
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:ING3-IVNS1ABP (FusionGDB2 ID:39742) |
Fusion Gene Summary for ING3-IVNS1ABP |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: ING3-IVNS1ABP | Fusion gene ID: 39742 | Hgene | Tgene | Gene symbol | ING3 | IVNS1ABP | Gene ID | 54556 | 10625 |
| Gene name | inhibitor of growth family member 3 | influenza virus NS1A binding protein | |
| Synonyms | Eaf4|ING2|MEAF4|p47ING3 | ARA3|FLARA3|HSPC068|KLHL39|ND1|NS-1|NS1-BP|NS1BP | |
| Cytomap | 7q31.31 | 1q25.3 | |
| Type of gene | protein-coding | protein-coding | |
| Description | inhibitor of growth protein 3 | influenza virus NS1A-binding proteinNCX downstream gene 1NS1-binding proteinaryl hydrocarbon receptor-associated 3aryl hydrocarbon receptor-associated protein 3kelch-like family member 39kelch-like protein 39 | |
| Modification date | 20200313 | 20200320 | |
| UniProtAcc | Q9NXR8 | Q9Y6Y0 | |
| Ensembl transtripts involved in fusion gene | ENST00000315870, ENST00000339121, ENST00000431467, ENST00000445699, | ENST00000392007, ENST00000459929, ENST00000367497, ENST00000367498, | |
| Fusion gene scores | * DoF score | 6 X 1 X 3=18 | 17 X 9 X 9=1377 |
| # samples | 6 | 18 | |
| ** MAII score | log2(6/18*10)=1.73696559416621 effective Gene in Pan-Cancer Fusion Genes (eGinPCFGs). DoF>8 and MAII>0 | log2(18/1377*10)=-2.93545974780529 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: ING3 [Title/Abstract] AND IVNS1ABP [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | ING3(120595678)-IVNS1ABP(185274775), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | ING3-IVNS1ABP seems lost the major protein functional domain in Hgene partner, which is a epigenetic factor due to the frame-shifted ORF. ING3-IVNS1ABP seems lost the major protein functional domain in Hgene partner, which is a essential gene due to the frame-shifted ORF. ING3-IVNS1ABP seems lost the major protein functional domain in Hgene partner, which is a tumor suppressor due to the frame-shifted ORF. ING3-IVNS1ABP seems lost the major protein functional domain in Tgene partner, which is a essential gene due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | ING3 | GO:0043065 | positive regulation of apoptotic process | 16387653 |
| Hgene | ING3 | GO:0043967 | histone H4 acetylation | 14966270|16387653 |
| Hgene | ING3 | GO:0043968 | histone H2A acetylation | 14966270|16387653 |
 Fusion gene breakpoints across ING3 (5'-gene) Fusion gene breakpoints across ING3 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
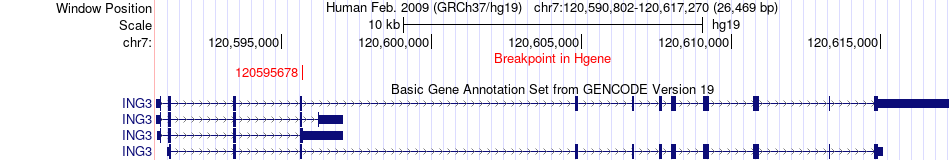 |
 Fusion gene breakpoints across IVNS1ABP (3'-gene) Fusion gene breakpoints across IVNS1ABP (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
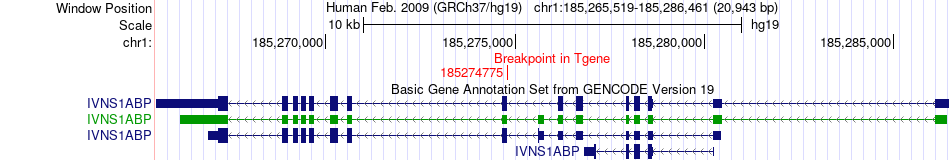 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | LUSC | TCGA-22-5489 | ING3 | chr7 | 120595678 | + | IVNS1ABP | chr1 | 185274775 | - |
Top |
Fusion Gene ORF analysis for ING3-IVNS1ABP |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-5UTR | ENST00000315870 | ENST00000392007 | ING3 | chr7 | 120595678 | + | IVNS1ABP | chr1 | 185274775 | - |
| 5CDS-5UTR | ENST00000315870 | ENST00000459929 | ING3 | chr7 | 120595678 | + | IVNS1ABP | chr1 | 185274775 | - |
| 5CDS-5UTR | ENST00000339121 | ENST00000392007 | ING3 | chr7 | 120595678 | + | IVNS1ABP | chr1 | 185274775 | - |
| 5CDS-5UTR | ENST00000339121 | ENST00000459929 | ING3 | chr7 | 120595678 | + | IVNS1ABP | chr1 | 185274775 | - |
| 5CDS-5UTR | ENST00000431467 | ENST00000392007 | ING3 | chr7 | 120595678 | + | IVNS1ABP | chr1 | 185274775 | - |
| 5CDS-5UTR | ENST00000431467 | ENST00000459929 | ING3 | chr7 | 120595678 | + | IVNS1ABP | chr1 | 185274775 | - |
| 5CDS-5UTR | ENST00000445699 | ENST00000392007 | ING3 | chr7 | 120595678 | + | IVNS1ABP | chr1 | 185274775 | - |
| 5CDS-5UTR | ENST00000445699 | ENST00000459929 | ING3 | chr7 | 120595678 | + | IVNS1ABP | chr1 | 185274775 | - |
| 5CDS-intron | ENST00000315870 | ENST00000367497 | ING3 | chr7 | 120595678 | + | IVNS1ABP | chr1 | 185274775 | - |
| 5CDS-intron | ENST00000339121 | ENST00000367497 | ING3 | chr7 | 120595678 | + | IVNS1ABP | chr1 | 185274775 | - |
| 5CDS-intron | ENST00000431467 | ENST00000367497 | ING3 | chr7 | 120595678 | + | IVNS1ABP | chr1 | 185274775 | - |
| 5CDS-intron | ENST00000445699 | ENST00000367497 | ING3 | chr7 | 120595678 | + | IVNS1ABP | chr1 | 185274775 | - |
| Frame-shift | ENST00000445699 | ENST00000367498 | ING3 | chr7 | 120595678 | + | IVNS1ABP | chr1 | 185274775 | - |
| In-frame | ENST00000315870 | ENST00000367498 | ING3 | chr7 | 120595678 | + | IVNS1ABP | chr1 | 185274775 | - |
| In-frame | ENST00000339121 | ENST00000367498 | ING3 | chr7 | 120595678 | + | IVNS1ABP | chr1 | 185274775 | - |
| In-frame | ENST00000431467 | ENST00000367498 | ING3 | chr7 | 120595678 | + | IVNS1ABP | chr1 | 185274775 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000315870 | ING3 | chr7 | 120595678 | + | ENST00000367498 | IVNS1ABP | chr1 | 185274775 | - | 3334 | 415 | 112 | 1686 | 524 |
| ENST00000339121 | ING3 | chr7 | 120595678 | + | ENST00000367498 | IVNS1ABP | chr1 | 185274775 | - | 3320 | 401 | 98 | 1672 | 524 |
| ENST00000431467 | ING3 | chr7 | 120595678 | + | ENST00000367498 | IVNS1ABP | chr1 | 185274775 | - | 3206 | 287 | 41 | 1558 | 505 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000315870 | ENST00000367498 | ING3 | chr7 | 120595678 | + | IVNS1ABP | chr1 | 185274775 | - | 0.000145946 | 0.9998541 |
| ENST00000339121 | ENST00000367498 | ING3 | chr7 | 120595678 | + | IVNS1ABP | chr1 | 185274775 | - | 0.000147317 | 0.99985266 |
| ENST00000431467 | ENST00000367498 | ING3 | chr7 | 120595678 | + | IVNS1ABP | chr1 | 185274775 | - | 0.000204035 | 0.999796 |
Top |
Fusion Genomic Features for ING3-IVNS1ABP |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
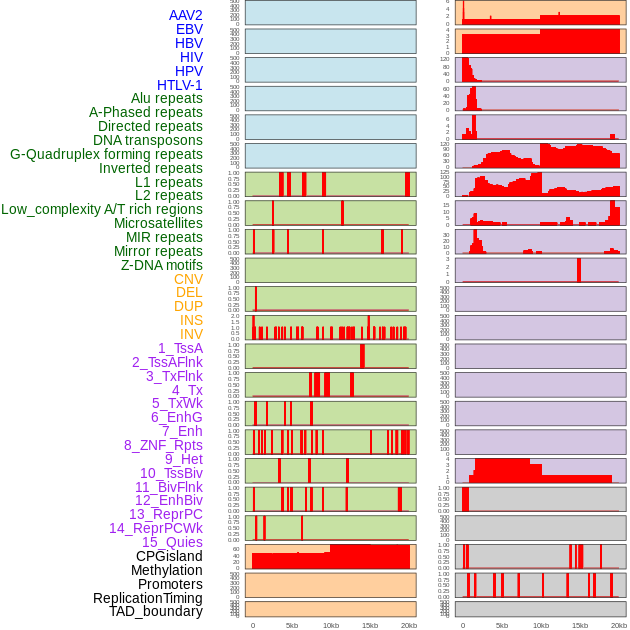 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for ING3-IVNS1ABP |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr7:120595678/chr1:185274775) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| ING3 | IVNS1ABP |
| FUNCTION: Component of the NuA4 histone acetyltransferase (HAT) complex which is involved in transcriptional activation of select genes principally by acetylation of nucleosomal histones H4 and H2A. This modification may both alter nucleosome - DNA interactions and promote interaction of the modified histones with other proteins which positively regulate transcription. This complex may be required for the activation of transcriptional programs associated with oncogene and proto-oncogene mediated growth induction, tumor suppressor mediated growth arrest and replicative senescence, apoptosis, and DNA repair. NuA4 may also play a direct role in DNA repair when directly recruited to sites of DNA damage. Component of a SWR1-like complex that specifically mediates the removal of histone H2A.Z/H2AZ1 from the nucleosome. {ECO:0000269|PubMed:12545155, ECO:0000269|PubMed:14966270, ECO:0000269|PubMed:24463511}. | FUNCTION: Involved in many cell functions, including pre-mRNA splicing, the aryl hydrocarbon receptor (AHR) pathway, F-actin organization and protein ubiquitination. Plays a role in the dynamic organization of the actin skeleton as a stabilizer of actin filaments by association with F-actin through Kelch repeats (By similarity). Protects cells from cell death induced by actin destabilization (By similarity). Functions as modifier of the AHR/Aryl hydrocarbon receptor pathway increasing the concentration of AHR available to activate transcription (PubMed:16582008). In addition, functions as a negative regulator of BCR(KLHL20) E3 ubiquitin ligase complex to prevent ubiquitin-mediated proteolysis of PML and DAPK1, two tumor suppressors (PubMed:25619834). Inhibits pre-mRNA splicing (in vitro) (PubMed:9696811). {ECO:0000250|UniProtKB:Q920Q8, ECO:0000269|PubMed:16582008, ECO:0000269|PubMed:25619834, ECO:0000269|PubMed:9696811}.; FUNCTION: (Microbial infection) Involved in the alternative splicing of influenza A virus M1 mRNA through interaction with HNRNPK, thereby facilitating the generation of viral M2 protein. {ECO:0000269|PubMed:23825951, ECO:0000269|PubMed:9696811}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | IVNS1ABP | chr7:120595678 | chr1:185274775 | ENST00000367498 | 6 | 15 | 369_415 | 219 | 643.0 | Repeat | Note=Kelch 1 | |
| Tgene | IVNS1ABP | chr7:120595678 | chr1:185274775 | ENST00000367498 | 6 | 15 | 416_463 | 219 | 643.0 | Repeat | Note=Kelch 2 | |
| Tgene | IVNS1ABP | chr7:120595678 | chr1:185274775 | ENST00000367498 | 6 | 15 | 465_512 | 219 | 643.0 | Repeat | Note=Kelch 3 | |
| Tgene | IVNS1ABP | chr7:120595678 | chr1:185274775 | ENST00000367498 | 6 | 15 | 513_559 | 219 | 643.0 | Repeat | Note=Kelch 4 | |
| Tgene | IVNS1ABP | chr7:120595678 | chr1:185274775 | ENST00000367498 | 6 | 15 | 560_606 | 219 | 643.0 | Repeat | Note=Kelch 5 | |
| Tgene | IVNS1ABP | chr7:120595678 | chr1:185274775 | ENST00000367498 | 6 | 15 | 608_642 | 219 | 643.0 | Repeat | Note=Kelch 6 |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | ING3 | chr7:120595678 | chr1:185274775 | ENST00000315870 | + | 4 | 12 | 313_329 | 89 | 419.0 | Compositional bias | Note=Poly-Ser |
| Hgene | ING3 | chr7:120595678 | chr1:185274775 | ENST00000339121 | + | 4 | 5 | 313_329 | 89 | 93.0 | Compositional bias | Note=Poly-Ser |
| Hgene | ING3 | chr7:120595678 | chr1:185274775 | ENST00000445699 | + | 1 | 4 | 313_329 | 0 | 100.0 | Compositional bias | Note=Poly-Ser |
| Hgene | ING3 | chr7:120595678 | chr1:185274775 | ENST00000315870 | + | 4 | 12 | 360_409 | 89 | 419.0 | Zinc finger | PHD-type |
| Hgene | ING3 | chr7:120595678 | chr1:185274775 | ENST00000339121 | + | 4 | 5 | 360_409 | 89 | 93.0 | Zinc finger | PHD-type |
| Hgene | ING3 | chr7:120595678 | chr1:185274775 | ENST00000445699 | + | 1 | 4 | 360_409 | 0 | 100.0 | Zinc finger | PHD-type |
| Tgene | IVNS1ABP | chr7:120595678 | chr1:185274775 | ENST00000367498 | 6 | 15 | 134_233 | 219 | 643.0 | Domain | Note=BACK | |
| Tgene | IVNS1ABP | chr7:120595678 | chr1:185274775 | ENST00000367498 | 6 | 15 | 32_99 | 219 | 643.0 | Domain | BTB |
Top |
Fusion Gene Sequence for ING3-IVNS1ABP |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >39742_39742_1_ING3-IVNS1ABP_ING3_chr7_120595678_ENST00000315870_IVNS1ABP_chr1_185274775_ENST00000367498_length(transcript)=3334nt_BP=415nt TTTTTTTTTTTTTCTTTTTTTTTTTTTGCCGGAGTCGAGCGGGTGCTGCTAGCGGAGGCGCCATATTGGAGGGGACAAAACTCCGGCGAC AGCGAGTGACACAAATAAACCCCTGGACCCCCTTGTTCCCTCAGCTCTAAGGGCCGCGATGTTGTACCTAGAAGACTATCTGGAAATGAT TGAGCAGCTTCCTATGGATCTGCGGGACCGCTTCACGGAAATGCGCGAGATGGACCTGCAGGTGCAGAATGCAATGGATCAACTAGAACA AAGAGTCAGTGAATTCTTTATGAATGCAAAGAAAAATAAACCTGAGTGGAGGGAAGAGCAAATGGCATCCATCAAAAAAGACTACTATAA AGCTTTGGAAGATGCAGATGAGAAGGTTCAGTTGGCAAACCAGATATATGACTTGGTTCAAACCTTGTACTACTCAGCTGATCACAAGCT GCTTGATGGGAACCTACTAGATGGACAGGCTGAGGTGTTTGGCAGTGATGATGACCACATTCAGTTTGTGCAGAAAAAGCCACCACGTGA GAATGGCCATAAGCAGATAAGTAGCAGTTCAACTGGATGTCTCTCTTCTCCAAATGCTACAGTACAAAGCCCTAAGCATGAGTGGAAAAT CGTTGCTTCAGAAAAGACTTCAAATAACACTTACTTGTGCCTGGCTGTGCTGGATGGTATATTCTGTGTCATTTTTCTTCATGGGAGAAA CAGCCCACAGAGCTCACCAACAAGTACTCCAAAACTAAGTAAGAGTTTAAGCTTTGAGATGCAACAAGATGAGCTAATCGAAAAGCCCAT GTCTCCTATGCAGTACGCACGATCTGGTCTGGGAACAGCAGAGATGAATGGCAAACTCATAGCTGCAGGTGGCTATAACAGAGAGGAATG TCTTCGAACAGTCGAATGCTATAATCCACATACAGATCACTGGTCCTTTCTTGCTCCCATGAGAACACCAAGAGCCCGATTTCAAATGGC TGTACTCATGGGCCAGCTCTATGTGGTAGGTGGATCAAATGGCCACTCAGATGACCTGAGTTGTGGAGAGATGTATGATTCAAACATAGA TGACTGGATTCCTGTTCCAGAATTGAGAACTAACCGTTGTAATGCAGGAGTGTGTGCTCTGAATGGAAAGTTATACATCGTTGGTGGCTC TGATCCATATGGTCAAAAAGGACTGAAAAATTGTGATGTATTTGATCCTGTAACAAAGTTGTGGACAAGCTGTGCCCCTCTTAACATTCG GAGACACCAGTCTGCAGTCTGTGAGCTTGGTGGTTATTTGTACATAATCGGAGGTGCAGAATCTTGGAATTGTCTGAACACAGTAGAACG ATACAATCCTGAAAATAATACCTGGACTTTAATTGCACCCATGAATGTGGCTAGGCGAGGAGCTGGAGTGGCTGTTCTTAATGGAAAACT GTTTGTATGTGGTGGCTTTGATGGTTCTCATGCCATCAGTTGTGTGGAAATGTATGATCCAACTAGAAATGAATGGAAGATGATGGGAAA TATGACTTCACCAAGGAGCAATGCTGGGATTGCAACTGTAGGGAACACCATTTATGCAGTGGGAGGATTCGATGGCAATGAATTTCTGAA TACGGTGGAAGTCTATAACCTTGAGTCAAATGAATGGAGCCCCTATACAAAGATTTTCCAGTTTTAACAAATTTAAGACCCTCTCAAACT AACAGGCTTAGTGATGTAATTATGGTTAGTAGAGGTACACTTGTGAATAAAGAGGGTGGGTGGGTATAGATGTTGCTAACAGCAACACAA AGCTTTTGCATATTGCATACTATTAAACATGCTGTACATACTTTTTGGGTTTATTTGGAAAGGAATGCAAAGATGAAGGTCTGTTTTGTG TACTTTTAAGACTTTGGTTATTTTACTTTTTGGAAAAGAATAAACCAAGAATTGATTGGGCACATCATTTCAAGAAGTCCCCTCTCCTCC ACATTTGTTTTGCCAATTTGCACATTAAATGACTCTTCCCTCAAATGTGTACTATGGGGTAAAAGGGGTAGGGTTTAAAGATGTAGACAG TTGGGTTTTTTAAGGGCCCTTTTTCAATAACTGGAACACTCTATAACAAAGGATACTTATTTAAATAGATGACATTGACTATTTTTGTTT TTATTAAAAGGAAGCTTACATGCCTACCAATATTTAATCTTTTATGATTGCCTTTTTATAACTTTTTATATTCTCAGCAGAGTGCTTTAC CAATTGAAGTAAAATGTGGCAGGCTGGAGTTATTGAAGCAGAGTGGCAGTCTTCAGTTTGCAGAGTAGGGGTCTGTCTTTTAAACTCTGA GTGCAAACTTCAGAGTTCTTGCCTTGGCTGCAGTTTTTTTCCTTCAAGAATGCAGTACTAACATTTATTTGAGTGGAGTTACTGAACAGT AACATAGCTGTGATTTTTGGTATTTGAAACACTGGTTTTAAATATTTTGACTTGTTGAGGGTATGTTTTATATAGCAAGACATTATATAG CAGTAAAAAATGGTGTTTTATCTTCTATATAATTCCTGTTTTTATTATTAACAAAACAGTCCTAAATAGCAGCCCTCAATTGTGAAAAAA TTTACTTTAAACTACATTAGGTTGTGAATGCAGGTTTTATCAGAACTATGTTTTTGTTCAGTTTATCTGTTCATATGGATAAATATTGGT TGGGATGACTTGGTGTCTAATGTGTAGTGCTACACACCTAACTTATGGGGCCAAAATAGCATGTCCTAATGCTTGCTGCTGATTTAAACA CATTAAAGGTACTTTGCAGGAAATCCTTGCACCATGGGATTAATATCCAATTGCTGCTTGTACACTCATTCATTACTAAAAGTTTTGAGA AATTTTTTTTTCCAGTAATGAGCTTAAGAAATTTGTGGAAAATAACTCACCTGGCATCTTACATCTGAAATAAGGAATGATATAAGGTTT TTTTTTCTCACAGAAGATGAAGCACACAGGAACCTAATGGGCCAACTGGGATGAGGTGACTATTCTGAGATGACTATTCAGTGGCTAACT TGGGTTAGGAAGAAAATAATTAGGTATTTTCTCCAAATGTTCACTGGTACTCTGCCACTTTATTTCTCTCATCTGTTACACAAAGAACCA CCAGGAAAGCAAATCAGTTTGGTTGGTAACTCTGTAATTCCTAACTATCACTGGTTTGGTTCTGGACTAAAACTACATTGACAGATTGAA TTTGCCTAATATGATGACTGTTTTTAATATGGATCTGTATGTGTTCTATTCAGCACAAGGAAATAAAATTTTAGTTGAGGATTCAGCACT >39742_39742_1_ING3-IVNS1ABP_ING3_chr7_120595678_ENST00000315870_IVNS1ABP_chr1_185274775_ENST00000367498_length(amino acids)=524AA_BP=101 MDPLVPSALRAAMLYLEDYLEMIEQLPMDLRDRFTEMREMDLQVQNAMDQLEQRVSEFFMNAKKNKPEWREEQMASIKKDYYKALEDADE KVQLANQIYDLVQTLYYSADHKLLDGNLLDGQAEVFGSDDDHIQFVQKKPPRENGHKQISSSSTGCLSSPNATVQSPKHEWKIVASEKTS NNTYLCLAVLDGIFCVIFLHGRNSPQSSPTSTPKLSKSLSFEMQQDELIEKPMSPMQYARSGLGTAEMNGKLIAAGGYNREECLRTVECY NPHTDHWSFLAPMRTPRARFQMAVLMGQLYVVGGSNGHSDDLSCGEMYDSNIDDWIPVPELRTNRCNAGVCALNGKLYIVGGSDPYGQKG LKNCDVFDPVTKLWTSCAPLNIRRHQSAVCELGGYLYIIGGAESWNCLNTVERYNPENNTWTLIAPMNVARRGAGVAVLNGKLFVCGGFD -------------------------------------------------------------- >39742_39742_2_ING3-IVNS1ABP_ING3_chr7_120595678_ENST00000339121_IVNS1ABP_chr1_185274775_ENST00000367498_length(transcript)=3320nt_BP=401nt TTTTTTTTTTTTTGCCGGAGTCGAGCGGGTGCTGCTAGCGGAGGCGCCATATTGGAGGGGACAAAACTCCGGCGACAGCGAGTGACACAA ATAAACCCCTGGACCCCCTTGTTCCCTCAGCTCTAAGGGCCGCGATGTTGTACCTAGAAGACTATCTGGAAATGATTGAGCAGCTTCCTA TGGATCTGCGGGACCGCTTCACGGAAATGCGCGAGATGGACCTGCAGGTGCAGAATGCAATGGATCAACTAGAACAAAGAGTCAGTGAAT TCTTTATGAATGCAAAGAAAAATAAACCTGAGTGGAGGGAAGAGCAAATGGCATCCATCAAAAAAGACTACTATAAAGCTTTGGAAGATG CAGATGAGAAGGTTCAGTTGGCAAACCAGATATATGACTTGGTTCAAACCTTGTACTACTCAGCTGATCACAAGCTGCTTGATGGGAACC TACTAGATGGACAGGCTGAGGTGTTTGGCAGTGATGATGACCACATTCAGTTTGTGCAGAAAAAGCCACCACGTGAGAATGGCCATAAGC AGATAAGTAGCAGTTCAACTGGATGTCTCTCTTCTCCAAATGCTACAGTACAAAGCCCTAAGCATGAGTGGAAAATCGTTGCTTCAGAAA AGACTTCAAATAACACTTACTTGTGCCTGGCTGTGCTGGATGGTATATTCTGTGTCATTTTTCTTCATGGGAGAAACAGCCCACAGAGCT CACCAACAAGTACTCCAAAACTAAGTAAGAGTTTAAGCTTTGAGATGCAACAAGATGAGCTAATCGAAAAGCCCATGTCTCCTATGCAGT ACGCACGATCTGGTCTGGGAACAGCAGAGATGAATGGCAAACTCATAGCTGCAGGTGGCTATAACAGAGAGGAATGTCTTCGAACAGTCG AATGCTATAATCCACATACAGATCACTGGTCCTTTCTTGCTCCCATGAGAACACCAAGAGCCCGATTTCAAATGGCTGTACTCATGGGCC AGCTCTATGTGGTAGGTGGATCAAATGGCCACTCAGATGACCTGAGTTGTGGAGAGATGTATGATTCAAACATAGATGACTGGATTCCTG TTCCAGAATTGAGAACTAACCGTTGTAATGCAGGAGTGTGTGCTCTGAATGGAAAGTTATACATCGTTGGTGGCTCTGATCCATATGGTC AAAAAGGACTGAAAAATTGTGATGTATTTGATCCTGTAACAAAGTTGTGGACAAGCTGTGCCCCTCTTAACATTCGGAGACACCAGTCTG CAGTCTGTGAGCTTGGTGGTTATTTGTACATAATCGGAGGTGCAGAATCTTGGAATTGTCTGAACACAGTAGAACGATACAATCCTGAAA ATAATACCTGGACTTTAATTGCACCCATGAATGTGGCTAGGCGAGGAGCTGGAGTGGCTGTTCTTAATGGAAAACTGTTTGTATGTGGTG GCTTTGATGGTTCTCATGCCATCAGTTGTGTGGAAATGTATGATCCAACTAGAAATGAATGGAAGATGATGGGAAATATGACTTCACCAA GGAGCAATGCTGGGATTGCAACTGTAGGGAACACCATTTATGCAGTGGGAGGATTCGATGGCAATGAATTTCTGAATACGGTGGAAGTCT ATAACCTTGAGTCAAATGAATGGAGCCCCTATACAAAGATTTTCCAGTTTTAACAAATTTAAGACCCTCTCAAACTAACAGGCTTAGTGA TGTAATTATGGTTAGTAGAGGTACACTTGTGAATAAAGAGGGTGGGTGGGTATAGATGTTGCTAACAGCAACACAAAGCTTTTGCATATT GCATACTATTAAACATGCTGTACATACTTTTTGGGTTTATTTGGAAAGGAATGCAAAGATGAAGGTCTGTTTTGTGTACTTTTAAGACTT TGGTTATTTTACTTTTTGGAAAAGAATAAACCAAGAATTGATTGGGCACATCATTTCAAGAAGTCCCCTCTCCTCCACATTTGTTTTGCC AATTTGCACATTAAATGACTCTTCCCTCAAATGTGTACTATGGGGTAAAAGGGGTAGGGTTTAAAGATGTAGACAGTTGGGTTTTTTAAG GGCCCTTTTTCAATAACTGGAACACTCTATAACAAAGGATACTTATTTAAATAGATGACATTGACTATTTTTGTTTTTATTAAAAGGAAG CTTACATGCCTACCAATATTTAATCTTTTATGATTGCCTTTTTATAACTTTTTATATTCTCAGCAGAGTGCTTTACCAATTGAAGTAAAA TGTGGCAGGCTGGAGTTATTGAAGCAGAGTGGCAGTCTTCAGTTTGCAGAGTAGGGGTCTGTCTTTTAAACTCTGAGTGCAAACTTCAGA GTTCTTGCCTTGGCTGCAGTTTTTTTCCTTCAAGAATGCAGTACTAACATTTATTTGAGTGGAGTTACTGAACAGTAACATAGCTGTGAT TTTTGGTATTTGAAACACTGGTTTTAAATATTTTGACTTGTTGAGGGTATGTTTTATATAGCAAGACATTATATAGCAGTAAAAAATGGT GTTTTATCTTCTATATAATTCCTGTTTTTATTATTAACAAAACAGTCCTAAATAGCAGCCCTCAATTGTGAAAAAATTTACTTTAAACTA CATTAGGTTGTGAATGCAGGTTTTATCAGAACTATGTTTTTGTTCAGTTTATCTGTTCATATGGATAAATATTGGTTGGGATGACTTGGT GTCTAATGTGTAGTGCTACACACCTAACTTATGGGGCCAAAATAGCATGTCCTAATGCTTGCTGCTGATTTAAACACATTAAAGGTACTT TGCAGGAAATCCTTGCACCATGGGATTAATATCCAATTGCTGCTTGTACACTCATTCATTACTAAAAGTTTTGAGAAATTTTTTTTTCCA GTAATGAGCTTAAGAAATTTGTGGAAAATAACTCACCTGGCATCTTACATCTGAAATAAGGAATGATATAAGGTTTTTTTTTCTCACAGA AGATGAAGCACACAGGAACCTAATGGGCCAACTGGGATGAGGTGACTATTCTGAGATGACTATTCAGTGGCTAACTTGGGTTAGGAAGAA AATAATTAGGTATTTTCTCCAAATGTTCACTGGTACTCTGCCACTTTATTTCTCTCATCTGTTACACAAAGAACCACCAGGAAAGCAAAT CAGTTTGGTTGGTAACTCTGTAATTCCTAACTATCACTGGTTTGGTTCTGGACTAAAACTACATTGACAGATTGAATTTGCCTAATATGA >39742_39742_2_ING3-IVNS1ABP_ING3_chr7_120595678_ENST00000339121_IVNS1ABP_chr1_185274775_ENST00000367498_length(amino acids)=524AA_BP=101 MDPLVPSALRAAMLYLEDYLEMIEQLPMDLRDRFTEMREMDLQVQNAMDQLEQRVSEFFMNAKKNKPEWREEQMASIKKDYYKALEDADE KVQLANQIYDLVQTLYYSADHKLLDGNLLDGQAEVFGSDDDHIQFVQKKPPRENGHKQISSSSTGCLSSPNATVQSPKHEWKIVASEKTS NNTYLCLAVLDGIFCVIFLHGRNSPQSSPTSTPKLSKSLSFEMQQDELIEKPMSPMQYARSGLGTAEMNGKLIAAGGYNREECLRTVECY NPHTDHWSFLAPMRTPRARFQMAVLMGQLYVVGGSNGHSDDLSCGEMYDSNIDDWIPVPELRTNRCNAGVCALNGKLYIVGGSDPYGQKG LKNCDVFDPVTKLWTSCAPLNIRRHQSAVCELGGYLYIIGGAESWNCLNTVERYNPENNTWTLIAPMNVARRGAGVAVLNGKLFVCGGFD -------------------------------------------------------------- >39742_39742_3_ING3-IVNS1ABP_ING3_chr7_120595678_ENST00000431467_IVNS1ABP_chr1_185274775_ENST00000367498_length(transcript)=3206nt_BP=287nt GTATTTCGCCGTTGGCCCCGCCCCTCTGACGGACTCTCCCTTTGACAGTGATTGAGCAGCTTCCTATGGATCTGCGGGACCGCTTCACGG AAATGCGCGAGATGGACCTGCAGGTGCAGAATGCAATGGATCAACTAGAACAAAGAGTCAGTGAATTCTTTATGAATGCAAAGAAAAATA AACCTGAGTGGAGGGAAGAGCAAATGGCATCCATCAAAAAAGACTACTATAAAGCTTTGGAAGATGCAGATGAGAAGGTTCAGTTGGCAA ACCAGATATATGACTTGGTTCAAACCTTGTACTACTCAGCTGATCACAAGCTGCTTGATGGGAACCTACTAGATGGACAGGCTGAGGTGT TTGGCAGTGATGATGACCACATTCAGTTTGTGCAGAAAAAGCCACCACGTGAGAATGGCCATAAGCAGATAAGTAGCAGTTCAACTGGAT GTCTCTCTTCTCCAAATGCTACAGTACAAAGCCCTAAGCATGAGTGGAAAATCGTTGCTTCAGAAAAGACTTCAAATAACACTTACTTGT GCCTGGCTGTGCTGGATGGTATATTCTGTGTCATTTTTCTTCATGGGAGAAACAGCCCACAGAGCTCACCAACAAGTACTCCAAAACTAA GTAAGAGTTTAAGCTTTGAGATGCAACAAGATGAGCTAATCGAAAAGCCCATGTCTCCTATGCAGTACGCACGATCTGGTCTGGGAACAG CAGAGATGAATGGCAAACTCATAGCTGCAGGTGGCTATAACAGAGAGGAATGTCTTCGAACAGTCGAATGCTATAATCCACATACAGATC ACTGGTCCTTTCTTGCTCCCATGAGAACACCAAGAGCCCGATTTCAAATGGCTGTACTCATGGGCCAGCTCTATGTGGTAGGTGGATCAA ATGGCCACTCAGATGACCTGAGTTGTGGAGAGATGTATGATTCAAACATAGATGACTGGATTCCTGTTCCAGAATTGAGAACTAACCGTT GTAATGCAGGAGTGTGTGCTCTGAATGGAAAGTTATACATCGTTGGTGGCTCTGATCCATATGGTCAAAAAGGACTGAAAAATTGTGATG TATTTGATCCTGTAACAAAGTTGTGGACAAGCTGTGCCCCTCTTAACATTCGGAGACACCAGTCTGCAGTCTGTGAGCTTGGTGGTTATT TGTACATAATCGGAGGTGCAGAATCTTGGAATTGTCTGAACACAGTAGAACGATACAATCCTGAAAATAATACCTGGACTTTAATTGCAC CCATGAATGTGGCTAGGCGAGGAGCTGGAGTGGCTGTTCTTAATGGAAAACTGTTTGTATGTGGTGGCTTTGATGGTTCTCATGCCATCA GTTGTGTGGAAATGTATGATCCAACTAGAAATGAATGGAAGATGATGGGAAATATGACTTCACCAAGGAGCAATGCTGGGATTGCAACTG TAGGGAACACCATTTATGCAGTGGGAGGATTCGATGGCAATGAATTTCTGAATACGGTGGAAGTCTATAACCTTGAGTCAAATGAATGGA GCCCCTATACAAAGATTTTCCAGTTTTAACAAATTTAAGACCCTCTCAAACTAACAGGCTTAGTGATGTAATTATGGTTAGTAGAGGTAC ACTTGTGAATAAAGAGGGTGGGTGGGTATAGATGTTGCTAACAGCAACACAAAGCTTTTGCATATTGCATACTATTAAACATGCTGTACA TACTTTTTGGGTTTATTTGGAAAGGAATGCAAAGATGAAGGTCTGTTTTGTGTACTTTTAAGACTTTGGTTATTTTACTTTTTGGAAAAG AATAAACCAAGAATTGATTGGGCACATCATTTCAAGAAGTCCCCTCTCCTCCACATTTGTTTTGCCAATTTGCACATTAAATGACTCTTC CCTCAAATGTGTACTATGGGGTAAAAGGGGTAGGGTTTAAAGATGTAGACAGTTGGGTTTTTTAAGGGCCCTTTTTCAATAACTGGAACA CTCTATAACAAAGGATACTTATTTAAATAGATGACATTGACTATTTTTGTTTTTATTAAAAGGAAGCTTACATGCCTACCAATATTTAAT CTTTTATGATTGCCTTTTTATAACTTTTTATATTCTCAGCAGAGTGCTTTACCAATTGAAGTAAAATGTGGCAGGCTGGAGTTATTGAAG CAGAGTGGCAGTCTTCAGTTTGCAGAGTAGGGGTCTGTCTTTTAAACTCTGAGTGCAAACTTCAGAGTTCTTGCCTTGGCTGCAGTTTTT TTCCTTCAAGAATGCAGTACTAACATTTATTTGAGTGGAGTTACTGAACAGTAACATAGCTGTGATTTTTGGTATTTGAAACACTGGTTT TAAATATTTTGACTTGTTGAGGGTATGTTTTATATAGCAAGACATTATATAGCAGTAAAAAATGGTGTTTTATCTTCTATATAATTCCTG TTTTTATTATTAACAAAACAGTCCTAAATAGCAGCCCTCAATTGTGAAAAAATTTACTTTAAACTACATTAGGTTGTGAATGCAGGTTTT ATCAGAACTATGTTTTTGTTCAGTTTATCTGTTCATATGGATAAATATTGGTTGGGATGACTTGGTGTCTAATGTGTAGTGCTACACACC TAACTTATGGGGCCAAAATAGCATGTCCTAATGCTTGCTGCTGATTTAAACACATTAAAGGTACTTTGCAGGAAATCCTTGCACCATGGG ATTAATATCCAATTGCTGCTTGTACACTCATTCATTACTAAAAGTTTTGAGAAATTTTTTTTTCCAGTAATGAGCTTAAGAAATTTGTGG AAAATAACTCACCTGGCATCTTACATCTGAAATAAGGAATGATATAAGGTTTTTTTTTCTCACAGAAGATGAAGCACACAGGAACCTAAT GGGCCAACTGGGATGAGGTGACTATTCTGAGATGACTATTCAGTGGCTAACTTGGGTTAGGAAGAAAATAATTAGGTATTTTCTCCAAAT GTTCACTGGTACTCTGCCACTTTATTTCTCTCATCTGTTACACAAAGAACCACCAGGAAAGCAAATCAGTTTGGTTGGTAACTCTGTAAT TCCTAACTATCACTGGTTTGGTTCTGGACTAAAACTACATTGACAGATTGAATTTGCCTAATATGATGACTGTTTTTAATATGGATCTGT >39742_39742_3_ING3-IVNS1ABP_ING3_chr7_120595678_ENST00000431467_IVNS1ABP_chr1_185274775_ENST00000367498_length(amino acids)=505AA_BP=82 MTVIEQLPMDLRDRFTEMREMDLQVQNAMDQLEQRVSEFFMNAKKNKPEWREEQMASIKKDYYKALEDADEKVQLANQIYDLVQTLYYSA DHKLLDGNLLDGQAEVFGSDDDHIQFVQKKPPRENGHKQISSSSTGCLSSPNATVQSPKHEWKIVASEKTSNNTYLCLAVLDGIFCVIFL HGRNSPQSSPTSTPKLSKSLSFEMQQDELIEKPMSPMQYARSGLGTAEMNGKLIAAGGYNREECLRTVECYNPHTDHWSFLAPMRTPRAR FQMAVLMGQLYVVGGSNGHSDDLSCGEMYDSNIDDWIPVPELRTNRCNAGVCALNGKLYIVGGSDPYGQKGLKNCDVFDPVTKLWTSCAP LNIRRHQSAVCELGGYLYIIGGAESWNCLNTVERYNPENNTWTLIAPMNVARRGAGVAVLNGKLFVCGGFDGSHAISCVEMYDPTRNEWK -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for ING3-IVNS1ABP |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for ING3-IVNS1ABP |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for ING3-IVNS1ABP |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
