
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:IP6K2-COL7A1 (FusionGDB2 ID:40019) |
Fusion Gene Summary for IP6K2-COL7A1 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: IP6K2-COL7A1 | Fusion gene ID: 40019 | Hgene | Tgene | Gene symbol | IP6K2 | COL7A1 | Gene ID | 51447 | 1294 |
| Gene name | inositol hexakisphosphate kinase 2 | collagen type VII alpha 1 chain | |
| Synonyms | IHPK2|InsP6K2|PIUS | EBD1|EBDCT|EBR1|NDNC8 | |
| Cytomap | 3p21.31 | 3p21.31 | |
| Type of gene | protein-coding | protein-coding | |
| Description | inositol hexakisphosphate kinase 2ATP:1D-myo-inositol-hexakisphosphate phosphotransferaseinositol hexaphosphate kinase 2insp6 kinase 2pi uptake stimulator | collagen alpha-1(VII) chainLC collagencollagen VII, alpha-1 polypeptidecollagen, type VII, alpha 1long-chain collagen | |
| Modification date | 20200327 | 20200329 | |
| UniProtAcc | Q9UHH9 | Q02388 | |
| Ensembl transtripts involved in fusion gene | ENST00000431721, ENST00000443964, ENST00000446860, ENST00000450045, ENST00000436134, ENST00000328631, ENST00000340879, ENST00000413298, ENST00000417896, ENST00000432678, ENST00000449610, ENST00000453202, | ENST00000470076, ENST00000328333, ENST00000454817, | |
| Fusion gene scores | * DoF score | 13 X 12 X 7=1092 | 5 X 8 X 4=160 |
| # samples | 20 | 6 | |
| ** MAII score | log2(20/1092*10)=-2.44890095114513 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(6/160*10)=-1.41503749927884 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: IP6K2 [Title/Abstract] AND COL7A1 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | IP6K2(48752822)-COL7A1(48623408), # samples:2 | ||
| Anticipated loss of major functional domain due to fusion event. | IP6K2-COL7A1 seems lost the major protein functional domain in Hgene partner, which is a essential gene due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | IP6K2 | GO:0043647 | inositol phosphate metabolic process | 30624931 |
| Hgene | IP6K2 | GO:0046854 | phosphatidylinositol phosphorylation | 10574768 |
 Fusion gene breakpoints across IP6K2 (5'-gene) Fusion gene breakpoints across IP6K2 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
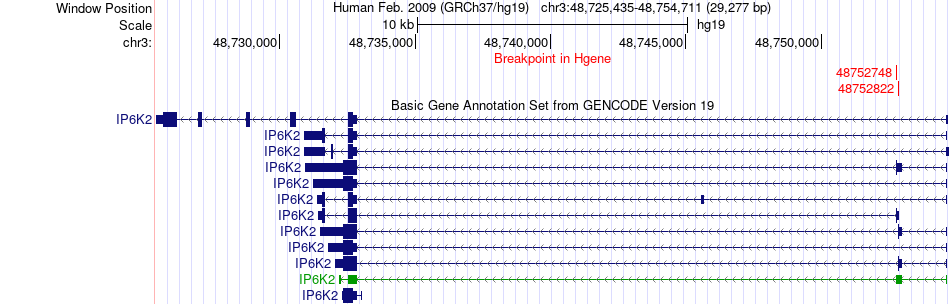 |
 Fusion gene breakpoints across COL7A1 (3'-gene) Fusion gene breakpoints across COL7A1 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
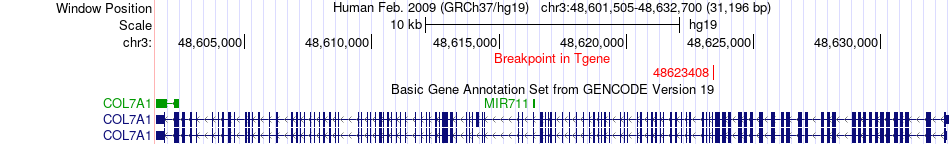 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | OV | TCGA-23-1110-01A | IP6K2 | chr3 | 48752748 | - | COL7A1 | chr3 | 48623408 | - |
| ChimerDB4 | OV | TCGA-23-1110-01A | IP6K2 | chr3 | 48752822 | - | COL7A1 | chr3 | 48623408 | - |
Top |
Fusion Gene ORF analysis for IP6K2-COL7A1 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-intron | ENST00000431721 | ENST00000470076 | IP6K2 | chr3 | 48752748 | - | COL7A1 | chr3 | 48623408 | - |
| 5CDS-intron | ENST00000443964 | ENST00000470076 | IP6K2 | chr3 | 48752822 | - | COL7A1 | chr3 | 48623408 | - |
| 5CDS-intron | ENST00000446860 | ENST00000470076 | IP6K2 | chr3 | 48752822 | - | COL7A1 | chr3 | 48623408 | - |
| 5CDS-intron | ENST00000450045 | ENST00000470076 | IP6K2 | chr3 | 48752748 | - | COL7A1 | chr3 | 48623408 | - |
| 5UTR-3CDS | ENST00000436134 | ENST00000328333 | IP6K2 | chr3 | 48752748 | - | COL7A1 | chr3 | 48623408 | - |
| 5UTR-3CDS | ENST00000436134 | ENST00000454817 | IP6K2 | chr3 | 48752748 | - | COL7A1 | chr3 | 48623408 | - |
| 5UTR-intron | ENST00000436134 | ENST00000470076 | IP6K2 | chr3 | 48752748 | - | COL7A1 | chr3 | 48623408 | - |
| Frame-shift | ENST00000431721 | ENST00000328333 | IP6K2 | chr3 | 48752748 | - | COL7A1 | chr3 | 48623408 | - |
| Frame-shift | ENST00000431721 | ENST00000454817 | IP6K2 | chr3 | 48752748 | - | COL7A1 | chr3 | 48623408 | - |
| Frame-shift | ENST00000443964 | ENST00000328333 | IP6K2 | chr3 | 48752822 | - | COL7A1 | chr3 | 48623408 | - |
| Frame-shift | ENST00000443964 | ENST00000454817 | IP6K2 | chr3 | 48752822 | - | COL7A1 | chr3 | 48623408 | - |
| Frame-shift | ENST00000446860 | ENST00000328333 | IP6K2 | chr3 | 48752822 | - | COL7A1 | chr3 | 48623408 | - |
| Frame-shift | ENST00000446860 | ENST00000454817 | IP6K2 | chr3 | 48752822 | - | COL7A1 | chr3 | 48623408 | - |
| In-frame | ENST00000450045 | ENST00000328333 | IP6K2 | chr3 | 48752748 | - | COL7A1 | chr3 | 48623408 | - |
| In-frame | ENST00000450045 | ENST00000454817 | IP6K2 | chr3 | 48752748 | - | COL7A1 | chr3 | 48623408 | - |
| intron-3CDS | ENST00000328631 | ENST00000328333 | IP6K2 | chr3 | 48752748 | - | COL7A1 | chr3 | 48623408 | - |
| intron-3CDS | ENST00000328631 | ENST00000328333 | IP6K2 | chr3 | 48752822 | - | COL7A1 | chr3 | 48623408 | - |
| intron-3CDS | ENST00000328631 | ENST00000454817 | IP6K2 | chr3 | 48752748 | - | COL7A1 | chr3 | 48623408 | - |
| intron-3CDS | ENST00000328631 | ENST00000454817 | IP6K2 | chr3 | 48752822 | - | COL7A1 | chr3 | 48623408 | - |
| intron-3CDS | ENST00000340879 | ENST00000328333 | IP6K2 | chr3 | 48752748 | - | COL7A1 | chr3 | 48623408 | - |
| intron-3CDS | ENST00000340879 | ENST00000328333 | IP6K2 | chr3 | 48752822 | - | COL7A1 | chr3 | 48623408 | - |
| intron-3CDS | ENST00000340879 | ENST00000454817 | IP6K2 | chr3 | 48752748 | - | COL7A1 | chr3 | 48623408 | - |
| intron-3CDS | ENST00000340879 | ENST00000454817 | IP6K2 | chr3 | 48752822 | - | COL7A1 | chr3 | 48623408 | - |
| intron-3CDS | ENST00000413298 | ENST00000328333 | IP6K2 | chr3 | 48752748 | - | COL7A1 | chr3 | 48623408 | - |
| intron-3CDS | ENST00000413298 | ENST00000328333 | IP6K2 | chr3 | 48752822 | - | COL7A1 | chr3 | 48623408 | - |
| intron-3CDS | ENST00000413298 | ENST00000454817 | IP6K2 | chr3 | 48752748 | - | COL7A1 | chr3 | 48623408 | - |
| intron-3CDS | ENST00000413298 | ENST00000454817 | IP6K2 | chr3 | 48752822 | - | COL7A1 | chr3 | 48623408 | - |
| intron-3CDS | ENST00000417896 | ENST00000328333 | IP6K2 | chr3 | 48752748 | - | COL7A1 | chr3 | 48623408 | - |
| intron-3CDS | ENST00000417896 | ENST00000328333 | IP6K2 | chr3 | 48752822 | - | COL7A1 | chr3 | 48623408 | - |
| intron-3CDS | ENST00000417896 | ENST00000454817 | IP6K2 | chr3 | 48752748 | - | COL7A1 | chr3 | 48623408 | - |
| intron-3CDS | ENST00000417896 | ENST00000454817 | IP6K2 | chr3 | 48752822 | - | COL7A1 | chr3 | 48623408 | - |
| intron-3CDS | ENST00000431721 | ENST00000328333 | IP6K2 | chr3 | 48752822 | - | COL7A1 | chr3 | 48623408 | - |
| intron-3CDS | ENST00000431721 | ENST00000454817 | IP6K2 | chr3 | 48752822 | - | COL7A1 | chr3 | 48623408 | - |
| intron-3CDS | ENST00000432678 | ENST00000328333 | IP6K2 | chr3 | 48752748 | - | COL7A1 | chr3 | 48623408 | - |
| intron-3CDS | ENST00000432678 | ENST00000328333 | IP6K2 | chr3 | 48752822 | - | COL7A1 | chr3 | 48623408 | - |
| intron-3CDS | ENST00000432678 | ENST00000454817 | IP6K2 | chr3 | 48752748 | - | COL7A1 | chr3 | 48623408 | - |
| intron-3CDS | ENST00000432678 | ENST00000454817 | IP6K2 | chr3 | 48752822 | - | COL7A1 | chr3 | 48623408 | - |
| intron-3CDS | ENST00000436134 | ENST00000328333 | IP6K2 | chr3 | 48752822 | - | COL7A1 | chr3 | 48623408 | - |
| intron-3CDS | ENST00000436134 | ENST00000454817 | IP6K2 | chr3 | 48752822 | - | COL7A1 | chr3 | 48623408 | - |
| intron-3CDS | ENST00000443964 | ENST00000328333 | IP6K2 | chr3 | 48752748 | - | COL7A1 | chr3 | 48623408 | - |
| intron-3CDS | ENST00000443964 | ENST00000454817 | IP6K2 | chr3 | 48752748 | - | COL7A1 | chr3 | 48623408 | - |
| intron-3CDS | ENST00000446860 | ENST00000328333 | IP6K2 | chr3 | 48752748 | - | COL7A1 | chr3 | 48623408 | - |
| intron-3CDS | ENST00000446860 | ENST00000454817 | IP6K2 | chr3 | 48752748 | - | COL7A1 | chr3 | 48623408 | - |
| intron-3CDS | ENST00000449610 | ENST00000328333 | IP6K2 | chr3 | 48752748 | - | COL7A1 | chr3 | 48623408 | - |
| intron-3CDS | ENST00000449610 | ENST00000328333 | IP6K2 | chr3 | 48752822 | - | COL7A1 | chr3 | 48623408 | - |
| intron-3CDS | ENST00000449610 | ENST00000454817 | IP6K2 | chr3 | 48752748 | - | COL7A1 | chr3 | 48623408 | - |
| intron-3CDS | ENST00000449610 | ENST00000454817 | IP6K2 | chr3 | 48752822 | - | COL7A1 | chr3 | 48623408 | - |
| intron-3CDS | ENST00000450045 | ENST00000328333 | IP6K2 | chr3 | 48752822 | - | COL7A1 | chr3 | 48623408 | - |
| intron-3CDS | ENST00000450045 | ENST00000454817 | IP6K2 | chr3 | 48752822 | - | COL7A1 | chr3 | 48623408 | - |
| intron-3CDS | ENST00000453202 | ENST00000328333 | IP6K2 | chr3 | 48752748 | - | COL7A1 | chr3 | 48623408 | - |
| intron-3CDS | ENST00000453202 | ENST00000328333 | IP6K2 | chr3 | 48752822 | - | COL7A1 | chr3 | 48623408 | - |
| intron-3CDS | ENST00000453202 | ENST00000454817 | IP6K2 | chr3 | 48752748 | - | COL7A1 | chr3 | 48623408 | - |
| intron-3CDS | ENST00000453202 | ENST00000454817 | IP6K2 | chr3 | 48752822 | - | COL7A1 | chr3 | 48623408 | - |
| intron-intron | ENST00000328631 | ENST00000470076 | IP6K2 | chr3 | 48752748 | - | COL7A1 | chr3 | 48623408 | - |
| intron-intron | ENST00000328631 | ENST00000470076 | IP6K2 | chr3 | 48752822 | - | COL7A1 | chr3 | 48623408 | - |
| intron-intron | ENST00000340879 | ENST00000470076 | IP6K2 | chr3 | 48752748 | - | COL7A1 | chr3 | 48623408 | - |
| intron-intron | ENST00000340879 | ENST00000470076 | IP6K2 | chr3 | 48752822 | - | COL7A1 | chr3 | 48623408 | - |
| intron-intron | ENST00000413298 | ENST00000470076 | IP6K2 | chr3 | 48752748 | - | COL7A1 | chr3 | 48623408 | - |
| intron-intron | ENST00000413298 | ENST00000470076 | IP6K2 | chr3 | 48752822 | - | COL7A1 | chr3 | 48623408 | - |
| intron-intron | ENST00000417896 | ENST00000470076 | IP6K2 | chr3 | 48752748 | - | COL7A1 | chr3 | 48623408 | - |
| intron-intron | ENST00000417896 | ENST00000470076 | IP6K2 | chr3 | 48752822 | - | COL7A1 | chr3 | 48623408 | - |
| intron-intron | ENST00000431721 | ENST00000470076 | IP6K2 | chr3 | 48752822 | - | COL7A1 | chr3 | 48623408 | - |
| intron-intron | ENST00000432678 | ENST00000470076 | IP6K2 | chr3 | 48752748 | - | COL7A1 | chr3 | 48623408 | - |
| intron-intron | ENST00000432678 | ENST00000470076 | IP6K2 | chr3 | 48752822 | - | COL7A1 | chr3 | 48623408 | - |
| intron-intron | ENST00000436134 | ENST00000470076 | IP6K2 | chr3 | 48752822 | - | COL7A1 | chr3 | 48623408 | - |
| intron-intron | ENST00000443964 | ENST00000470076 | IP6K2 | chr3 | 48752748 | - | COL7A1 | chr3 | 48623408 | - |
| intron-intron | ENST00000446860 | ENST00000470076 | IP6K2 | chr3 | 48752748 | - | COL7A1 | chr3 | 48623408 | - |
| intron-intron | ENST00000449610 | ENST00000470076 | IP6K2 | chr3 | 48752748 | - | COL7A1 | chr3 | 48623408 | - |
| intron-intron | ENST00000449610 | ENST00000470076 | IP6K2 | chr3 | 48752822 | - | COL7A1 | chr3 | 48623408 | - |
| intron-intron | ENST00000450045 | ENST00000470076 | IP6K2 | chr3 | 48752822 | - | COL7A1 | chr3 | 48623408 | - |
| intron-intron | ENST00000453202 | ENST00000470076 | IP6K2 | chr3 | 48752748 | - | COL7A1 | chr3 | 48623408 | - |
| intron-intron | ENST00000453202 | ENST00000470076 | IP6K2 | chr3 | 48752822 | - | COL7A1 | chr3 | 48623408 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000450045 | IP6K2 | chr3 | 48752748 | - | ENST00000454817 | COL7A1 | chr3 | 48623408 | - | 5452 | 103 | 163 | 5118 | 1651 |
| ENST00000450045 | IP6K2 | chr3 | 48752748 | - | ENST00000328333 | COL7A1 | chr3 | 48623408 | - | 5548 | 103 | 163 | 5214 | 1683 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000450045 | ENST00000454817 | IP6K2 | chr3 | 48752748 | - | COL7A1 | chr3 | 48623408 | - | 0.004818532 | 0.9951815 |
| ENST00000450045 | ENST00000328333 | IP6K2 | chr3 | 48752748 | - | COL7A1 | chr3 | 48623408 | - | 0.004890313 | 0.9951096 |
Top |
Fusion Genomic Features for IP6K2-COL7A1 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
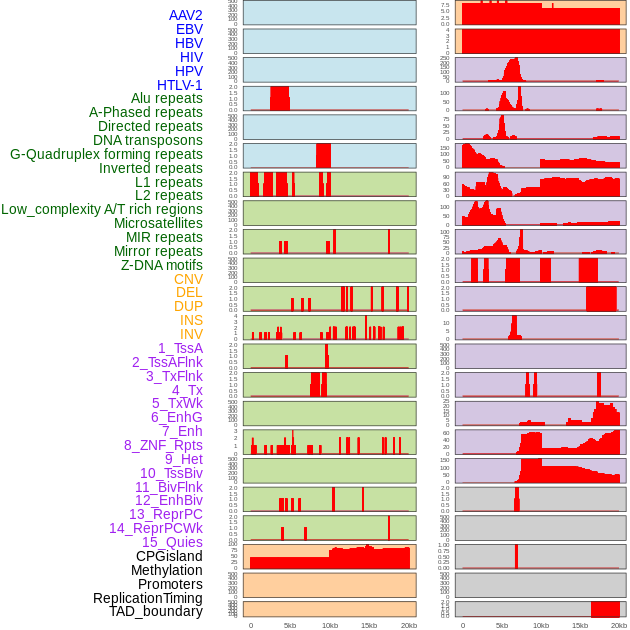 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for IP6K2-COL7A1 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr3:48752822/chr3:48623408) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| IP6K2 | COL7A1 |
| FUNCTION: Converts inositol hexakisphosphate (InsP6) to diphosphoinositol pentakisphosphate (InsP7/PP-InsP5). {ECO:0000269|PubMed:10574768, ECO:0000269|PubMed:30624931}. | FUNCTION: Stratified squamous epithelial basement membrane protein that forms anchoring fibrils which may contribute to epithelial basement membrane organization and adherence by interacting with extracellular matrix (ECM) proteins such as type IV collagen. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | COL7A1 | chr3:48752748 | chr3:48623408 | ENST00000328333 | 26 | 118 | 2872_2944 | 1241 | 2945.0 | Domain | BPTI/Kunitz inhibitor | |
| Tgene | COL7A1 | chr3:48752748 | chr3:48623408 | ENST00000454817 | 26 | 117 | 2872_2944 | 1241 | 2913.0 | Domain | BPTI/Kunitz inhibitor | |
| Tgene | COL7A1 | chr3:48752748 | chr3:48623408 | ENST00000328333 | 26 | 118 | 1334_1336 | 1241 | 2945.0 | Motif | Cell attachment site | |
| Tgene | COL7A1 | chr3:48752748 | chr3:48623408 | ENST00000328333 | 26 | 118 | 2008_2010 | 1241 | 2945.0 | Motif | Cell attachment site | |
| Tgene | COL7A1 | chr3:48752748 | chr3:48623408 | ENST00000328333 | 26 | 118 | 2553_2555 | 1241 | 2945.0 | Motif | Cell attachment site | |
| Tgene | COL7A1 | chr3:48752748 | chr3:48623408 | ENST00000454817 | 26 | 117 | 1334_1336 | 1241 | 2913.0 | Motif | Cell attachment site | |
| Tgene | COL7A1 | chr3:48752748 | chr3:48623408 | ENST00000454817 | 26 | 117 | 2008_2010 | 1241 | 2913.0 | Motif | Cell attachment site | |
| Tgene | COL7A1 | chr3:48752748 | chr3:48623408 | ENST00000454817 | 26 | 117 | 2553_2555 | 1241 | 2913.0 | Motif | Cell attachment site | |
| Tgene | COL7A1 | chr3:48752748 | chr3:48623408 | ENST00000328333 | 26 | 118 | 1254_1477 | 1241 | 2945.0 | Region | Note=Interrupted collagenous region | |
| Tgene | COL7A1 | chr3:48752748 | chr3:48623408 | ENST00000328333 | 26 | 118 | 1254_2784 | 1241 | 2945.0 | Region | Note=Triple-helical region | |
| Tgene | COL7A1 | chr3:48752748 | chr3:48623408 | ENST00000328333 | 26 | 118 | 2785_2944 | 1241 | 2945.0 | Region | Note=Nonhelical region (NC2) | |
| Tgene | COL7A1 | chr3:48752748 | chr3:48623408 | ENST00000454817 | 26 | 117 | 1254_1477 | 1241 | 2913.0 | Region | Note=Interrupted collagenous region | |
| Tgene | COL7A1 | chr3:48752748 | chr3:48623408 | ENST00000454817 | 26 | 117 | 1254_2784 | 1241 | 2913.0 | Region | Note=Triple-helical region | |
| Tgene | COL7A1 | chr3:48752748 | chr3:48623408 | ENST00000454817 | 26 | 117 | 2785_2944 | 1241 | 2913.0 | Region | Note=Nonhelical region (NC2) |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | IP6K2 | chr3:48752748 | chr3:48623408 | ENST00000328631 | - | 1 | 6 | 207_209 | 0 | 427.0 | Nucleotide binding | ATP |
| Hgene | IP6K2 | chr3:48752748 | chr3:48623408 | ENST00000340879 | - | 1 | 3 | 207_209 | 0 | 98.0 | Nucleotide binding | ATP |
| Hgene | IP6K2 | chr3:48752748 | chr3:48623408 | ENST00000413298 | - | 1 | 4 | 207_209 | 0 | 98.0 | Nucleotide binding | ATP |
| Hgene | IP6K2 | chr3:48752748 | chr3:48623408 | ENST00000431721 | - | 2 | 3 | 207_209 | 11 | 186.0 | Nucleotide binding | ATP |
| Hgene | IP6K2 | chr3:48752748 | chr3:48623408 | ENST00000432678 | - | 1 | 4 | 207_209 | 0 | 221.66666666666666 | Nucleotide binding | ATP |
| Hgene | IP6K2 | chr3:48752748 | chr3:48623408 | ENST00000446860 | - | 1 | 3 | 207_209 | 0 | 189.0 | Nucleotide binding | ATP |
| Hgene | IP6K2 | chr3:48752748 | chr3:48623408 | ENST00000328631 | - | 1 | 6 | 236_243 | 0 | 427.0 | Region | Substrate binding |
| Hgene | IP6K2 | chr3:48752748 | chr3:48623408 | ENST00000340879 | - | 1 | 3 | 236_243 | 0 | 98.0 | Region | Substrate binding |
| Hgene | IP6K2 | chr3:48752748 | chr3:48623408 | ENST00000413298 | - | 1 | 4 | 236_243 | 0 | 98.0 | Region | Substrate binding |
| Hgene | IP6K2 | chr3:48752748 | chr3:48623408 | ENST00000431721 | - | 2 | 3 | 236_243 | 11 | 186.0 | Region | Substrate binding |
| Hgene | IP6K2 | chr3:48752748 | chr3:48623408 | ENST00000432678 | - | 1 | 4 | 236_243 | 0 | 221.66666666666666 | Region | Substrate binding |
| Hgene | IP6K2 | chr3:48752748 | chr3:48623408 | ENST00000446860 | - | 1 | 3 | 236_243 | 0 | 189.0 | Region | Substrate binding |
| Tgene | COL7A1 | chr3:48752748 | chr3:48623408 | ENST00000328333 | 26 | 118 | 1054_1229 | 1241 | 2945.0 | Domain | VWFA 2 | |
| Tgene | COL7A1 | chr3:48752748 | chr3:48623408 | ENST00000328333 | 26 | 118 | 234_329 | 1241 | 2945.0 | Domain | Fibronectin type-III 1 | |
| Tgene | COL7A1 | chr3:48752748 | chr3:48623408 | ENST00000328333 | 26 | 118 | 330_416 | 1241 | 2945.0 | Domain | Fibronectin type-III 2 | |
| Tgene | COL7A1 | chr3:48752748 | chr3:48623408 | ENST00000328333 | 26 | 118 | 38_211 | 1241 | 2945.0 | Domain | VWFA 1 | |
| Tgene | COL7A1 | chr3:48752748 | chr3:48623408 | ENST00000328333 | 26 | 118 | 417_507 | 1241 | 2945.0 | Domain | Fibronectin type-III 3 | |
| Tgene | COL7A1 | chr3:48752748 | chr3:48623408 | ENST00000328333 | 26 | 118 | 510_597 | 1241 | 2945.0 | Domain | Fibronectin type-III 4 | |
| Tgene | COL7A1 | chr3:48752748 | chr3:48623408 | ENST00000328333 | 26 | 118 | 600_687 | 1241 | 2945.0 | Domain | Fibronectin type-III 5 | |
| Tgene | COL7A1 | chr3:48752748 | chr3:48623408 | ENST00000328333 | 26 | 118 | 688_775 | 1241 | 2945.0 | Domain | Fibronectin type-III 6 | |
| Tgene | COL7A1 | chr3:48752748 | chr3:48623408 | ENST00000328333 | 26 | 118 | 778_866 | 1241 | 2945.0 | Domain | Fibronectin type-III 7 | |
| Tgene | COL7A1 | chr3:48752748 | chr3:48623408 | ENST00000328333 | 26 | 118 | 869_957 | 1241 | 2945.0 | Domain | Fibronectin type-III 8 | |
| Tgene | COL7A1 | chr3:48752748 | chr3:48623408 | ENST00000328333 | 26 | 118 | 958_1051 | 1241 | 2945.0 | Domain | Fibronectin type-III 9 | |
| Tgene | COL7A1 | chr3:48752748 | chr3:48623408 | ENST00000454817 | 26 | 117 | 1054_1229 | 1241 | 2913.0 | Domain | VWFA 2 | |
| Tgene | COL7A1 | chr3:48752748 | chr3:48623408 | ENST00000454817 | 26 | 117 | 234_329 | 1241 | 2913.0 | Domain | Fibronectin type-III 1 | |
| Tgene | COL7A1 | chr3:48752748 | chr3:48623408 | ENST00000454817 | 26 | 117 | 330_416 | 1241 | 2913.0 | Domain | Fibronectin type-III 2 | |
| Tgene | COL7A1 | chr3:48752748 | chr3:48623408 | ENST00000454817 | 26 | 117 | 38_211 | 1241 | 2913.0 | Domain | VWFA 1 | |
| Tgene | COL7A1 | chr3:48752748 | chr3:48623408 | ENST00000454817 | 26 | 117 | 417_507 | 1241 | 2913.0 | Domain | Fibronectin type-III 3 | |
| Tgene | COL7A1 | chr3:48752748 | chr3:48623408 | ENST00000454817 | 26 | 117 | 510_597 | 1241 | 2913.0 | Domain | Fibronectin type-III 4 | |
| Tgene | COL7A1 | chr3:48752748 | chr3:48623408 | ENST00000454817 | 26 | 117 | 600_687 | 1241 | 2913.0 | Domain | Fibronectin type-III 5 | |
| Tgene | COL7A1 | chr3:48752748 | chr3:48623408 | ENST00000454817 | 26 | 117 | 688_775 | 1241 | 2913.0 | Domain | Fibronectin type-III 6 | |
| Tgene | COL7A1 | chr3:48752748 | chr3:48623408 | ENST00000454817 | 26 | 117 | 778_866 | 1241 | 2913.0 | Domain | Fibronectin type-III 7 | |
| Tgene | COL7A1 | chr3:48752748 | chr3:48623408 | ENST00000454817 | 26 | 117 | 869_957 | 1241 | 2913.0 | Domain | Fibronectin type-III 8 | |
| Tgene | COL7A1 | chr3:48752748 | chr3:48623408 | ENST00000454817 | 26 | 117 | 958_1051 | 1241 | 2913.0 | Domain | Fibronectin type-III 9 | |
| Tgene | COL7A1 | chr3:48752748 | chr3:48623408 | ENST00000328333 | 26 | 118 | 1170_1172 | 1241 | 2945.0 | Motif | Cell attachment site | |
| Tgene | COL7A1 | chr3:48752748 | chr3:48623408 | ENST00000454817 | 26 | 117 | 1170_1172 | 1241 | 2913.0 | Motif | Cell attachment site | |
| Tgene | COL7A1 | chr3:48752748 | chr3:48623408 | ENST00000328333 | 26 | 118 | 17_1253 | 1241 | 2945.0 | Region | Note=Nonhelical region (NC1) | |
| Tgene | COL7A1 | chr3:48752748 | chr3:48623408 | ENST00000454817 | 26 | 117 | 17_1253 | 1241 | 2913.0 | Region | Note=Nonhelical region (NC1) |
Top |
Fusion Gene Sequence for IP6K2-COL7A1 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >40019_40019_1_IP6K2-COL7A1_IP6K2_chr3_48752748_ENST00000450045_COL7A1_chr3_48623408_ENST00000328333_length(transcript)=5548nt_BP=103nt CCCTTCCAAGAGGGATGCCAGAGCCTTCTGTAAGCTCCTGAGATGTCACTGGTATCTAGGCAACAGGGATGAGCCTGAACCTCCCTGAGG CCAGCTTACTGAGCCCCGGCCAGAGCCCTGCCCAGTGTATTGTCCAAAGGGCCAGAAGGGGGAACCTGGAGAGATGGGCCTGAGAGGACA AGTTGGGCCTCCTGGCGACCCTGGCCTCCCGGGCAGGACCGGTGCTCCCGGCCCCCAGGGGCCCCCTGGAAGTGCCACTGCCAAGGGCGA GAGGGGCTTCCCTGGAGCAGATGGGCGTCCAGGCAGCCCTGGCCGCGCCGGGAATCCTGGGACCCCTGGAGCCCCTGGCCTAAAGGGCTC TCCAGGGTTGCCTGGCCCTCGTGGGGACCCGGGAGAGCGAGGACCTCGAGGCCCAAAGGGGGAGCCGGGGGCTCCCGGACAAGTCATCGG AGGTGAAGGACCTGGGCTTCCTGGGCGGAAAGGGGACCCTGGACCATCGGGCCCCCCTGGACCTCGTGGACCACTGGGGGACCCAGGACC CCGTGGCCCCCCAGGGCTTCCTGGAACAGCCATGAAGGGTGACAAAGGCGATCGTGGGGAGCGGGGTCCCCCTGGACCAGGTGAAGGTGG CATTGCTCCTGGGGAGCCTGGGCTGCCGGGTCTTCCCGGAAGCCCTGGACCCCAAGGCCCCGTTGGCCCCCCTGGAAAGAAAGGAGAAAA AGGTGACTCTGAGGATGGAGCTCCAGGCCTCCCAGGACAACCTGGGTCTCCGGGTGAGCAGGGCCCACGGGGACCTCCTGGAGCTATTGG CCCCAAAGGTGACCGGGGCTTTCCAGGGCCCCTGGGTGAGGCTGGAGAGAAGGGCGAACGTGGACCCCCAGGCCCAGCGGGATCCCGGGG GCTGCCAGGGGTTGCTGGACGTCCTGGAGCCAAGGGTCCTGAAGGGCCACCAGGACCCACTGGCCGCCAAGGAGAGAAGGGGGAGCCTGG TCGCCCTGGGGACCCTGCAGTGGTGGGACCTGCTGTTGCTGGACCCAAAGGAGAAAAGGGAGATGTGGGGCCCGCTGGGCCCAGAGGAGC TACCGGAGTCCAAGGGGAACGGGGCCCACCCGGCTTGGTTCTTCCTGGAGACCCTGGCCCCAAGGGAGACCCTGGAGACCGGGGTCCCAT TGGCCTTACTGGCAGAGCAGGACCCCCAGGTGACTCAGGGCCTCCTGGAGAGAAGGGAGACCCTGGGCGGCCTGGCCCCCCAGGACCTGT TGGCCCCCGAGGACGAGATGGTGAAGTTGGAGAGAAAGGTGACGAGGGTCCTCCGGGTGACCCGGGTTTGCCTGGAAAAGCAGGCGAGCG TGGCCTTCGGGGGGCACCTGGAGTTCGGGGGCCTGTGGGTGAAAAGGGAGACCAGGGAGATCCTGGAGAGGATGGACGAAATGGCAGCCC TGGATCATCTGGACCCAAGGGTGACCGTGGGGAGCCGGGTCCCCCAGGACCCCCGGGACGGCTGGTAGACACAGGACCTGGAGCCAGAGA GAAGGGAGAGCCTGGGGACCGCGGACAAGAGGGTCCTCGAGGGCCCAAGGGTGATCCTGGCCTCCCTGGAGCCCCTGGGGAAAGGGGCAT TGAAGGGTTTCGGGGACCCCCAGGCCCACAGGGGGACCCAGGTGTCCGAGGCCCAGCAGGAGAAAAGGGTGACCGGGGTCCCCCTGGGCT GGATGGCCGGAGCGGACTGGATGGGAAACCAGGAGCCGCTGGGCCCTCTGGGCCGAATGGTGCTGCAGGCAAAGCTGGGGACCCAGGGAG AGACGGGCTTCCAGGCCTCCGTGGAGAACAGGGCCTCCCTGGCCCCTCTGGTCCCCCTGGATTACCGGGAAAGCCAGGCGAGGATGGCAA ACCTGGCCTGAATGGAAAAAACGGAGAACCTGGGGACCCTGGAGAAGACGGGAGGAAGGGAGAGAAAGGAGATTCAGGCGCCTCTGGGAG AGAAGGTCGTGATGGCCCCAAGGGTGAGCGTGGAGCTCCTGGTATCCTTGGACCCCAGGGGCCTCCAGGCCTCCCAGGGCCAGTGGGCCC TCCTGGCCAGGGTTTTCCTGGTGTCCCAGGAGGCACGGGCCCCAAGGGTGACCGTGGGGAGACTGGATCCAAAGGGGAGCAGGGCCTCCC TGGAGAGCGTGGCCTGCGAGGAGAGCCTGGAAGTGTGCCGAATGTGGATCGGTTGCTGGAAACTGCTGGCATCAAGGCATCTGCCCTGCG GGAGATCGTGGAGACCTGGGATGAGAGCTCTGGTAGCTTCCTGCCTGTGCCCGAACGGCGTCGAGGCCCCAAGGGGGACTCAGGCGAACA GGGCCCCCCAGGCAAGGAGGGCCCCATCGGCTTTCCTGGAGAACGCGGGCTGAAGGGCGACCGTGGAGACCCTGGCCCTCAGGGGCCACC TGGTCTGGCCCTTGGGGAGAGGGGCCCCCCCGGGCCTTCCGGCCTTGCCGGGGAGCCTGGAAAGCCTGGTATTCCCGGGCTCCCAGGCAG GGCTGGGGGTGTGGGAGAGGCAGGAAGGCCAGGAGAGAGGGGAGAACGGGGAGAGAAAGGAGAACGTGGAGAACAGGGCAGAGATGGCCC TCCTGGACTCCCTGGAACCCCTGGGCCCCCCGGACCCCCTGGCCCCAAGGTGTCTGTGGATGAGCCAGGTCCTGGACTCTCTGGAGAACA GGGACCCCCTGGACTCAAGGGTGCTAAGGGGGAGCCGGGCAGCAATGGTGACCAAGGTCCCAAAGGAGACAGGGGTGTGCCAGGCATCAA AGGAGACCGGGGAGAGCCTGGACCGAGGGGTCAGGACGGCAACCCGGGTCTACCAGGAGAGCGTGGTATGGCTGGGCCTGAAGGGAAGCC GGGTCTGCAGGGTCCAAGAGGCCCCCCTGGCCCAGTGGGTGGTCATGGAGACCCTGGACCACCTGGTGCCCCGGGTCTTGCTGGCCCTGC AGGACCCCAAGGACCTTCTGGCCTGAAGGGGGAGCCTGGAGAGACAGGACCTCCAGGACGGGGCCTGACTGGACCTACTGGAGCTGTGGG ACTTCCTGGACCCCCCGGCCCTTCAGGCCTTGTGGGTCCACAGGGGTCTCCAGGTTTGCCTGGACAAGTGGGGGAGACAGGGAAGCCGGG AGCCCCAGGTCGAGATGGTGCCAGTGGAAAAGATGGAGACAGAGGGAGCCCTGGTGTGCCAGGGTCACCAGGTCTGCCTGGCCCTGTCGG ACCTAAAGGAGAACCTGGCCCCACGGGGGCCCCTGGACAGGCTGTGGTCGGGCTCCCTGGAGCAAAGGGAGAGAAGGGAGCCCCTGGAGG CCTTGCTGGAGACCTGGTGGGTGAGCCGGGAGCCAAAGGTGACCGAGGACTGCCAGGGCCGCGAGGCGAGAAGGGTGAAGCTGGCCGTGC AGGGGAGCCCGGAGACCCTGGGGAAGATGGTCAGAAAGGGGCTCCAGGACCCAAAGGTTTCAAGGGTGACCCAGGAGTCGGGGTCCCGGG CTCCCCTGGGCCTCCTGGCCCTCCAGGTGTGAAGGGAGATCTGGGCCTCCCTGGCCTGCCCGGTGCTCCTGGTGTTGTTGGGTTCCCGGG TCAGACAGGCCCTCGAGGAGAGATGGGTCAGCCAGGCCCTAGTGGAGAGCGGGGTCTGGCAGGCCCCCCAGGGAGAGAAGGAATCCCAGG ACCCCTGGGGCCACCTGGACCACCGGGGTCAGTGGGACCACCTGGGGCCTCTGGACTCAAAGGAGACAAGGGAGACCCTGGAGTAGGGCT GCCTGGGCCCCGAGGCGAGCGTGGGGAGCCAGGCATCCGGGGTGAAGATGGCCGCCCCGGCCAGGAGGGACCCCGAGGACTCACGGGGCC CCCTGGCAGCAGGGGAGAGCGTGGGGAGAAGGGTGATGTTGGGAGTGCAGGACTAAAGGGTGACAAGGGAGACTCAGCTGTGATCCTGGG GCCTCCAGGCCCACGGGGTGCCAAGGGGGACATGGGTGAACGAGGGCCTCGGGGCTTGGATGGTGACAAAGGACCTCGGGGAGACAATGG GGACCCTGGTGACAAGGGCAGCAAGGGAGAGCCTGGTGACAAGGGCTCAGCCGGGTTGCCAGGACTGCGTGGACTCCTGGGACCCCAGGG TCAACCTGGTGCAGCAGGGATCCCTGGTGACCCGGGATCCCCAGGAAAGGATGGAGTGCCTGGTATCCGAGGAGAAAAAGGAGATGTTGG CTTCATGGGTCCCCGGGGCCTCAAGGGTGAACGGGGAGTGAAGGGAGCCTGTGGCCTTGATGGAGAGAAGGGAGACAAGGGAGAAGCTGG TCCCCCAGGCCGCCCCGGGCTGGCAGGACACAAAGGAGAGATGGGGGAGCCTGGTGTGCCGGGCCAGTCGGGGGCCCCTGGCAAGGAGGG CCTGATCGGTCCCAAGGGTGACCGAGGCTTTGACGGGCAGCCAGGCCCCAAGGGTGACCAGGGCGAGAAAGGGGAGCGGGGAACCCCAGG AATTGGGGGCTTCCCAGGCCCCAGTGGAAATGATGGCTCTGCTGGTCCCCCAGGGCCACCTGGCAGTGTTGGTCCCAGAGGCCCCGAAGG ACTTCAGGGCCAGAAGGGTGAGCGAGGTCCCCCCGGAGAGAGAGTGGTGGGGGCTCCTGGGGTCCCTGGAGCTCCTGGCGAGAGAGGGGA GCAGGGGCGGCCAGGGCCTGCCGGTCCTCGAGGCGAGAAGGGAGAAGCTGCACTGACGGAGGATGACATCCGGGGCTTTGTGCGCCAAGA GATGAGTCAGCACTGTGCCTGCCAGGGCCAGTTCATCGCATCTGGATCACGACCCCTCCCTAGTTATGCTGCAGACACTGCCGGCTCCCA GCTCCATGCTGTGCCTGTGCTCCGCGTCTCTCATGCAGAGGAGGAAGAGCGGGTACCCCCTGAGGATGATGAGTACTCTGAATACTCCGA GTATTCTGTGGAGGAGTACCAGGACCCTGAAGCTCCTTGGGATAGTGATGACCCCTGTTCCCTGCCACTGGATGAGGGCTCCTGCACTGC CTACACCCTGCGCTGGTACCATCGGGCTGTGACAGGCAGCACAGAGGCCTGTCACCCTTTTGTCTATGGTGGCTGTGGAGGGAATGCCAA CCGTTTTGGGACCCGTGAGGCCTGCGAGCGCCGCTGCCCACCCCGGGTGGTCCAGAGCCAGGGGACAGGTACTGCCCAGGACTGAGGCCC AGATAATGAGCTGAGATTCAGCATCCCCTGGAGGAGTCGGGGTCTCAGCAGAACCCCACTGTCCCTCCCCTTGGTGCTAGAGGCTTGTGT GCACGTGAGCGTGCGTGTGCACGTCCGTTATTTCAGTGACTTGGTCCCGTGGGTCTAGCCTTCCCCCCTGTGGACAAACCCCCATTGTGG CTCCTGCCACCCTGGCAGATGACTCACTGTGGGGGGGTGGCTGTGGGCAGTGAGCGGATGTGACTGGCGTCTGACCCGCCCCTTGACCCA >40019_40019_1_IP6K2-COL7A1_IP6K2_chr3_48752748_ENST00000450045_COL7A1_chr3_48623408_ENST00000328333_length(amino acids)=1683AA_BP=241 MGLRGQVGPPGDPGLPGRTGAPGPQGPPGSATAKGERGFPGADGRPGSPGRAGNPGTPGAPGLKGSPGLPGPRGDPGERGPRGPKGEPGA PGQVIGGEGPGLPGRKGDPGPSGPPGPRGPLGDPGPRGPPGLPGTAMKGDKGDRGERGPPGPGEGGIAPGEPGLPGLPGSPGPQGPVGPP GKKGEKGDSEDGAPGLPGQPGSPGEQGPRGPPGAIGPKGDRGFPGPLGEAGEKGERGPPGPAGSRGLPGVAGRPGAKGPEGPPGPTGRQG EKGEPGRPGDPAVVGPAVAGPKGEKGDVGPAGPRGATGVQGERGPPGLVLPGDPGPKGDPGDRGPIGLTGRAGPPGDSGPPGEKGDPGRP GPPGPVGPRGRDGEVGEKGDEGPPGDPGLPGKAGERGLRGAPGVRGPVGEKGDQGDPGEDGRNGSPGSSGPKGDRGEPGPPGPPGRLVDT GPGAREKGEPGDRGQEGPRGPKGDPGLPGAPGERGIEGFRGPPGPQGDPGVRGPAGEKGDRGPPGLDGRSGLDGKPGAAGPSGPNGAAGK AGDPGRDGLPGLRGEQGLPGPSGPPGLPGKPGEDGKPGLNGKNGEPGDPGEDGRKGEKGDSGASGREGRDGPKGERGAPGILGPQGPPGL PGPVGPPGQGFPGVPGGTGPKGDRGETGSKGEQGLPGERGLRGEPGSVPNVDRLLETAGIKASALREIVETWDESSGSFLPVPERRRGPK GDSGEQGPPGKEGPIGFPGERGLKGDRGDPGPQGPPGLALGERGPPGPSGLAGEPGKPGIPGLPGRAGGVGEAGRPGERGERGEKGERGE QGRDGPPGLPGTPGPPGPPGPKVSVDEPGPGLSGEQGPPGLKGAKGEPGSNGDQGPKGDRGVPGIKGDRGEPGPRGQDGNPGLPGERGMA GPEGKPGLQGPRGPPGPVGGHGDPGPPGAPGLAGPAGPQGPSGLKGEPGETGPPGRGLTGPTGAVGLPGPPGPSGLVGPQGSPGLPGQVG ETGKPGAPGRDGASGKDGDRGSPGVPGSPGLPGPVGPKGEPGPTGAPGQAVVGLPGAKGEKGAPGGLAGDLVGEPGAKGDRGLPGPRGEK GEAGRAGEPGDPGEDGQKGAPGPKGFKGDPGVGVPGSPGPPGPPGVKGDLGLPGLPGAPGVVGFPGQTGPRGEMGQPGPSGERGLAGPPG REGIPGPLGPPGPPGSVGPPGASGLKGDKGDPGVGLPGPRGERGEPGIRGEDGRPGQEGPRGLTGPPGSRGERGEKGDVGSAGLKGDKGD SAVILGPPGPRGAKGDMGERGPRGLDGDKGPRGDNGDPGDKGSKGEPGDKGSAGLPGLRGLLGPQGQPGAAGIPGDPGSPGKDGVPGIRG EKGDVGFMGPRGLKGERGVKGACGLDGEKGDKGEAGPPGRPGLAGHKGEMGEPGVPGQSGAPGKEGLIGPKGDRGFDGQPGPKGDQGEKG ERGTPGIGGFPGPSGNDGSAGPPGPPGSVGPRGPEGLQGQKGERGPPGERVVGAPGVPGAPGERGEQGRPGPAGPRGEKGEAALTEDDIR GFVRQEMSQHCACQGQFIASGSRPLPSYAADTAGSQLHAVPVLRVSHAEEEERVPPEDDEYSEYSEYSVEEYQDPEAPWDSDDPCSLPLD -------------------------------------------------------------- >40019_40019_2_IP6K2-COL7A1_IP6K2_chr3_48752748_ENST00000450045_COL7A1_chr3_48623408_ENST00000454817_length(transcript)=5452nt_BP=103nt CCCTTCCAAGAGGGATGCCAGAGCCTTCTGTAAGCTCCTGAGATGTCACTGGTATCTAGGCAACAGGGATGAGCCTGAACCTCCCTGAGG CCAGCTTACTGAGCCCCGGCCAGAGCCCTGCCCAGTGTATTGTCCAAAGGGCCAGAAGGGGGAACCTGGAGAGATGGGCCTGAGAGGACA AGTTGGGCCTCCTGGCGACCCTGGCCTCCCGGGCAGGACCGGTGCTCCCGGCCCCCAGGGGCCCCCTGGAAGTGCCACTGCCAAGGGCGA GAGGGGCTTCCCTGGAGCAGATGGGCGTCCAGGCAGCCCTGGCCGCGCCGGGAATCCTGGGACCCCTGGAGCCCCTGGCCTAAAGGGCTC TCCAGGGTTGCCTGGCCCTCGTGGGGACCCGGGAGAGCGAGGACCTCGAGGCCCAAAGGGGGAGCCGGGGGCTCCCGGACAAGTCATCGG AGGTGAAGGACCTGGGCTTCCTGGGCGGAAAGGGGACCCTGGACCATCGGGCCCCCCTGGACCTCGTGGACCACTGGGGGACCCAGGACC CCGTGGCCCCCCAGGGCTTCCTGGAACAGCCATGAAGGGTGACAAAGGCGATCGTGGGGAGCGGGGTCCCCCTGGACCAGGTGAAGGTGG CATTGCTCCTGGGGAGCCTGGGCTGCCGGGTCTTCCCGGAAGCCCTGGACCCCAAGGCCCCGTTGGCCCCCCTGGAAAGAAAGGAGAAAA AGGTGACTCTGAGGATGGAGCTCCAGGCCTCCCAGGACAACCTGGGTCTCCGGGTGAGCAGGGCCCACGGGGACCTCCTGGAGCTATTGG CCCCAAAGGTGACCGGGGCTTTCCAGGGCCCCTGGGTGAGGCTGGAGAGAAGGGCGAACGTGGACCCCCAGGCCCAGCGGGATCCCGGGG GCTGCCAGGGGTTGCTGGACGTCCTGGAGCCAAGGGTCCTGAAGGGCCACCAGGACCCACTGGCCGCCAAGGAGAGAAGGGGGAGCCTGG TCGCCCTGGGGACCCTGCAGTGGTGGGACCTGCTGTTGCTGGACCCAAAGGAGAAAAGGGAGATGTGGGGCCCGCTGGGCCCAGAGGAGC TACCGGAGTCCAAGGGGAACGGGGCCCACCCGGCTTGGTTCTTCCTGGAGACCCTGGCCCCAAGGGAGACCCTGGAGACCGGGGTCCCAT TGGCCTTACTGGCAGAGCAGGACCCCCAGGTGACTCAGGGCCTCCTGGAGAGAAGGGAGACCCTGGGCGGCCTGGCCCCCCAGGACCTGT TGGCCCCCGAGGACGAGATGGTGAAGTTGGAGAGAAAGGTGACGAGGGTCCTCCGGGTGACCCGGGTTTGCCTGGAAAAGCAGGCGAGCG TGGCCTTCGGGGGGCACCTGGAGTTCGGGGGCCTGTGGGTGAAAAGGGAGACCAGGGAGATCCTGGAGAGGATGGACGAAATGGCAGCCC TGGATCATCTGGACCCAAGGGTGACCGTGGGGAGCCGGGTCCCCCAGGACCCCCGGGACGGCTGGTAGACACAGGACCTGGAGCCAGAGA GAAGGGAGAGCCTGGGGACCGCGGACAAGAGGGTCCTCGAGGGCCCAAGGGTGATCCTGGCCTCCCTGGAGCCCCTGGGGAAAGGGGCAT TGAAGGGTTTCGGGGACCCCCAGGCCCACAGGGGGACCCAGGTGTCCGAGGCCCAGCAGGAGAAAAGGGTGACCGGGGTCCCCCTGGGCT GGATGGCCGGAGCGGACTGGATGGGAAACCAGGAGCCGCTGGGCCCTCTGGGCCGAATGGTGCTGCAGGCAAAGCTGGGGACCCAGGGAG AGACGGGCTTCCAGGCCTCCGTGGAGAACAGGGCCTCCCTGGCCCCTCTGGTCCCCCTGGATTACCGGGAAAGCCAGGCGAGGATGGCAA ACCTGGCCTGAATGGAAAAAACGGAGAACCTGGGGACCCTGGAGAAGACGGGAGGAAGGGAGAGAAAGGAGATTCAGGCGCCTCTGGGAG AGAAGGTTTTCCTGGTGTCCCAGGAGGCACGGGCCCCAAGGGTGACCGTGGGGAGACTGGATCCAAAGGGGAGCAGGGCCTCCCTGGAGA GCGTGGCCTGCGAGGAGAGCCTGGAAGTGTGCCGAATGTGGATCGGTTGCTGGAAACTGCTGGCATCAAGGCATCTGCCCTGCGGGAGAT CGTGGAGACCTGGGATGAGAGCTCTGGTAGCTTCCTGCCTGTGCCCGAACGGCGTCGAGGCCCCAAGGGGGACTCAGGCGAACAGGGCCC CCCAGGCAAGGAGGGCCCCATCGGCTTTCCTGGAGAACGCGGGCTGAAGGGCGACCGTGGAGACCCTGGCCCTCAGGGGCCACCTGGTCT GGCCCTTGGGGAGAGGGGCCCCCCCGGGCCTTCCGGCCTTGCCGGGGAGCCTGGAAAGCCTGGTATTCCCGGGCTCCCAGGCAGGGCTGG GGGTGTGGGAGAGGCAGGAAGGCCAGGAGAGAGGGGAGAACGGGGAGAGAAAGGAGAACGTGGAGAACAGGGCAGAGATGGCCCTCCTGG ACTCCCTGGAACCCCTGGGCCCCCCGGACCCCCTGGCCCCAAGGTGTCTGTGGATGAGCCAGGTCCTGGACTCTCTGGAGAACAGGGACC CCCTGGACTCAAGGGTGCTAAGGGGGAGCCGGGCAGCAATGGTGACCAAGGTCCCAAAGGAGACAGGGGTGTGCCAGGCATCAAAGGAGA CCGGGGAGAGCCTGGACCGAGGGGTCAGGACGGCAACCCGGGTCTACCAGGAGAGCGTGGTATGGCTGGGCCTGAAGGGAAGCCGGGTCT GCAGGGTCCAAGAGGCCCCCCTGGCCCAGTGGGTGGTCATGGAGACCCTGGACCACCTGGTGCCCCGGGTCTTGCTGGCCCTGCAGGACC CCAAGGACCTTCTGGCCTGAAGGGGGAGCCTGGAGAGACAGGACCTCCAGGACGGGGCCTGACTGGACCTACTGGAGCTGTGGGACTTCC TGGACCCCCCGGCCCTTCAGGCCTTGTGGGTCCACAGGGGTCTCCAGGTTTGCCTGGACAAGTGGGGGAGACAGGGAAGCCGGGAGCCCC AGGTCGAGATGGTGCCAGTGGAAAAGATGGAGACAGAGGGAGCCCTGGTGTGCCAGGGTCACCAGGTCTGCCTGGCCCTGTCGGACCTAA AGGAGAACCTGGCCCCACGGGGGCCCCTGGACAGGCTGTGGTCGGGCTCCCTGGAGCAAAGGGAGAGAAGGGAGCCCCTGGAGGCCTTGC TGGAGACCTGGTGGGTGAGCCGGGAGCCAAAGGTGACCGAGGACTGCCAGGGCCGCGAGGCGAGAAGGGTGAAGCTGGCCGTGCAGGGGA GCCCGGAGACCCTGGGGAAGATGGTCAGAAAGGGGCTCCAGGACCCAAAGGTTTCAAGGGTGACCCAGGAGTCGGGGTCCCGGGCTCCCC TGGGCCTCCTGGCCCTCCAGGTGTGAAGGGAGATCTGGGCCTCCCTGGCCTGCCCGGTGCTCCTGGTGTTGTTGGGTTCCCGGGTCAGAC AGGCCCTCGAGGAGAGATGGGTCAGCCAGGCCCTAGTGGAGAGCGGGGTCTGGCAGGCCCCCCAGGGAGAGAAGGAATCCCAGGACCCCT GGGGCCACCTGGACCACCGGGGTCAGTGGGACCACCTGGGGCCTCTGGACTCAAAGGAGACAAGGGAGACCCTGGAGTAGGGCTGCCTGG GCCCCGAGGCGAGCGTGGGGAGCCAGGCATCCGGGGTGAAGATGGCCGCCCCGGCCAGGAGGGACCCCGAGGACTCACGGGGCCCCCTGG CAGCAGGGGAGAGCGTGGGGAGAAGGGTGATGTTGGGAGTGCAGGACTAAAGGGTGACAAGGGAGACTCAGCTGTGATCCTGGGGCCTCC AGGCCCACGGGGTGCCAAGGGGGACATGGGTGAACGAGGGCCTCGGGGCTTGGATGGTGACAAAGGACCTCGGGGAGACAATGGGGACCC TGGTGACAAGGGCAGCAAGGGAGAGCCTGGTGACAAGGGCTCAGCCGGGTTGCCAGGACTGCGTGGACTCCTGGGACCCCAGGGTCAACC TGGTGCAGCAGGGATCCCTGGTGACCCGGGATCCCCAGGAAAGGATGGAGTGCCTGGTATCCGAGGAGAAAAAGGAGATGTTGGCTTCAT GGGTCCCCGGGGCCTCAAGGGTGAACGGGGAGTGAAGGGAGCCTGTGGCCTTGATGGAGAGAAGGGAGACAAGGGAGAAGCTGGTCCCCC AGGCCGCCCCGGGCTGGCAGGACACAAAGGAGAGATGGGGGAGCCTGGTGTGCCGGGCCAGTCGGGGGCCCCTGGCAAGGAGGGCCTGAT CGGTCCCAAGGGTGACCGAGGCTTTGACGGGCAGCCAGGCCCCAAGGGTGACCAGGGCGAGAAAGGGGAGCGGGGAACCCCAGGAATTGG GGGCTTCCCAGGCCCCAGTGGAAATGATGGCTCTGCTGGTCCCCCAGGGCCACCTGGCAGTGTTGGTCCCAGAGGCCCCGAAGGACTTCA GGGCCAGAAGGGTGAGCGAGGTCCCCCCGGAGAGAGAGTGGTGGGGGCTCCTGGGGTCCCTGGAGCTCCTGGCGAGAGAGGGGAGCAGGG GCGGCCAGGGCCTGCCGGTCCTCGAGGCGAGAAGGGAGAAGCTGCACTGACGGAGGATGACATCCGGGGCTTTGTGCGCCAAGAGATGAG TCAGCACTGTGCCTGCCAGGGCCAGTTCATCGCATCTGGATCACGACCCCTCCCTAGTTATGCTGCAGACACTGCCGGCTCCCAGCTCCA TGCTGTGCCTGTGCTCCGCGTCTCTCATGCAGAGGAGGAAGAGCGGGTACCCCCTGAGGATGATGAGTACTCTGAATACTCCGAGTATTC TGTGGAGGAGTACCAGGACCCTGAAGCTCCTTGGGATAGTGATGACCCCTGTTCCCTGCCACTGGATGAGGGCTCCTGCACTGCCTACAC CCTGCGCTGGTACCATCGGGCTGTGACAGGCAGCACAGAGGCCTGTCACCCTTTTGTCTATGGTGGCTGTGGAGGGAATGCCAACCGTTT TGGGACCCGTGAGGCCTGCGAGCGCCGCTGCCCACCCCGGGTGGTCCAGAGCCAGGGGACAGGTACTGCCCAGGACTGAGGCCCAGATAA TGAGCTGAGATTCAGCATCCCCTGGAGGAGTCGGGGTCTCAGCAGAACCCCACTGTCCCTCCCCTTGGTGCTAGAGGCTTGTGTGCACGT GAGCGTGCGTGTGCACGTCCGTTATTTCAGTGACTTGGTCCCGTGGGTCTAGCCTTCCCCCCTGTGGACAAACCCCCATTGTGGCTCCTG CCACCCTGGCAGATGACTCACTGTGGGGGGGTGGCTGTGGGCAGTGAGCGGATGTGACTGGCGTCTGACCCGCCCCTTGACCCAAGCCTG >40019_40019_2_IP6K2-COL7A1_IP6K2_chr3_48752748_ENST00000450045_COL7A1_chr3_48623408_ENST00000454817_length(amino acids)=1651AA_BP=45 MGLRGQVGPPGDPGLPGRTGAPGPQGPPGSATAKGERGFPGADGRPGSPGRAGNPGTPGAPGLKGSPGLPGPRGDPGERGPRGPKGEPGA PGQVIGGEGPGLPGRKGDPGPSGPPGPRGPLGDPGPRGPPGLPGTAMKGDKGDRGERGPPGPGEGGIAPGEPGLPGLPGSPGPQGPVGPP GKKGEKGDSEDGAPGLPGQPGSPGEQGPRGPPGAIGPKGDRGFPGPLGEAGEKGERGPPGPAGSRGLPGVAGRPGAKGPEGPPGPTGRQG EKGEPGRPGDPAVVGPAVAGPKGEKGDVGPAGPRGATGVQGERGPPGLVLPGDPGPKGDPGDRGPIGLTGRAGPPGDSGPPGEKGDPGRP GPPGPVGPRGRDGEVGEKGDEGPPGDPGLPGKAGERGLRGAPGVRGPVGEKGDQGDPGEDGRNGSPGSSGPKGDRGEPGPPGPPGRLVDT GPGAREKGEPGDRGQEGPRGPKGDPGLPGAPGERGIEGFRGPPGPQGDPGVRGPAGEKGDRGPPGLDGRSGLDGKPGAAGPSGPNGAAGK AGDPGRDGLPGLRGEQGLPGPSGPPGLPGKPGEDGKPGLNGKNGEPGDPGEDGRKGEKGDSGASGREGFPGVPGGTGPKGDRGETGSKGE QGLPGERGLRGEPGSVPNVDRLLETAGIKASALREIVETWDESSGSFLPVPERRRGPKGDSGEQGPPGKEGPIGFPGERGLKGDRGDPGP QGPPGLALGERGPPGPSGLAGEPGKPGIPGLPGRAGGVGEAGRPGERGERGEKGERGEQGRDGPPGLPGTPGPPGPPGPKVSVDEPGPGL SGEQGPPGLKGAKGEPGSNGDQGPKGDRGVPGIKGDRGEPGPRGQDGNPGLPGERGMAGPEGKPGLQGPRGPPGPVGGHGDPGPPGAPGL AGPAGPQGPSGLKGEPGETGPPGRGLTGPTGAVGLPGPPGPSGLVGPQGSPGLPGQVGETGKPGAPGRDGASGKDGDRGSPGVPGSPGLP GPVGPKGEPGPTGAPGQAVVGLPGAKGEKGAPGGLAGDLVGEPGAKGDRGLPGPRGEKGEAGRAGEPGDPGEDGQKGAPGPKGFKGDPGV GVPGSPGPPGPPGVKGDLGLPGLPGAPGVVGFPGQTGPRGEMGQPGPSGERGLAGPPGREGIPGPLGPPGPPGSVGPPGASGLKGDKGDP GVGLPGPRGERGEPGIRGEDGRPGQEGPRGLTGPPGSRGERGEKGDVGSAGLKGDKGDSAVILGPPGPRGAKGDMGERGPRGLDGDKGPR GDNGDPGDKGSKGEPGDKGSAGLPGLRGLLGPQGQPGAAGIPGDPGSPGKDGVPGIRGEKGDVGFMGPRGLKGERGVKGACGLDGEKGDK GEAGPPGRPGLAGHKGEMGEPGVPGQSGAPGKEGLIGPKGDRGFDGQPGPKGDQGEKGERGTPGIGGFPGPSGNDGSAGPPGPPGSVGPR GPEGLQGQKGERGPPGERVVGAPGVPGAPGERGEQGRPGPAGPRGEKGEAALTEDDIRGFVRQEMSQHCACQGQFIASGSRPLPSYAADT AGSQLHAVPVLRVSHAEEEERVPPEDDEYSEYSEYSVEEYQDPEAPWDSDDPCSLPLDEGSCTAYTLRWYHRAVTGSTEACHPFVYGGCG -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for IP6K2-COL7A1 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for IP6K2-COL7A1 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for IP6K2-COL7A1 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
