAD1 | chr8 | 57890608 | - | TCF12 | chr15 | 57355947 | + |
| 5CDS-intron | ENST00000262644 | ENST00000452095 | IMPAD1 | chr8 | 57890608 | - | TCF12 | chr15 | 57355947 | + |
| 5CDS-intron | ENST00000262644 | ENST00000537840 | IMPAD1 | chr8 | 57890608 | - | TCF12 | chr15 | 57355947 | + |
| 5CDS-intron | ENST00000262644 | ENST00000543579 | IMPAD1 | chr8 | 57890608 | - | TCF12 | chr15 | 57355947 | + |
| 5CDS-intron | ENST00000262644 | ENST00000557843 | IMPAD1 | chr8 | 57890608 | - | TCF12 | chr15 | 57355947 | + |
| 5CDS-intron | ENST00000262644 | ENST00000559703 | IMPAD1 | chr8 | 57890608 | - | TCF12 | chr15 | 57355947 | + |
| 5CDS-intron | ENST00000262644 | ENST00000559710 | IMPAD1 | chr8 | 57890608 | - | TCF12 | chr15 | 57355947 | + |
| 5CDS-intron | ENST00000262644 | ENST00000560764 | IMPAD1 | chr8 | 57890608 | - | TCF12 | chr15 | 57355947 | + |
| In-frame | ENST00000262644 | ENST00000267811 | IMPAD1 | chr8 | 57890608 | - | TCF12 | chr15 | 57355947 | + |
| In-frame | ENST00000262644 | ENST00000438423 | IMPAD1 | chr8 | 57890608 | - | TCF12 | chr15 | 57355947 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000262644 | IMPAD1 | chr8 | 57890608 | - | ENST00000438423 | TCF12 | chr15 | 57355947 | + | 5247 | 905 | 187 | 2877 | 896 |
| ENST00000262644 | IMPAD1 | chr8 | 57890608 | - | ENST00000267811 | TCF12 | chr15 | 57355947 | + | 6514 | 905 | 187 | 2805 | 872 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000262644 | ENST00000438423 | IMPAD1 | chr8 | 57890608 | - | TCF12 | chr15 | 57355947 | + | 0.000691723 | 0.9993082 |
| ENST00000262644 | ENST00000267811 | IMPAD1 | chr8 | 57890608 | - | TCF12 | chr15 | 57355947 | + | 0.000463761 | 0.9995363 |
Top |
Fusion Genomic Features for IMPAD1-TCF12 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| IMPAD1 | chr8 | 57890608 | - | TCF12 | chr15 | 57355947 | + | 3.32E-07 | 0.99999964 |
| IMPAD1 | chr8 | 57890608 | - | TCF12 | chr15 | 57355947 | + | 3.32E-07 | 0.99999964 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
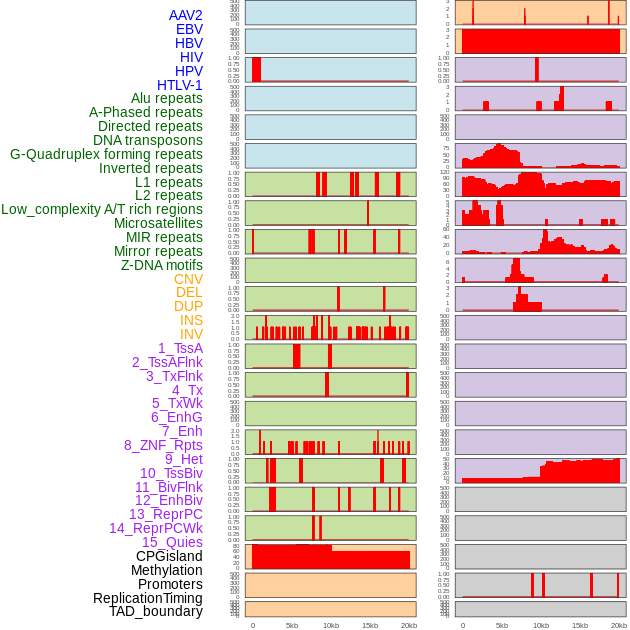 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
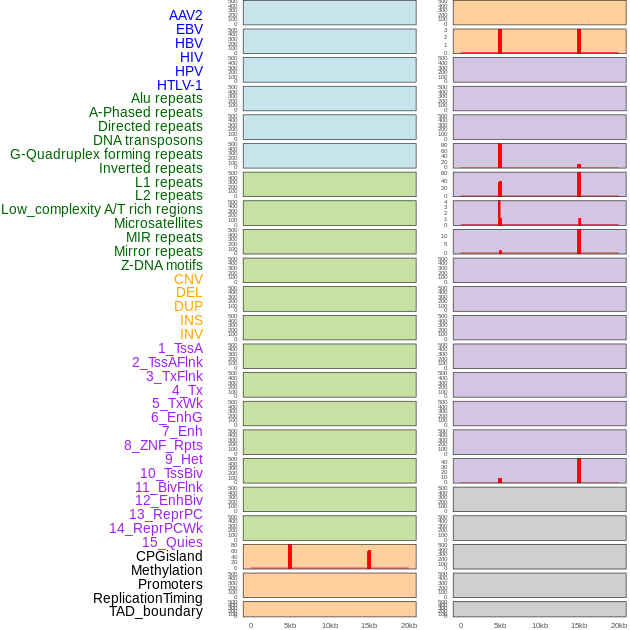 |
Top |
Fusion Protein Features for IMPAD1-TCF12 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr8:57890608/chr15:57355947) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| . | . |
| FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. | FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | IMPAD1 | chr8:57890608 | chr15:57355947 | ENST00000262644 | - | 3 | 5 | 53_56 | 215 | 360.0 | Compositional bias | Note=Poly-Ala |
| Hgene | IMPAD1 | chr8:57890608 | chr15:57355947 | ENST00000262644 | - | 3 | 5 | 1_12 | 215 | 360.0 | Topological domain | Cytoplasmic |
| Hgene | IMPAD1 | chr8:57890608 | chr15:57355947 | ENST00000262644 | - | 3 | 5 | 13_33 | 215 | 360.0 | Transmembrane | Helical |
| Tgene | TCF12 | chr8:57890608 | chr15:57355947 | ENST00000267811 | 2 | 20 | 577_630 | 49 | 1821.3333333333333 | Domain | bHLH | |
| Tgene | TCF12 | chr8:57890608 | chr15:57355947 | ENST00000333725 | 2 | 21 | 577_630 | 49 | 1420.0 | Domain | bHLH | |
| Tgene | TCF12 | chr8:57890608 | chr15:57355947 | ENST00000343827 | 0 | 13 | 577_630 | 0 | 1254.6666666666667 | Domain | bHLH | |
| Tgene | TCF12 | chr8:57890608 | chr15:57355947 | ENST00000438423 | 2 | 21 | 577_630 | 49 | 1398.3333333333333 | Domain | bHLH | |
| Tgene | TCF12 | chr8:57890608 | chr15:57355947 | ENST00000557843 | 2 | 20 | 577_630 | 49 | 1301.0 | Domain | bHLH | |
| Tgene | TCF12 | chr8:57890608 | chr15:57355947 | ENST00000343827 | 0 | 13 | 19_27 | 0 | 1254.6666666666667 | Motif | Note=9aaTAD | |
| Tgene | TCF12 | chr8:57890608 | chr15:57355947 | ENST00000267811 | 2 | 20 | 119_140 | 49 | 1821.3333333333333 | Region | Note=Leucine-zipper | |
| Tgene | TCF12 | chr8:57890608 | chr15:57355947 | ENST00000267811 | 2 | 20 | 632_655 | 49 | 1821.3333333333333 | Region | Note=Class A specific domain | |
| Tgene | TCF12 | chr8:57890608 | chr15:57355947 | ENST00000333725 | 2 | 21 | 119_140 | 49 | 1420.0 | Region | Note=Leucine-zipper | |
| Tgene | TCF12 | chr8:57890608 | chr15:57355947 | ENST00000333725 | 2 | 21 | 632_655 | 49 | 1420.0 | Region | Note=Class A specific domain | |
| Tgene | TCF12 | chr8:57890608 | chr15:57355947 | ENST00000343827 | 0 | 13 | 119_140 | 0 | 1254.6666666666667 | Region | Note=Leucine-zipper | |
| Tgene | TCF12 | chr8:57890608 | chr15:57355947 | ENST00000343827 | 0 | 13 | 632_655 | 0 | 1254.6666666666667 | Region | Note=Class A specific domain | |
| Tgene | TCF12 | chr8:57890608 | chr15:57355947 | ENST00000438423 | 2 | 21 | 119_140 | 49 | 1398.3333333333333 | Region | Note=Leucine-zipper | |
| Tgene | TCF12 | chr8:57890608 | chr15:57355947 | ENST00000438423 | 2 | 21 | 632_655 | 49 | 1398.3333333333333 | Region | Note=Class A specific domain | |
| Tgene | TCF12 | chr8:57890608 | chr15:57355947 | ENST00000557843 | 2 | 20 | 119_140 | 49 | 1301.0 | Region | Note=Leucine-zipper | |
| Tgene | TCF12 | chr8:57890608 | chr15:57355947 | ENST00000557843 | 2 | 20 | 632_655 | 49 | 1301.0 | Region | Note=Class A specific domain |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | IMPAD1 | chr8:57890608 | chr15:57355947 | ENST00000262644 | - | 3 | 5 | 34_359 | 215 | 360.0 | Topological domain | Lumenal |
| Tgene | TCF12 | chr8:57890608 | chr15:57355947 | ENST00000267811 | 2 | 20 | 19_27 | 49 | 1821.3333333333333 | Motif | Note=9aaTAD | |
| Tgene | TCF12 | chr8:57890608 | chr15:57355947 | ENST00000333725 | 2 | 21 | 19_27 | 49 | 1420.0 | Motif | Note=9aaTAD | |
| Tgene | TCF12 | chr8:57890608 | chr15:57355947 | ENST00000438423 | 2 | 21 | 19_27 | 49 | 1398.3333333333333 | Motif | Note=9aaTAD | |
| Tgene | TCF12 | chr8:57890608 | chr15:57355947 | ENST00000557843 | 2 | 20 | 19_27 | 49 | 1301.0 | Motif | Note=9aaTAD |
Top |
Fusion Gene Sequence for IMPAD1-TCF12 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >39694_39694_1_IMPAD1-TCF12_IMPAD1_chr8_57890608_ENST00000262644_TCF12_chr15_57355947_ENST00000267811_length(transcript)=6514nt_BP=905nt GGAAGTCGGACCAGCCGGCCGGCGGAAGAACCTAGAGCGCGCTGCCTGGCGAGTCAGGCGCGCGGGGCGGCGTTGGTGGTCTTCGCGGCG CAACTCGGCCTTTCCTGGGAGGGAGTGATGGGGCGCACCGGGGCCGGGGAGCGGGCGCCAGTGTAGCCCGCGCGGCGCCTGGCCCGGAGC GCGGCGGCTGCGGCGGCGGCGGCGGCGGGCGCTGGAGGCCTGTGAGAGCCGCAGCCCGGAGCGCCCGGCTTCCCACGCCATGGCCCCCAT GGGCATCCGCCTTTCCCCACTGGGGGTGGCAGTGTTTTGCCTGCTGGGGCTCGGCGTGCTCTACCACCTCTACTCGGGCTTCTTGGCCGG CCGCTTCAGCCTCTTCGGCCTGGGCGGCGAGCCTGGCGGCGGCGCGGCGGGGCCCGCGGCCGCGGCCGATGGGGGCACCGTGGACTTGCG CGAGATGCTGGCTGTGTCAGTGCTGGCCGCAGTCCGCGGCGGCGACGAGGTGAGGCGCGTCCGCGAGAGCAACGTCCTCCACGAGAAGTC CAAGGGGAAGACGCGCGAGGGAGCCGAGGACAAGATGACCAGCGGCGACGTGCTGTCCAACCGCAAGATGTTCTACCTGCTCAAGACCGC CTTCCCCAGCGTCCAGATTAATACTGAGGAACACGTGGATGCAGCTGATCAGGAGGTTATCTTGTGGGATCATAAGATTCCTGAGGATAT CCTAAAGGAAGTAACTACTCCTAAAGAGGTACCAGCAGAAAGTGTTACTGTCTGGATTGACCCACTTGATGCTACACAGGAATATACAGA GGATCTTCGAAAGTACGTCACTACTATGGTGTGTGTGGCTGTAAATGGTAAACCCATGCTAGGAGTTATACATAAGCCATTTTCCGAATA TACAGGTATTGATGAAAGAGGAGGTACAACATCTTGGGGAACAAGTGGTCAACCAAGTCCTTCCTATGATTCATCTAGAGGTTTTACAGA CAGCCCTCATTACAGTGATCACTTGAATGACAGTCGATTAGGAGCCCATGAAGGCTTGTCCCCAACACCTTTCATGAACTCAAATCTGAT GGGAAAAACATCAGAGAGAGGCTCATTTTCCCTGTACAGCAGAGATACTGGATTACCAGGCTGTCAATCTAGTCTCCTGAGACAAGATCT GGGGCTTGGGAGCCCAGCACAGCTATCTTCTTCAGGAAAACCTGGGACAGCATACTATTCATTCTCTGCTACAAGTTCCAGGAGGAGACC ACTCCATGACTCTGCAGCGCTTGATCCCTTGCAAGCAAAAAAAGTCAGAAAGGTGCCTCCTGGTTTGCCTTCTTCTGTATATGCACCATC CCCAAATTCAGATGATTTCAACCGTGAATCTCCTAGTTATCCATCTCCTAAGCCACCAACCAGTATGTTCGCTAGCACTTTCTTTATGCA AGATGGGACCCACAATTCTTCTGACCTTTGGAGTTCATCAAATGGGATGAGCCAGCCTGGTTTTGGTGGAATTCTGGGGACCTCCACTTC CCACATGTCTCAATCCAGTAGTTATGGCAACCTTCATTCACATGACCGCTTGAGTTATCCTCCACACTCAGTTTCACCAACAGACATAAA CACGAGTCTTCCACCAATGTCCAGCTTTCATCGCGGCAGTACCAGCAGTTCACCTTACGTTGCTGCCTCACACACTCCTCCCATCAATGG ATCAGACAGCATTCTAGGAACCAGAGGGAATGCTGCTGGAAGCTCACAGACAGGTGATGCACTTGGAAAGGCTTTGGCATCTATTTATTC TCCTGACCATACCAGCAGTAGTTTTCCGTCAAATCCATCAACACCAGTTGGATCACCTTCACCTCTCACAGGTACCAGTCAGTGGCCAAG ACCTGGAGGGCAAGCACCTTCATCCCCAAGCTATGAAAACTCACTCCACTCCCTGCAGTCTCGAATGGAGGATCGTTTAGACAGACTGGA TGATGCAATCCATGTGCTGCGGAACCATGCTGTGGGACCTTCCACCAGTTTGCCTGCTGGTCACAGTGATATACATAGTTTATTGGGACC ATCCCATAATGCACCAATTGGAAGCCTCAATTCAAACTATGGAGGATCAAGCCTTGTTGCAAGCAGTCGATCAGCTTCAATGGTTGGAAC TCATCGGGAAGACTCTGTCAGTCTCAATGGCAATCATTCAGTCCTGTCTAGTACAGTCACTACTTCAAGCACAGACCTGAACCATAAAAC ACAAGAAAATTATAGAGGTGGCTTGCAAAGTCAGTCTGGAACTGTTGTTACAACAGAAATCAAGACTGAAAACAAAGAAAAGGATGAAAA CCTTCATGAACCTCCTTCATCAGATGACATGAAGTCAGATGATGAATCCTCCCAAAAAGATATCAAGGTTTCATCTAGAGGCAGAACAAG CAGTACTAATGAAGATGAGGATTTGAACCCTGAACAGAAGATAGAAAGGGAGAAGGAGAGGCGGATGGCTAACAATGCCAGAGAACGCTT ACGCGTGCGGGATATTAATGAAGCATTCAAAGAGCTTGGCCGAATGTGTCAGCTTCACTTGAAGAGTGAAAAACCCCAAACAAAACTCCT TATTCTTCATCAAGCCGTGGCAGTCATCCTTAGTCTAGAACAGCAAGTCAGAGAGAGGAACCTTAACCCCAAAGCAGCCTGCCTTAAGAG AAGGGAAGAAGAAAAAGTTTCTGCCGTATCGGCAGAGCCGCCAACCACACTGCCAGGAACCCATCCTGGGCTTAGTGAAACTACCAACCC TATGGGTCATATGTAAACATCAGCCAGTTCCAGAGTTATCAGTAGGCTAGATAGAAGGTGACCTCTCCTCATAAGGACTTGGACAACTCA GATTATCTGAAGACACAAACCTGACAGGAGGGAGAAGAAAAAACAAAACACTTGAACCAAGAAACTCAAATGTAATCCTACGATCAAAGC AACTGGTCAACACTTCCATCAGAAGTGAAGATAGGAAGCTCATCAGATAGAACATCAGCCCATGAGATGTTTGCAACAAATCTTTTGTTG CAAGCAGTGTGTCGCTTCTGCACAATCAGAGACTGTCTCGATCTCTCCACTCACCGTGGAAGTTGCCTTGTGCCTAAACTGAATTGACAA ATGCATTGTAACTACAAATTTTATTTATTGTTATGAAACTGTAAGGTCTACATATAAAGGGAAAAAGTTAATGTGGAAAGCTGATCTACA CTCAGCTGATGCCAGCATACATTAAAGCGGTTCACGTGCAGAGAACAAAGCAGTGACAACCATTGGCCCTTAGCATTCCCGGCATACCTA TTAGTGTCTTAAAAAGGAAGGGAAAAGTCTTTTGTTGCCCTCTCCTATCCTCTTGCCATATGAATAGCGTTTTCCATGAAATAGGAAAAT ATTACTTGGTATAGCATTTCTCTTGCTCTCATTTTTTGATTTATTTTTATTTTCTCTTTGTGGGTGTTATATTTGATCTCTAAATCTGAA CAGTTTATGGTCACAGTCCAGCCTCCTCCGTGCAGCCCTGTGTGCTTTGCACATTTACCTTACAGTGGTAAGCAGAGACCATCTGTGACC ATAGCCTAGCTAGCATTTTAAAAGGGGAAATTTTGTTCTCTAGGTTTTCCCCCAAATAAACATTGCTTTATTTCTAATAATAACCAAGAC TTTTCAAGCTTCTAGATCTCATAGGAAAGCTTGTAATAGCAAAATTGTAAATTACAAGGGAAGAATCTACTTTTTAGAAATCGCTTTGTT TTCCAAGCAGTAAGTACTACATACAGTACTTGTAAAGTGTTAGCTGTAAGTAAGCACAAAATACATTTAAAATACAAAGACGATTTTTTC AGGCTGTGATTATGGTGAACATAACAAAACCCAGTAGTCACCAAGGCAGGTAGTGTGATAAATGAACACACCACTCTGAGGCTAATTACC TAATGGAATACAAGAGCAATGGTCACCCGTATTTCCTTATCCTAGCCTTTATTTCTCTGTCATTTGGATGGCTGGTCAATGGGGAAGAAT TGAGTGGGTGATTTAATCAACTGCAAACCATCTGCCCCTGTCCCAAAATGATGAGCCAGATTAGCATTAAACCAGTACTTGTCAGTCCAT CTTAATACTGTTCATTAAGGCACTCTCTGTCTCTAATCCTTAGGAGTTGTTTTAAAAGACATAATCACTTTGAACTTCCATGAAACCTGT CTTCCACCACAACAACCCTGGGAGAGAAAAACATGCTAAAGGAGGTATCTTGGCTTAATAATTCCTTATAGCCAATATCAACAGTGGCAA TCAGCACACAGAGGAAAGGACCCAAATCACTATGTAGCTTAAAGATTTCTGTTAATTTGAAAGAACAAAAACAAGACAGAACTTCTGGTA CTCTAATCAGGATGATTCCTAACAAGTCAGTCATTTGTGAACTTAGTGGACTTTTTGGTTACTTTAATTTGCATATATTCTCCAGTTACA TCGGACTCTATCTGTGGCCTTGTTCTTCATTTCAGTGTTAATCAGCTAAACAGAAGTTGTTGCTTATGATGTGTGAGTGAACATATGCCA CTGCCTGGCCTTTTTTTCTTCAGAGCTTGTTGTCTTTTTCGCTATATTAGACTTTGCAGTATGCCCAGAAGCTTTCCTTCATAAAATAGA AAGAAAAAAACATTTGGCTTATTTTTCACTGTAGCTAGTCTTTTATACAATAATCTTGTAAGAAAATTTCTTGAATTCTAAATATTACTC TTTCTAGATTTTTGAAATCAAAAAGTTTTCAGTAAAAAGTTTCTTACTTTATTTTATTATATTAGGTAGTAAAAAATGTAGGGTTATTTA CCATAACCTGTTCATTAATATCAGAAATTTACAATAGCATTTTAAGACCATAGTAGGATTCTAGCATACCGTGTAGTACCTATGGAGTAT TGTAAGAGCTAATTGTTGGAGATGAATTGCTTCTCATCTTGTTCTCCAGTTTCCATTGTTGGTTTATTGCAGATTTGTATCCTGTGTCAA ATTCAAGGTATTATTGATAAACCTTTTCAACCAGCAGCAAGAAGTTCAAATTTTTTTCTGTCACTGTAACAGAAAACACAATATGTATAT AACATTTATGTAGCAATAAATGTGCCATCTTTTTTTTAACACAGTAAAATAGTGAGTTTTTTACATTTCTCTTTCTCAAATAATAATGTA TTTTGTTTTATTTTCTCCATCTCATTCGTCCCAGAAACACTCACACTGCTTTTCCTAACTGCATTACCGACATTATCTGGGAAACCCTTC AGGACAGAATCAGGCTTGTGGAGCTAAGTTGGCAATCTGGTCTAGAGCTTCTCTAGCTTGTGCTTTTCTCCTCTTGCCCTCACTACTGAC GGTGGCCTTTTAACCTTTTCCTAAAGATTGACCAAACAGCAACTAGTAGTTATAGAAAATCTACTCATTTGTAGATACAGAGAAAAATGA AGAAGATGGAAAAAGACTCAAAAGAGGCTTTTTAAGTTATTCTTCAAAGCACTTTTCACATTTCCCCATACCCTTTCTCACAAAAAAAGT GTCATAATTAAGTAATGGTATTGTTTACTGTTTAAAAGTTAAAAGATCAAAAATTTGCTTTTATCCCAGTTTTTAACCACAAAAAAAAGC GTAGGGATTATCCATGAGGACTTCATGCCAAGCAAGAACCTCAAACAAACTAGACAAACTTTTTTTTTGACAGTGAATGACTTTTTGTAG GACCTGTGCGTGCGAAACCCATGGCAATTGTCACATCCTCTTGGTATGCTGGCAGATTGCTTCTCTTGGTGAATTATGAAATCCACTGTT CACATTGGGTGCCTAACAGAACATTTTGCTTCTTGTGGGATTTAGTGAAAACTATTAAACTTGTTAAGTTGATTTTATACAAAACGATAA ATAAAAAGCTCTAAGAAGAAAATGTATAATCTTAGAGCTGAAATAAAATATAGAGACCATCTAGTAAATGACCTCATTAATATATCTGTG AAAACTGAGACTCAGATTGTATGTCTCTAAGAACACATAATTAGTAACAGATCAAGACACTTAAAACTTTCCCTACAAAACCTCCCTGCC TTGACTTTCTCTTTCTCTTTGCAGATTTCTAGGCCGCTTCTGCTCAGTGTCTTCATTTTCTTCCATATTTGTTTTATTTCATTTTTCTTT TCTATAGTTCATGTTTTCTTTTTCCTTGGAAACTCAAAATTTAAACACACGTCGTGTGTGTGTGTGTGTGTGTGTCTGTGTGTGTGTGAC TTAAAGAATCTTAAGCTTTGGCATTAAATAGTCCTCGATTCAAATCTAAGCTCAACATCTGATTAACTTCATTTTCCTATCTGAAAAATG GAGATAACATTAGAATTGTGTAAGTATTGAATGAAACAATGTATGGAAAGCTCTTATGGTTCTTGTCACCTAAGAAGTACTTAATAAATG >39694_39694_1_IMPAD1-TCF12_IMPAD1_chr8_57890608_ENST00000262644_TCF12_chr15_57355947_ENST00000267811_length(amino acids)=872AA_BP=239 MRRRRRRALEACESRSPERPASHAMAPMGIRLSPLGVAVFCLLGLGVLYHLYSGFLAGRFSLFGLGGEPGGGAAGPAAAADGGTVDLREM LAVSVLAAVRGGDEVRRVRESNVLHEKSKGKTREGAEDKMTSGDVLSNRKMFYLLKTAFPSVQINTEEHVDAADQEVILWDHKIPEDILK EVTTPKEVPAESVTVWIDPLDATQEYTEDLRKYVTTMVCVAVNGKPMLGVIHKPFSEYTGIDERGGTTSWGTSGQPSPSYDSSRGFTDSP HYSDHLNDSRLGAHEGLSPTPFMNSNLMGKTSERGSFSLYSRDTGLPGCQSSLLRQDLGLGSPAQLSSSGKPGTAYYSFSATSSRRRPLH DSAALDPLQAKKVRKVPPGLPSSVYAPSPNSDDFNRESPSYPSPKPPTSMFASTFFMQDGTHNSSDLWSSSNGMSQPGFGGILGTSTSHM SQSSSYGNLHSHDRLSYPPHSVSPTDINTSLPPMSSFHRGSTSSSPYVAASHTPPINGSDSILGTRGNAAGSSQTGDALGKALASIYSPD HTSSSFPSNPSTPVGSPSPLTGTSQWPRPGGQAPSSPSYENSLHSLQSRMEDRLDRLDDAIHVLRNHAVGPSTSLPAGHSDIHSLLGPSH NAPIGSLNSNYGGSSLVASSRSASMVGTHREDSVSLNGNHSVLSSTVTTSSTDLNHKTQENYRGGLQSQSGTVVTTEIKTENKEKDENLH EPPSSDDMKSDDESSQKDIKVSSRGRTSSTNEDEDLNPEQKIEREKERRMANNARERLRVRDINEAFKELGRMCQLHLKSEKPQTKLLIL -------------------------------------------------------------- >39694_39694_2_IMPAD1-TCF12_IMPAD1_chr8_57890608_ENST00000262644_TCF12_chr15_57355947_ENST00000438423_length(transcript)=5247nt_BP=905nt GGAAGTCGGACCAGCCGGCCGGCGGAAGAACCTAGAGCGCGCTGCCTGGCGAGTCAGGCGCGCGGGGCGGCGTTGGTGGTCTTCGCGGCG CAACTCGGCCTTTCCTGGGAGGGAGTGATGGGGCGCACCGGGGCCGGGGAGCGGGCGCCAGTGTAGCCCGCGCGGCGCCTGGCCCGGAGC GCGGCGGCTGCGGCGGCGGCGGCGGCGGGCGCTGGAGGCCTGTGAGAGCCGCAGCCCGGAGCGCCCGGCTTCCCACGCCATGGCCCCCAT GGGCATCCGCCTTTCCCCACTGGGGGTGGCAGTGTTTTGCCTGCTGGGGCTCGGCGTGCTCTACCACCTCTACTCGGGCTTCTTGGCCGG CCGCTTCAGCCTCTTCGGCCTGGGCGGCGAGCCTGGCGGCGGCGCGGCGGGGCCCGCGGCCGCGGCCGATGGGGGCACCGTGGACTTGCG CGAGATGCTGGCTGTGTCAGTGCTGGCCGCAGTCCGCGGCGGCGACGAGGTGAGGCGCGTCCGCGAGAGCAACGTCCTCCACGAGAAGTC CAAGGGGAAGACGCGCGAGGGAGCCGAGGACAAGATGACCAGCGGCGACGTGCTGTCCAACCGCAAGATGTTCTACCTGCTCAAGACCGC CTTCCCCAGCGTCCAGATTAATACTGAGGAACACGTGGATGCAGCTGATCAGGAGGTTATCTTGTGGGATCATAAGATTCCTGAGGATAT CCTAAAGGAAGTAACTACTCCTAAAGAGGTACCAGCAGAAAGTGTTACTGTCTGGATTGACCCACTTGATGCTACACAGGAATATACAGA GGATCTTCGAAAGTACGTCACTACTATGGTGTGTGTGGCTGTAAATGGTAAACCCATGCTAGGAGTTATACATAAGCCATTTTCCGAATA TACAGGTATTGATGAAAGAGGAGGTACAACATCTTGGGGAACAAGTGGTCAACCAAGTCCTTCCTATGATTCATCTAGAGGTTTTACAGA CAGCCCTCATTACAGTGATCACTTGAATGACAGTCGATTAGGAGCCCATGAAGGCTTGTCCCCAACACCTTTCATGAACTCAAATCTGAT GGGAAAAACATCAGAGAGAGGCTCATTTTCCCTGTACAGCAGAGATACTGGATTACCAGGCTGTCAATCTAGTCTCCTGAGACAAGATCT GGGGCTTGGGAGCCCAGCACAGCTATCTTCTTCAGGAAAACCTGGGACAGCATACTATTCATTCTCTGCTACAAGTTCCAGGAGGAGACC ACTCCATGACTCTGCAGCGCTTGATCCCTTGCAAGCAAAAAAAGTCAGAAAGGTGCCTCCTGGTTTGCCTTCTTCTGTATATGCACCATC CCCAAATTCAGATGATTTCAACCGTGAATCTCCTAGTTATCCATCTCCTAAGCCACCAACCAGTATGTTCGCTAGCACTTTCTTTATGCA AGATGGGACCCACAATTCTTCTGACCTTTGGAGTTCATCAAATGGGATGAGCCAGCCTGGTTTTGGTGGAATTCTGGGGACCTCCACTTC CCACATGTCTCAATCCAGTAGTTATGGCAACCTTCATTCACATGACCGCTTGAGTTATCCTCCACACTCAGTTTCACCAACAGACATAAA CACGAGTCTTCCACCAATGTCCAGCTTTCATCGCGGCAGTACCAGCAGTTCACCTTACGTTGCTGCCTCACACACTCCTCCCATCAATGG ATCAGACAGCATTCTAGGAACCAGAGGGAATGCTGCTGGAAGCTCACAGACAGGTGATGCACTTGGAAAGGCTTTGGCATCTATTTATTC TCCTGACCATACCAGCAGTAGTTTTCCGTCAAATCCATCAACACCAGTTGGATCACCTTCACCTCTCACAGGTACCAGTCAGTGGCCAAG ACCTGGAGGGCAAGCACCTTCATCCCCAAGCTATGAAAACTCACTCCACTCCCTGAAAAATCGAGTTGAGCAGCAACTTCACGAGCATTT GCAAGATGCAATGTCCTTCTTAAAGGATGTCTGTGAGCAGTCTCGAATGGAGGATCGTTTAGACAGACTGGATGATGCAATCCATGTGCT GCGGAACCATGCTGTGGGACCTTCCACCAGTTTGCCTGCTGGTCACAGTGATATACATAGTTTATTGGGACCATCCCATAATGCACCAAT TGGAAGCCTCAATTCAAACTATGGAGGATCAAGCCTTGTTGCAAGCAGTCGATCAGCTTCAATGGTTGGAACTCATCGGGAAGACTCTGT CAGTCTCAATGGCAATCATTCAGTCCTGTCTAGTACAGTCACTACTTCAAGCACAGACCTGAACCATAAAACACAAGAAAATTATAGAGG TGGCTTGCAAAGTCAGTCTGGAACTGTTGTTACAACAGAAATCAAGACTGAAAACAAAGAAAAGGATGAAAACCTTCATGAACCTCCTTC ATCAGATGACATGAAGTCAGATGATGAATCCTCCCAAAAAGATATCAAGGTTTCATCTAGAGGCAGAACAAGCAGTACTAATGAAGATGA GGATTTGAACCCTGAACAGAAGATAGAAAGGGAGAAGGAGAGGCGGATGGCTAACAATGCCAGAGAACGCTTACGCGTGCGGGATATTAA TGAAGCATTCAAAGAGCTTGGCCGAATGTGTCAGCTTCACTTGAAGAGTGAAAAACCCCAAACAAAACTCCTTATTCTTCATCAAGCCGT GGCAGTCATCCTTAGTCTAGAACAGCAAGTCAGAGAGAGGAACCTTAACCCCAAAGCAGCCTGCCTTAAGAGAAGGGAAGAAGAAAAAGT TTCTGCCGTATCGGCAGAGCCGCCAACCACACTGCCAGGAACCCATCCTGGGCTTAGTGAAACTACCAACCCTATGGGTCATATGTAAAC ATCAGCCAGTTCCAGAGTTATCAGTAGGCTAGATAGAAGGTGACCTCTCCTCATAAGGACTTGGACAACTCAGATTATCTGAAGACACAA ACCTGACAGGAGGGAGAAGAAAAAACAAAACACTTGAACCAAGAAACTCAAATGTAATCCTACGATCAAAGCAACTGGTCAACACTTCCA TCAGAAGTGAAGATAGGAAGCTCATCAGATAGAACATCAGCCCATGAGATGTTTGCAACAAATCTTTTGTTGCAAGCAGTGTGTCGCTTC TGCACAATCAGAGACTGTCTCGATCTCTCCACTCACCGTGGAAGTTGCCTTGTGCCTAAACTGAATTGACAAATGCATTGTAACTACAAA TTTTATTTATTGTTATGAAACTGTAAGGTCTACATATAAAGGGAAAAAGTTAATGTGGAAAGCTGATCTACACTCAGCTGATGCCAGCAT ACATTAAAGCGGTTCACGTGCAGAGAACAAAGCAGTGACAACCATTGGCCCTTAGCATTCCCGGCATACCTATTAGTGTCTTAAAAAGGA AGGGAAAAGTCTTTTGTTGCCCTCTCCTATCCTCTTGCCATATGAATAGCGTTTTCCATGAAATAGGAAAATATTACTTGGTATAGCATT TCTCTTGCTCTCATTTTTTGATTTATTTTTATTTTCTCTTTGTGGGTGTTATATTTGATCTCTAAATCTGAACAGTTTATGGTCACAGTC CAGCCTCCTCCGTGCAGCCCTGTGTGCTTTGCACATTTACCTTACAGTGGTAAGCAGAGACCATCTGTGACCATAGCCTAGCTAGCATTT TAAAAGGGGAAATTTTGTTCTCTAGGTTTTCCCCCAAATAAACATTGCTTTATTTCTAATAATAACCAAGACTTTTCAAGCTTCTAGATC TCATAGGAAAGCTTGTAATAGCAAAATTGTAAATTACAAGGGAAGAATCTACTTTTTAGAAATCGCTTTGTTTTCCAAGCAGTAAGTACT ACATACAGTACTTGTAAAGTGTTAGCTGTAAGTAAGCACAAAATACATTTAAAATACAAAGACGATTTTTTCAGGCTGTGATTATGGTGA ACATAACAAAACCCAGTAGTCACCAAGGCAGGTAGTGTGATAAATGAACACACCACTCTGAGGCTAATTACCTAATGGAATACAAGAGCA ATGGTCACCCGTATTTCCTTATCCTAGCCTTTATTTCTCTGTCATTTGGATGGCTGGTCAATGGGGAAGAATTGAGTGGGTGATTTAATC AACTGCAAACCATCTGCCCCTGTCCCAAAATGATGAGCCAGATTAGCATTAAACCAGTACTTGTCAGTCCATCTTAATACTGTTCATTAA GGCACTCTCTGTCTCTAATCCTTAGGAGTTGTTTTAAAAGACATAATCACTTTGAACTTCCATGAAACCTGTCTTCCACCACAACAACCC TGGGAGAGAAAAACATGCTAAAGGAGGTATCTTGGCTTAATAATTCCTTATAGCCAATATCAACAGTGGCAATCAGCACACAGAGGAAAG GACCCAAATCACTATGTAGCTTAAAGATTTCTGTTAATTTGAAAGAACAAAAACAAGACAGAACTTCTGGTACTCTAATCAGGATGATTC CTAACAAGTCAGTCATTTGTGAACTTAGTGGACTTTTTGGTTACTTTAATTTGCATATATTCTCCAGTTACATCGGACTCTATCTGTGGC CTTGTTCTTCATTTCAGTGTTAATCAGCTAAACAGAAGTTGTTGCTTATGATGTGTGAGTGAACATATGCCACTGCCTGGCCTTTTTTTC TTCAGAGCTTGTTGTCTTTTTCGCTATATTAGACTTTGCAGTATGCCCAGAAGCTTTCCTTCATAAAATAGAAAGAAAAAAACATTTGGC TTATTTTTCACTGTAGCTAGTCTTTTATACAATAATCTTGTAAGAAAATTTCTTGAATTCTAAATATTACTCTTTCTAGATTTTTGAAAT CAAAAAGTTTTCAGTAAAAAGTTTCTTACTTTATTTTATTATATTAGGTAGTAAAAAATGTAGGGTTATTTACCATAACCTGTTCATTAA TATCAGAAATTTACAATAGCATTTTAAGACCATAGTAGGATTCTAGCATACCGTGTAGTACCTATGGAGTATTGTAAGAGCTAATTGTTG GAGATGAATTGCTTCTCATCTTGTTCTCCAGTTTCCATTGTTGGTTTATTGCAGATTTGTATCCTGTGTCAAATTCAAGGTATTATTGAT AAACCTTTTCAACCAGCAGCAAGAAGTTCAAATTTTTTTCTGTCACTGTAACAGAAAACACAATATGTATATAACATTTATGTAGCAATA >39694_39694_2_IMPAD1-TCF12_IMPAD1_chr8_57890608_ENST00000262644_TCF12_chr15_57355947_ENST00000438423_length(amino acids)=896AA_BP=239 MRRRRRRALEACESRSPERPASHAMAPMGIRLSPLGVAVFCLLGLGVLYHLYSGFLAGRFSLFGLGGEPGGGAAGPAAAADGGTVDLREM LAVSVLAAVRGGDEVRRVRESNVLHEKSKGKTREGAEDKMTSGDVLSNRKMFYLLKTAFPSVQINTEEHVDAADQEVILWDHKIPEDILK EVTTPKEVPAESVTVWIDPLDATQEYTEDLRKYVTTMVCVAVNGKPMLGVIHKPFSEYTGIDERGGTTSWGTSGQPSPSYDSSRGFTDSP HYSDHLNDSRLGAHEGLSPTPFMNSNLMGKTSERGSFSLYSRDTGLPGCQSSLLRQDLGLGSPAQLSSSGKPGTAYYSFSATSSRRRPLH DSAALDPLQAKKVRKVPPGLPSSVYAPSPNSDDFNRESPSYPSPKPPTSMFASTFFMQDGTHNSSDLWSSSNGMSQPGFGGILGTSTSHM SQSSSYGNLHSHDRLSYPPHSVSPTDINTSLPPMSSFHRGSTSSSPYVAASHTPPINGSDSILGTRGNAAGSSQTGDALGKALASIYSPD HTSSSFPSNPSTPVGSPSPLTGTSQWPRPGGQAPSSPSYENSLHSLKNRVEQQLHEHLQDAMSFLKDVCEQSRMEDRLDRLDDAIHVLRN HAVGPSTSLPAGHSDIHSLLGPSHNAPIGSLNSNYGGSSLVASSRSASMVGTHREDSVSLNGNHSVLSSTVTTSSTDLNHKTQENYRGGL QSQSGTVVTTEIKTENKEKDENLHEPPSSDDMKSDDESSQKDIKVSSRGRTSSTNEDEDLNPEQKIEREKERRMANNARERLRVRDINEA -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for IMPAD1-TCF12 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
| Tgene | TCF12 | chr8:57890608 | chr15:57355947 | ENST00000267811 | 2 | 20 | 182_196 | 49.333333333333336 | 1821.3333333333333 | RUNX1T1 | |
| Tgene | TCF12 | chr8:57890608 | chr15:57355947 | ENST00000333725 | 2 | 21 | 182_196 | 49.333333333333336 | 1420.0 | RUNX1T1 | |
| Tgene | TCF12 | chr8:57890608 | chr15:57355947 | ENST00000343827 | 0 | 13 | 182_196 | 0 | 1254.6666666666667 | RUNX1T1 | |
| Tgene | TCF12 | chr8:57890608 | chr15:57355947 | ENST00000438423 | 2 | 21 | 182_196 | 49.333333333333336 | 1398.3333333333333 | RUNX1T1 | |
| Tgene | TCF12 | chr8:57890608 | chr15:57355947 | ENST00000557843 | 2 | 20 | 182_196 | 49.333333333333336 | 1301.0 | RUNX1T1 |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for IMPAD1-TCF12 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for IMPAD1-TCF12 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
