
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:INTS12-CXXC4 (FusionGDB2 ID:39913) |
Fusion Gene Summary for INTS12-CXXC4 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: INTS12-CXXC4 | Fusion gene ID: 39913 | Hgene | Tgene | Gene symbol | INTS12 | CXXC4 | Gene ID | 57117 | 80319 |
| Gene name | integrator complex subunit 12 | CXXC finger protein 4 | |
| Synonyms | INT12|PHF22|SBBI22 | IDAX | |
| Cytomap | 4q24 | 4q24 | |
| Type of gene | protein-coding | protein-coding | |
| Description | integrator complex subunit 12PHD finger protein 22hypothetical nuclear factor SBBI22 | CXXC-type zinc finger protein 4CXXC finger 4Dvl-binding protein IDAX (inhibition of the Dvl and Axin complex)inhibition of the Dvl and axin complex protein | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | Q96CB8 | Q9H2H0 | |
| Ensembl transtripts involved in fusion gene | ENST00000340139, ENST00000394735, ENST00000451321, | ENST00000426831, ENST00000466963, ENST00000394767, | |
| Fusion gene scores | * DoF score | 2 X 3 X 2=12 | 2 X 3 X 2=12 |
| # samples | 3 | 3 | |
| ** MAII score | log2(3/12*10)=1.32192809488736 effective Gene in Pan-Cancer Fusion Genes (eGinPCFGs). DoF>8 and MAII>0 | log2(3/12*10)=1.32192809488736 effective Gene in Pan-Cancer Fusion Genes (eGinPCFGs). DoF>8 and MAII>0 | |
| Context | PubMed: INTS12 [Title/Abstract] AND CXXC4 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | INTS12(106613133)-CXXC4(105393523), # samples:2 | ||
| Anticipated loss of major functional domain due to fusion event. | INTS12-CXXC4 seems lost the major protein functional domain in Hgene partner, which is a essential gene due to the frame-shifted ORF. INTS12-CXXC4 seems lost the major protein functional domain in Tgene partner, which is a transcription factor due to the frame-shifted ORF. INTS12-CXXC4 seems lost the major protein functional domain in Tgene partner, which is a tumor suppressor due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | INTS12 | GO:0016180 | snRNA processing | 16239144 |
| Tgene | CXXC4 | GO:0030178 | negative regulation of Wnt signaling pathway | 11113207 |
 Fusion gene breakpoints across INTS12 (5'-gene) Fusion gene breakpoints across INTS12 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
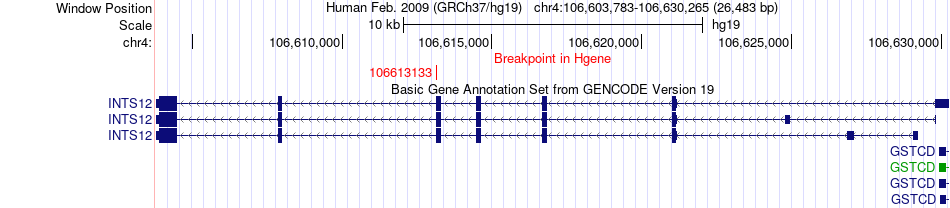 |
 Fusion gene breakpoints across CXXC4 (3'-gene) Fusion gene breakpoints across CXXC4 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
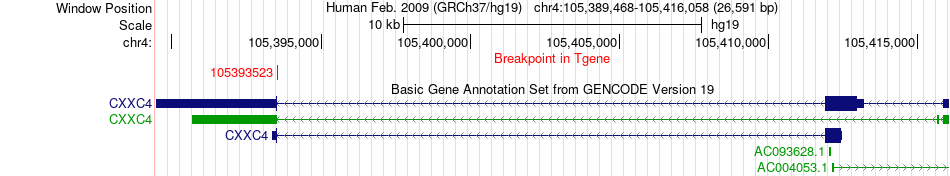 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | HNSC | TCGA-CN-5364-01A | INTS12 | chr4 | 106613133 | - | CXXC4 | chr4 | 105393523 | - |
Top |
Fusion Gene ORF analysis for INTS12-CXXC4 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-5UTR | ENST00000340139 | ENST00000426831 | INTS12 | chr4 | 106613133 | - | CXXC4 | chr4 | 105393523 | - |
| 5CDS-5UTR | ENST00000340139 | ENST00000466963 | INTS12 | chr4 | 106613133 | - | CXXC4 | chr4 | 105393523 | - |
| 5CDS-5UTR | ENST00000394735 | ENST00000426831 | INTS12 | chr4 | 106613133 | - | CXXC4 | chr4 | 105393523 | - |
| 5CDS-5UTR | ENST00000394735 | ENST00000466963 | INTS12 | chr4 | 106613133 | - | CXXC4 | chr4 | 105393523 | - |
| 5CDS-5UTR | ENST00000451321 | ENST00000426831 | INTS12 | chr4 | 106613133 | - | CXXC4 | chr4 | 105393523 | - |
| 5CDS-5UTR | ENST00000451321 | ENST00000466963 | INTS12 | chr4 | 106613133 | - | CXXC4 | chr4 | 105393523 | - |
| Frame-shift | ENST00000394735 | ENST00000394767 | INTS12 | chr4 | 106613133 | - | CXXC4 | chr4 | 105393523 | - |
| In-frame | ENST00000340139 | ENST00000394767 | INTS12 | chr4 | 106613133 | - | CXXC4 | chr4 | 105393523 | - |
| In-frame | ENST00000451321 | ENST00000394767 | INTS12 | chr4 | 106613133 | - | CXXC4 | chr4 | 105393523 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000340139 | INTS12 | chr4 | 106613133 | - | ENST00000394767 | CXXC4 | chr4 | 105393523 | - | 4927 | 872 | 215 | 916 | 233 |
| ENST00000451321 | INTS12 | chr4 | 106613133 | - | ENST00000394767 | CXXC4 | chr4 | 105393523 | - | 5192 | 1137 | 336 | 1181 | 281 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000340139 | ENST00000394767 | INTS12 | chr4 | 106613133 | - | CXXC4 | chr4 | 105393523 | - | 0.000306739 | 0.9996933 |
| ENST00000451321 | ENST00000394767 | INTS12 | chr4 | 106613133 | - | CXXC4 | chr4 | 105393523 | - | 0.000307394 | 0.9996927 |
Top |
Fusion Genomic Features for INTS12-CXXC4 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
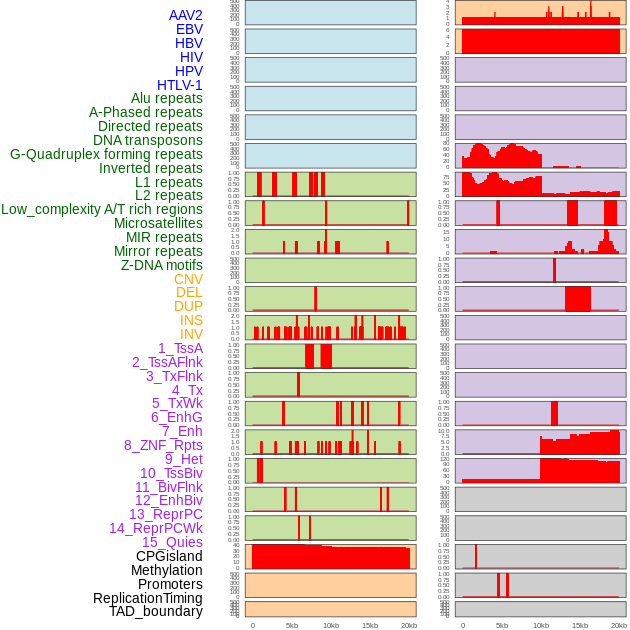 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for INTS12-CXXC4 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr4:106613133/chr4:105393523) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| INTS12 | CXXC4 |
| FUNCTION: Component of the Integrator complex, a complex involved in the small nuclear RNAs (snRNA) U1 and U2 transcription and in their 3'-box-dependent processing. The Integrator complex is associated with the C-terminal domain (CTD) of RNA polymerase II largest subunit (POLR2A) and is recruited to the U1 and U2 snRNAs genes (PubMed:16239144). Mediates recruitment of cytoplasmic dynein to the nuclear envelope, probably as component of the INT complex (PubMed:23904267). {ECO:0000269|PubMed:16239144, ECO:0000269|PubMed:23904267}. | FUNCTION: Acts as a negative regulator of the Wnt signaling pathway via its interaction with DVL1 (By similarity). Binds preferentially to DNA containing cytidine-phosphate-guanosine (CpG) dinucleotides over CpH (H=A, T, and C), hemimethylated-CpG and hemimethylated-hydroxymethyl-CpG (PubMed:29276034). {ECO:0000250|UniProtKB:Q6NXI8, ECO:0000269|PubMed:29276034}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | INTS12 | chr4:106613133 | chr4:105393523 | ENST00000340139 | - | 6 | 8 | 159_215 | 219 | 463.0 | Zinc finger | PHD-type |
| Hgene | INTS12 | chr4:106613133 | chr4:105393523 | ENST00000394735 | - | 6 | 8 | 159_215 | 219 | 463.0 | Zinc finger | PHD-type |
| Hgene | INTS12 | chr4:106613133 | chr4:105393523 | ENST00000451321 | - | 5 | 7 | 159_215 | 219 | 463.0 | Zinc finger | PHD-type |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | INTS12 | chr4:106613133 | chr4:105393523 | ENST00000340139 | - | 6 | 8 | 270_440 | 219 | 463.0 | Compositional bias | Note=Ser-rich |
| Hgene | INTS12 | chr4:106613133 | chr4:105393523 | ENST00000394735 | - | 6 | 8 | 270_440 | 219 | 463.0 | Compositional bias | Note=Ser-rich |
| Hgene | INTS12 | chr4:106613133 | chr4:105393523 | ENST00000451321 | - | 5 | 7 | 270_440 | 219 | 463.0 | Compositional bias | Note=Ser-rich |
| Tgene | CXXC4 | chr4:106613133 | chr4:105393523 | ENST00000426831 | 0 | 2 | 116_123 | 184 | 199.0 | Compositional bias | Note=Poly-Ser | |
| Tgene | CXXC4 | chr4:106613133 | chr4:105393523 | ENST00000426831 | 0 | 2 | 132_173 | 184 | 199.0 | Zinc finger | CXXC-type |
Top |
Fusion Gene Sequence for INTS12-CXXC4 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >39913_39913_1_INTS12-CXXC4_INTS12_chr4_106613133_ENST00000340139_CXXC4_chr4_105393523_ENST00000394767_length(transcript)=4927nt_BP=872nt AGGGACCACCGGGAACAGACGGATCGGCAGGGCGGGGCGGAACGGTGAGGCCATCTACTGAAACACAGAAGCAATTTTAAAAGAAAATGG AGGAAGTATAACTGATCTTTGCAGAAAGTGGGGAAGTGAGTTGACCAGAGAAGGATGAGAGAATTGCCAAGTTGTACTGAACGCCAAGCT GAGAATGGTGATTCTGAAGATAGAATGCGTTTGCAATGGCTGCTACTGTGAACTTGGAACTTGATCCCATTTTTTTGAAAGCACTAGGTT TCTTGCATTCAAAGAGTAAAGATTCTGCTGAAAAGCTAAAAGCACTGCTTGATGAATCTTTGGCTCGGGGCATTGATTCCAGTTACCGTC CATCTCAAAAGGATGTGGAGCCACCCAAAATTTCAAGCACAAAAAACATTTCCATTAAGCAAGAGCCCAAAATATCATCCAGTCTTCCTT CTGGTAATAATAATGGCAAGGTCCTCACAACTGAAAAGGTAAAGAAGGAAGCTGAAAAGAGACCTGCTGATAAAATGAAATCAGACATCA CTGAAGGAGTTGATATTCCAAAGAAACCTAGATTGGAGAAACCAGAAACACAGTCATCTCCCATTACTGTCCAAAGTAGCAAGGATTTAC CTATGGCTGACCTTTCCAGTTTTGAGGAGACCAGTGCTGATGATTTTGCCATGGAGATGGGATTGGCCTGCGTTGTTTGTAGGCAAATGA TGGTGGCATCTGGCAATCAATTAGTAGAATGTCAGGAGTGCCATAATCTCTACCACCGAGATTGTCATAAACCCCAGGTGACAGACAAGG AAGCGAATGACCCTCGCCTGGTGTGGTATTGTGCCCGATGTACCAGACAAATGAAAAGAATGAGAACACCTGTTCCCAGCGCTGAAGCAT TCCGATGGTTCTTTTAAAGCAGTAGTATATCTTATTTTCAAGGCATTTGGAAATGAAGGGCAAACTAATGTCTTGTTTTAAGAAACTGCT TAGTCCACCACTGAAGAAAATATCCAGAAATTATTTTCATTTTATGTATAGGGCTTTCTTCAAAAAAAAAAAAAAAGAGGAAAAGAAAAG AAAAAGACATAAAAATAATGTGAGAGCTTGGAGAATTGGCCAGTCTATTTACTTTCAATACGCTGATTCTTTCTTTGATGTAATTTAGCT ATAGTAGTGAAGTTGTTGTCTATTTTGAAAGTGGCTGTAAAAAATAAGTTTGGGTAAACCCCTGCTGTAAAATCATGTATCTTTGCAAAG TACATATCTATACTTCATTTTCAAATATATGTGTTTCAGTACTGTAAACTGTACAGATAGCAGCTTGTATTTTGTGTGTTTAGACACAAG GAGACAATCATGTCTGAGCATCTATGGAGATTAACAGTTTGTACACAACAGTATGGTTCTGCAAGTTAAATCTGGAGCAATAAATTTTAG CTTTAACTATTTTTTGCCAGTGGTTTAGAAGCAGCAACAGCACTGGCACCATTTTGCCATGCATCTTTCCATAGAGACTTGATGCCAAGT TTTACAAGACTAAAAGATTATGATGCATCCACCAATTACCTTCAGTTTTATTGTTATAAGAGGGGAAGATGTTATGAAAGTTTCAATTAA TCTTTTGAGCACTATATTAGAGTAGTGGTTATGCACTTGCATTGCTTACAAACGAGCTGTACAGAAGGGTATACCTCCCAAATACTTAGT GTAGTTGACTTGTCTTGGGTTGCACTGTAAGGCAGAGTACTCAGAGTAGTTGGAAAATGCAGAATCAGTTGTATAATTTTTTTTTATAAA AACGGTGTTTTTTAGGCTAAAGAATAAAGTATCATATCTTAGGAGGGGAAAATTTATAACTGACATTTTTTCCCCAGCCCAATATGGGCT GTATACGTTTTATTGTTTCTTAATTTTTTTCTTATTTTTTCTGTAGGAAAAAATCTTAATAAAAACTACCCCAATGTTTTAACTTCATGT ATGATATTAAATGGGTAGTTTTAACACTTGAATATGTTGAGGGCTTCTGCCTTGAGGCAGGGCTTGATATATTTTTAATTTACAAAGAAT TAAACATATTCATAAAAGTGAGGCATTTTATCTCTTTTTATTTTCTTTTCTCATAGCCACCCAGTTGGACTAGTGGTCTTCCCCTGGTCC TTCAGTAAAATGCTCCATTGCTGCCATTTCCATTGTTGTTACTTGATGCTTTCATTCCTGAGAAGGCAGGGCTACAGTCTCTGGAATTTC ATAAATGCATGACATACCCCTCCCCCCACAACCTACACACAAAGGATCTAATGCTTCATAGAACCTGTGCTACCTTTTCATGTTGGAGTT GGTTTTCTTATCACAGTCAGGGTTCTTAAGGCCGTCCCATCAGGACAAATATTTACCTTCCATTTTATTTCTGTTTGTCCCAATTATAAA GACTATTTTCAGTTTCAGGAGTAGAACAGGGTTTAGCAAAAATATGTAGAGTATCAAAGTTACCTACTGCAACTTTTTGTTCTTTGTCCA CTAGCCAGGTGATTTAACCCACTAAATCCATTAGCTTGCTGAAAAATGTTAGCAAAGTAAATCACGAGAAGAAAGATAATTTGAGAAGAG AAATGGTATGGTACAATGAAAGAAGCTGTAAAGATTAGGAAAGACTAGTAAGTGAATATTTTTAAAAATTTAGTTGTAGATTTCAATGGG ATACGATAGGACAGAAAAGATTTTTTAAAAAGCAGAAAGAGTGTTTCATGGTGAAAGTACTGGGGGAGGGTGGACAAAGCATGCACACAT GCCAATTTGAAAATCAAGTGTGACTTACCTCACGTAGAGTATGAATACATGGTCCACAGTTATGGTCAACAAGCGTTTTCAAGAAAAACT CATGATGTGTTACACCCATCCATATTTAGAGATGAGAATTAAATGATTACAAGCTAAAATGTGCTGTATACTGTATTAATTTTGAGATAG CTTTCTACTTGATGTATATTTCTTTGAGGGTTTCTATCTAGTTAGAATTTCAGCTTCTGCTGTGGGACCAATGAGAGCTGTATGAGTTTT TTTTTTTTTTTCCTTTTCCATTATTTTATTTTTTGTTTTTTGTTTACCAATTTGCTAATTAATTACAAGTAAGTGAAAACTTACTTTCAC ACTTGAATGATAAGCTTGCCTTGCTCTTTGGAAAATGATGATTGTTATTACAGAAGTGAAACAAACTTCTATTCAGGCTACAGTAGGCAT ACAATAACCTAAAGATCTCATAGAAGTATACTGAGAGTCTAAAATACATTCTAGTAGTGCATGTCTTGGTGTGCTTTTGTTTGTTTGCTT TCTAAAAGAACTAATGAATCTGTATATTGTAAACTGTGGTGTTTTACATGTCAATTCATTTTTTTAATGAAAGAAAATTTGTTGATTACT GAAATGAGAAAGTGTAGCTTTTCATTAATTCTCTTATGTTTTTTATTCTTGATATTGTTCTTTTATACCACCCACTTTTTAATGTTTTTG GTTAAACTCCATTTATATAATGTAGGATGCATATAAAGTTCTCATTGGTGATGTTGAAGAAAGAGATGACTCTTCTGGCCTTTCTCTAGT TTTCTTCCTCATATGTCCCTGTACTGAGAACTTCTAAAGCTTATCATTATAAACCTAAGGCAGTGATTTGAAAGTTTACATTTTTCATGA TTTTCAAATACCAATGAAATGTAACTTTTAAATATTCCTCACTGTGTGTTTTATATTCACTGTCAAGAAAATTCAGAATGTAGATTGTGT GGACAGCTATACACAACTTAATAATTATATATCAGATAAACTGAATAAAAAACTTTGTAAGAGTGGTCTACGCACACAGGATCTTGTCCT CCCTAGGCGTGGATAGAGAGATGCCATAATCTGCCAGCATTTGGGGAGATCATGGTAAATACTTGAGGCGTTGATACATAGTGCAGTGAG CCAATTTCCTTTTCAGTTGTCACTCAGCACCATAACATCACGAAGAACATCATTAATGCAAACCTCTCACTAAGCCTTCAGACCAAAGTG GCTTTCTGATGGACTCTGCCTGTCTGTCTACAAGTGGAGTCAGATTCAAAGTAGAAATGTATCCATCCTTGGGAAAGAAAATAGGAAGTC TTCCCCCACTGTAGGAGTATATAAAAATGTCTCCTGGTTGCTTAATTCCCAACACTCAATTTCCTTCTTCTAAACACATTTATAAGGAGC AAGAGATGGTGCACTTATTATGAATTAATATTAAGCAATCACTAAAAGAAAGGAATATTCTCTTCCTTTCAATCAGAAACCCTGATATGT AAGTTGTGCAATTAATGCTAAAGCATATAAAGTCTTTAAGCTGATAAGTATCTGTAATCAAATAATGAAAAAAGGAAAGCAGTTAAATGA ACACACTATTCAGGGTCAAATATGTTCAGCAAAAAGCCGTAGCGGGTCTTTGATCATGGTTAGATTCACAGTGATATCAAACAGTTGGTT CTCATGTAACAGCTAAATGTTTCAGTTTTTTTTTTTTTATTAAATTTGGCATTTCACACTGAGATCTGTATTCCTAGTGAATAAAAGACA GCTACTATATTAGGAGGGATTCCATTTTCACAATTGCAAAACAGATATTTCAGGAACATGGGCCACAAAATGTACTCCTTTCACAGTGTT CTCCTCTTCTGAACTGTGCAGTTATCTATTAAATTTTCTAATAGATATTTGTGTAAGGTGTATGTATGCTTGTGCAATTATTTAAAAAAC GGTTTTGGAAAACAGTGTATTTATTAAAGAAAATTACTTATGGGGCATGTACAAAACTGTAAAAAAAAAAAAAAAGTAAAAAAAATCACA >39913_39913_1_INTS12-CXXC4_INTS12_chr4_106613133_ENST00000340139_CXXC4_chr4_105393523_ENST00000394767_length(amino acids)=233AA_BP= MAATVNLELDPIFLKALGFLHSKSKDSAEKLKALLDESLARGIDSSYRPSQKDVEPPKISSTKNISIKQEPKISSSLPSGNNNGKVLTTE KVKKEAEKRPADKMKSDITEGVDIPKKPRLEKPETQSSPITVQSSKDLPMADLSSFEETSADDFAMEMGLACVVCRQMMVASGNQLVECQ -------------------------------------------------------------- >39913_39913_2_INTS12-CXXC4_INTS12_chr4_106613133_ENST00000451321_CXXC4_chr4_105393523_ENST00000394767_length(transcript)=5192nt_BP=1137nt CGCATCCTCAGCCCTGCAGACCCCCGAAGAAAAGACCAGTTCTTAGGGGGTCGCGGAGGCTGAGAAGCCGGCGGCCCGAGCGCCCTCTTT ACCTGGTCGCAGAGGCGCGGCCAGGCAGGGAGGGTGCGTGAGCGTGTCGGCGACCTGGGCCGCCTGCGCCTTCCCGCCCAGTCCGAGCTG GAATTCCAAGGCTGCCTCTGGGAGCTGCTGCCCGCAGACCACTCGCCTCCACAGACGCCAGGAGAAAAACGACAGCCACCTTATGTGGAA AATACTCGCGCCGGCCGTCATTCATAATCCGGCGGACCGGAAAAAGGGAAGTAACGTCACTTCCTACTGTCGCACTGCCAAACGTTTCCC CCGCCCCTACTGTGGGAACCTTTGTTTTCCTGCTTTCGGAGCCGGCCAGTGCGGGAACCGTTTCCGAAGGGACCACCGGGAACAGACGGA TCGGCAGGGCGGGGCGGAACGGCGTTTGCAATGGCTGCTACTGTGAACTTGGAACTTGATCCCATTTTTTTGAAAGCACTAGGTTTCTTG CATTCAAAGAGTAAAGATTCTGCTGAAAAGCTAAAAGCACTGCTTGATGAATCTTTGGCTCGGGGCATTGATTCCAGTTACCGTCCATCT CAAAAGGATGTGGAGCCACCCAAAATTTCAAGCACAAAAAACATTTCCATTAAGCAAGAGCCCAAAATATCATCCAGTCTTCCTTCTGGT AATAATAATGGCAAGGTCCTCACAACTGAAAAGGTAAAGAAGGAAGCTGAAAAGAGACCTGCTGATAAAATGAAATCAGACATCACTGAA GGAGTTGATATTCCAAAGAAACCTAGATTGGAGAAACCAGAAACACAGTCATCTCCCATTACTGTCCAAAGTAGCAAGGATTTACCTATG GCTGACCTTTCCAGTTTTGAGGAGACCAGTGCTGATGATTTTGCCATGGAGATGGGATTGGCCTGCGTTGTTTGTAGGCAAATGATGGTG GCATCTGGCAATCAATTAGTAGAATGTCAGGAGTGCCATAATCTCTACCACCGAGATTGTCATAAACCCCAGGTGACAGACAAGGAAGCG AATGACCCTCGCCTGGTGTGGTATTGTGCCCGATGTACCAGACAAATGAAAAGAATGAGAACACCTGTTCCCAGCGCTGAAGCATTCCGA TGGTTCTTTTAAAGCAGTAGTATATCTTATTTTCAAGGCATTTGGAAATGAAGGGCAAACTAATGTCTTGTTTTAAGAAACTGCTTAGTC CACCACTGAAGAAAATATCCAGAAATTATTTTCATTTTATGTATAGGGCTTTCTTCAAAAAAAAAAAAAAAGAGGAAAAGAAAAGAAAAA GACATAAAAATAATGTGAGAGCTTGGAGAATTGGCCAGTCTATTTACTTTCAATACGCTGATTCTTTCTTTGATGTAATTTAGCTATAGT AGTGAAGTTGTTGTCTATTTTGAAAGTGGCTGTAAAAAATAAGTTTGGGTAAACCCCTGCTGTAAAATCATGTATCTTTGCAAAGTACAT ATCTATACTTCATTTTCAAATATATGTGTTTCAGTACTGTAAACTGTACAGATAGCAGCTTGTATTTTGTGTGTTTAGACACAAGGAGAC AATCATGTCTGAGCATCTATGGAGATTAACAGTTTGTACACAACAGTATGGTTCTGCAAGTTAAATCTGGAGCAATAAATTTTAGCTTTA ACTATTTTTTGCCAGTGGTTTAGAAGCAGCAACAGCACTGGCACCATTTTGCCATGCATCTTTCCATAGAGACTTGATGCCAAGTTTTAC AAGACTAAAAGATTATGATGCATCCACCAATTACCTTCAGTTTTATTGTTATAAGAGGGGAAGATGTTATGAAAGTTTCAATTAATCTTT TGAGCACTATATTAGAGTAGTGGTTATGCACTTGCATTGCTTACAAACGAGCTGTACAGAAGGGTATACCTCCCAAATACTTAGTGTAGT TGACTTGTCTTGGGTTGCACTGTAAGGCAGAGTACTCAGAGTAGTTGGAAAATGCAGAATCAGTTGTATAATTTTTTTTTATAAAAACGG TGTTTTTTAGGCTAAAGAATAAAGTATCATATCTTAGGAGGGGAAAATTTATAACTGACATTTTTTCCCCAGCCCAATATGGGCTGTATA CGTTTTATTGTTTCTTAATTTTTTTCTTATTTTTTCTGTAGGAAAAAATCTTAATAAAAACTACCCCAATGTTTTAACTTCATGTATGAT ATTAAATGGGTAGTTTTAACACTTGAATATGTTGAGGGCTTCTGCCTTGAGGCAGGGCTTGATATATTTTTAATTTACAAAGAATTAAAC ATATTCATAAAAGTGAGGCATTTTATCTCTTTTTATTTTCTTTTCTCATAGCCACCCAGTTGGACTAGTGGTCTTCCCCTGGTCCTTCAG TAAAATGCTCCATTGCTGCCATTTCCATTGTTGTTACTTGATGCTTTCATTCCTGAGAAGGCAGGGCTACAGTCTCTGGAATTTCATAAA TGCATGACATACCCCTCCCCCCACAACCTACACACAAAGGATCTAATGCTTCATAGAACCTGTGCTACCTTTTCATGTTGGAGTTGGTTT TCTTATCACAGTCAGGGTTCTTAAGGCCGTCCCATCAGGACAAATATTTACCTTCCATTTTATTTCTGTTTGTCCCAATTATAAAGACTA TTTTCAGTTTCAGGAGTAGAACAGGGTTTAGCAAAAATATGTAGAGTATCAAAGTTACCTACTGCAACTTTTTGTTCTTTGTCCACTAGC CAGGTGATTTAACCCACTAAATCCATTAGCTTGCTGAAAAATGTTAGCAAAGTAAATCACGAGAAGAAAGATAATTTGAGAAGAGAAATG GTATGGTACAATGAAAGAAGCTGTAAAGATTAGGAAAGACTAGTAAGTGAATATTTTTAAAAATTTAGTTGTAGATTTCAATGGGATACG ATAGGACAGAAAAGATTTTTTAAAAAGCAGAAAGAGTGTTTCATGGTGAAAGTACTGGGGGAGGGTGGACAAAGCATGCACACATGCCAA TTTGAAAATCAAGTGTGACTTACCTCACGTAGAGTATGAATACATGGTCCACAGTTATGGTCAACAAGCGTTTTCAAGAAAAACTCATGA TGTGTTACACCCATCCATATTTAGAGATGAGAATTAAATGATTACAAGCTAAAATGTGCTGTATACTGTATTAATTTTGAGATAGCTTTC TACTTGATGTATATTTCTTTGAGGGTTTCTATCTAGTTAGAATTTCAGCTTCTGCTGTGGGACCAATGAGAGCTGTATGAGTTTTTTTTT TTTTTTCCTTTTCCATTATTTTATTTTTTGTTTTTTGTTTACCAATTTGCTAATTAATTACAAGTAAGTGAAAACTTACTTTCACACTTG AATGATAAGCTTGCCTTGCTCTTTGGAAAATGATGATTGTTATTACAGAAGTGAAACAAACTTCTATTCAGGCTACAGTAGGCATACAAT AACCTAAAGATCTCATAGAAGTATACTGAGAGTCTAAAATACATTCTAGTAGTGCATGTCTTGGTGTGCTTTTGTTTGTTTGCTTTCTAA AAGAACTAATGAATCTGTATATTGTAAACTGTGGTGTTTTACATGTCAATTCATTTTTTTAATGAAAGAAAATTTGTTGATTACTGAAAT GAGAAAGTGTAGCTTTTCATTAATTCTCTTATGTTTTTTATTCTTGATATTGTTCTTTTATACCACCCACTTTTTAATGTTTTTGGTTAA ACTCCATTTATATAATGTAGGATGCATATAAAGTTCTCATTGGTGATGTTGAAGAAAGAGATGACTCTTCTGGCCTTTCTCTAGTTTTCT TCCTCATATGTCCCTGTACTGAGAACTTCTAAAGCTTATCATTATAAACCTAAGGCAGTGATTTGAAAGTTTACATTTTTCATGATTTTC AAATACCAATGAAATGTAACTTTTAAATATTCCTCACTGTGTGTTTTATATTCACTGTCAAGAAAATTCAGAATGTAGATTGTGTGGACA GCTATACACAACTTAATAATTATATATCAGATAAACTGAATAAAAAACTTTGTAAGAGTGGTCTACGCACACAGGATCTTGTCCTCCCTA GGCGTGGATAGAGAGATGCCATAATCTGCCAGCATTTGGGGAGATCATGGTAAATACTTGAGGCGTTGATACATAGTGCAGTGAGCCAAT TTCCTTTTCAGTTGTCACTCAGCACCATAACATCACGAAGAACATCATTAATGCAAACCTCTCACTAAGCCTTCAGACCAAAGTGGCTTT CTGATGGACTCTGCCTGTCTGTCTACAAGTGGAGTCAGATTCAAAGTAGAAATGTATCCATCCTTGGGAAAGAAAATAGGAAGTCTTCCC CCACTGTAGGAGTATATAAAAATGTCTCCTGGTTGCTTAATTCCCAACACTCAATTTCCTTCTTCTAAACACATTTATAAGGAGCAAGAG ATGGTGCACTTATTATGAATTAATATTAAGCAATCACTAAAAGAAAGGAATATTCTCTTCCTTTCAATCAGAAACCCTGATATGTAAGTT GTGCAATTAATGCTAAAGCATATAAAGTCTTTAAGCTGATAAGTATCTGTAATCAAATAATGAAAAAAGGAAAGCAGTTAAATGAACACA CTATTCAGGGTCAAATATGTTCAGCAAAAAGCCGTAGCGGGTCTTTGATCATGGTTAGATTCACAGTGATATCAAACAGTTGGTTCTCAT GTAACAGCTAAATGTTTCAGTTTTTTTTTTTTTATTAAATTTGGCATTTCACACTGAGATCTGTATTCCTAGTGAATAAAAGACAGCTAC TATATTAGGAGGGATTCCATTTTCACAATTGCAAAACAGATATTTCAGGAACATGGGCCACAAAATGTACTCCTTTCACAGTGTTCTCCT CTTCTGAACTGTGCAGTTATCTATTAAATTTTCTAATAGATATTTGTGTAAGGTGTATGTATGCTTGTGCAATTATTTAAAAAACGGTTT TGGAAAACAGTGTATTTATTAAAGAAAATTACTTATGGGGCATGTACAAAACTGTAAAAAAAAAAAAAAAGTAAAAAAAATCACATTCAA >39913_39913_2_INTS12-CXXC4_INTS12_chr4_106613133_ENST00000451321_CXXC4_chr4_105393523_ENST00000394767_length(amino acids)=281AA_BP= MSHCQTFPPPLLWEPLFSCFRSRPVREPFPKGPPGTDGSAGRGGTAFAMAATVNLELDPIFLKALGFLHSKSKDSAEKLKALLDESLARG IDSSYRPSQKDVEPPKISSTKNISIKQEPKISSSLPSGNNNGKVLTTEKVKKEAEKRPADKMKSDITEGVDIPKKPRLEKPETQSSPITV QSSKDLPMADLSSFEETSADDFAMEMGLACVVCRQMMVASGNQLVECQECHNLYHRDCHKPQVTDKEANDPRLVWYCARCTRQMKRMRTP -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for INTS12-CXXC4 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
| Tgene | CXXC4 | chr4:106613133 | chr4:105393523 | ENST00000426831 | 0 | 2 | 161_166 | 184.0 | 199.0 | DVL1 |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for INTS12-CXXC4 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for INTS12-CXXC4 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
