
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:ITCH-MYL9 (FusionGDB2 ID:40358) |
Fusion Gene Summary for ITCH-MYL9 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: ITCH-MYL9 | Fusion gene ID: 40358 | Hgene | Tgene | Gene symbol | ITCH | MYL9 | Gene ID | 83737 | 10398 |
| Gene name | itchy E3 ubiquitin protein ligase | myosin light chain 9 | |
| Synonyms | ADMFD|AIF4|AIP4|NAPP1 | LC20|MLC-2C|MLC2|MRLC1|MYRL2 | |
| Cytomap | 20q11.22 | 20q11.23 | |
| Type of gene | protein-coding | protein-coding | |
| Description | E3 ubiquitin-protein ligase Itchy homologHECT-type E3 ubiquitin transferase Itchy homologNFE2-associated polypeptide 1atrophin-1 interacting protein 4itchy E3 ubiquitin protein ligase homolog | myosin regulatory light polypeptide 920 kDa myosin light chainepididymis secretory sperm binding proteinmyosin RLCmyosin regulatory light chain 1myosin regulatory light chain 2, smooth muscle isoformmyosin regulatory light chain 9myosin regulatory | |
| Modification date | 20200329 | 20200313 | |
| UniProtAcc | . | P24844 | |
| Ensembl transtripts involved in fusion gene | ENST00000262650, ENST00000374864, ENST00000535650, ENST00000483727, | ENST00000346786, ENST00000279022, | |
| Fusion gene scores | * DoF score | 27 X 16 X 18=7776 | 5 X 4 X 4=80 |
| # samples | 44 | 5 | |
| ** MAII score | log2(44/7776*10)=-4.14345279008112 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(5/80*10)=-0.678071905112638 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: ITCH [Title/Abstract] AND MYL9 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | ITCH(33080402)-MYL9(35176435), # samples:3 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | ITCH | GO:0006511 | ubiquitin-dependent protein catabolic process | 15678106 |
| Hgene | ITCH | GO:0016567 | protein ubiquitination | 17592138|25631046 |
| Hgene | ITCH | GO:0035519 | protein K29-linked ubiquitination | 17028573|18628966 |
| Hgene | ITCH | GO:0070534 | protein K63-linked ubiquitination | 18718448|19592251 |
| Hgene | ITCH | GO:0070936 | protein K48-linked ubiquitination | 19881509 |
 Fusion gene breakpoints across ITCH (5'-gene) Fusion gene breakpoints across ITCH (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
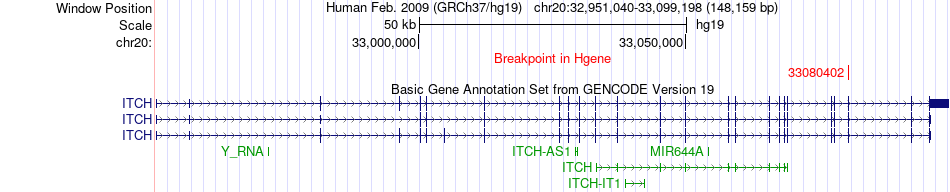 |
 Fusion gene breakpoints across MYL9 (3'-gene) Fusion gene breakpoints across MYL9 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
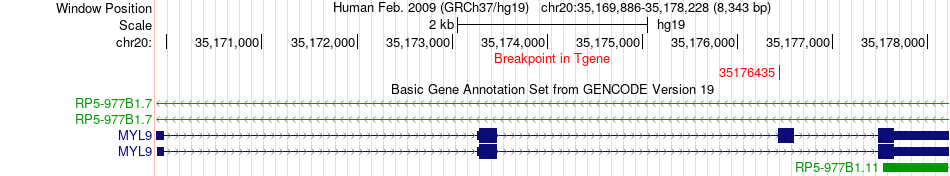 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | LUAD | TCGA-55-7815-01A | ITCH | chr20 | 33080402 | - | MYL9 | chr20 | 35176435 | + |
| ChimerDB4 | LUAD | TCGA-55-7815-01A | ITCH | chr20 | 33080402 | + | MYL9 | chr20 | 35176435 | + |
Top |
Fusion Gene ORF analysis for ITCH-MYL9 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-intron | ENST00000262650 | ENST00000346786 | ITCH | chr20 | 33080402 | + | MYL9 | chr20 | 35176435 | + |
| 5CDS-intron | ENST00000374864 | ENST00000346786 | ITCH | chr20 | 33080402 | + | MYL9 | chr20 | 35176435 | + |
| 5CDS-intron | ENST00000535650 | ENST00000346786 | ITCH | chr20 | 33080402 | + | MYL9 | chr20 | 35176435 | + |
| In-frame | ENST00000262650 | ENST00000279022 | ITCH | chr20 | 33080402 | + | MYL9 | chr20 | 35176435 | + |
| In-frame | ENST00000374864 | ENST00000279022 | ITCH | chr20 | 33080402 | + | MYL9 | chr20 | 35176435 | + |
| In-frame | ENST00000535650 | ENST00000279022 | ITCH | chr20 | 33080402 | + | MYL9 | chr20 | 35176435 | + |
| intron-3CDS | ENST00000483727 | ENST00000279022 | ITCH | chr20 | 33080402 | + | MYL9 | chr20 | 35176435 | + |
| intron-intron | ENST00000483727 | ENST00000346786 | ITCH | chr20 | 33080402 | + | MYL9 | chr20 | 35176435 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000374864 | ITCH | chr20 | 33080402 | + | ENST00000279022 | MYL9 | chr20 | 35176435 | + | 3540 | 2629 | 213 | 2963 | 916 |
| ENST00000535650 | ITCH | chr20 | 33080402 | + | ENST00000279022 | MYL9 | chr20 | 35176435 | + | 3357 | 2446 | 294 | 2780 | 828 |
| ENST00000262650 | ITCH | chr20 | 33080402 | + | ENST00000279022 | MYL9 | chr20 | 35176435 | + | 3586 | 2675 | 136 | 3009 | 957 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000374864 | ENST00000279022 | ITCH | chr20 | 33080402 | + | MYL9 | chr20 | 35176435 | + | 0.000396507 | 0.99960357 |
| ENST00000535650 | ENST00000279022 | ITCH | chr20 | 33080402 | + | MYL9 | chr20 | 35176435 | + | 0.000969524 | 0.9990305 |
| ENST00000262650 | ENST00000279022 | ITCH | chr20 | 33080402 | + | MYL9 | chr20 | 35176435 | + | 0.000406304 | 0.9995937 |
Top |
Fusion Genomic Features for ITCH-MYL9 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| ITCH | chr20 | 33080402 | + | MYL9 | chr20 | 35176434 | + | 6.49E-07 | 0.9999994 |
| ITCH | chr20 | 33080402 | + | MYL9 | chr20 | 35176434 | + | 6.49E-07 | 0.9999994 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
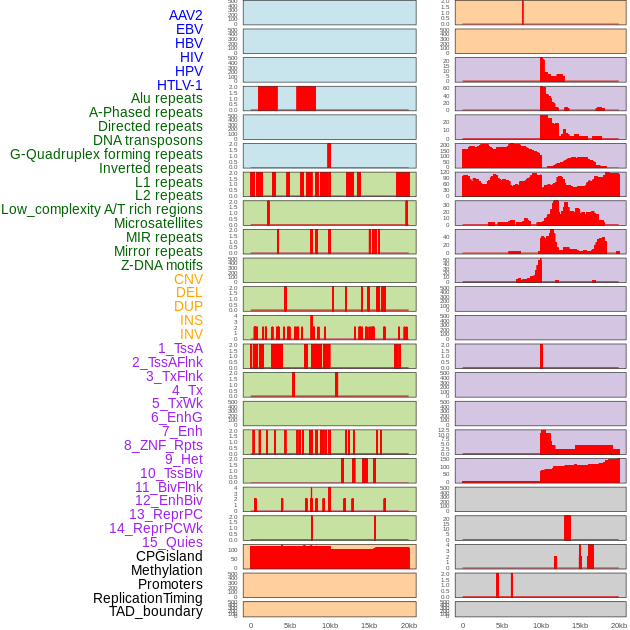 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
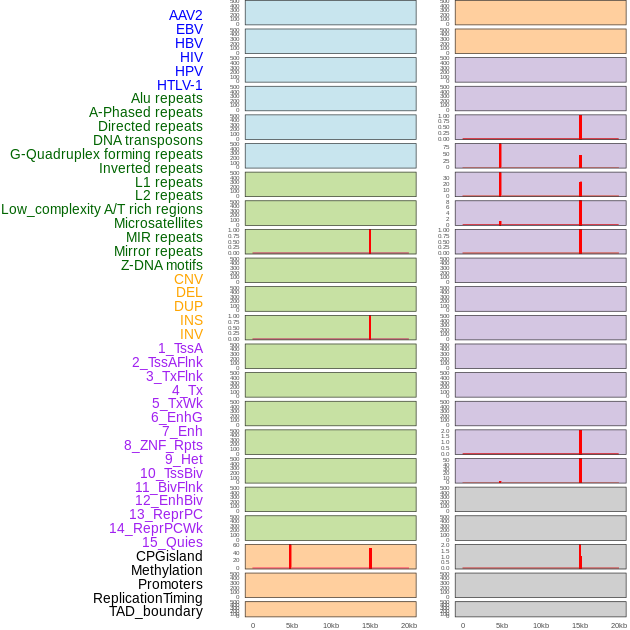 |
Top |
Fusion Protein Features for ITCH-MYL9 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr20:33080402/chr20:35176435) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| . | MYL9 |
| FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. | FUNCTION: Myosin regulatory subunit that plays an important role in regulation of both smooth muscle and nonmuscle cell contractile activity via its phosphorylation. Implicated in cytokinesis, receptor capping, and cell locomotion (PubMed:11942626, PubMed:2526655). In myoblasts, may regulate PIEZO1-dependent cortical actomyosin assembly involved in myotube formation (By similarity). {ECO:0000250|UniProtKB:Q9CQ19, ECO:0000269|PubMed:11942626, ECO:0000269|PubMed:2526655}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | ITCH | chr20:33080402 | chr20:35176435 | ENST00000262650 | + | 24 | 26 | 252_267 | 846 | 904.0 | Compositional bias | Note=Arg/Pro-rich (PRR domain) |
| Hgene | ITCH | chr20:33080402 | chr20:35176435 | ENST00000374864 | + | 23 | 25 | 252_267 | 805 | 863.0 | Compositional bias | Note=Arg/Pro-rich (PRR domain) |
| Hgene | ITCH | chr20:33080402 | chr20:35176435 | ENST00000535650 | + | 22 | 24 | 252_267 | 695 | 753.0 | Compositional bias | Note=Arg/Pro-rich (PRR domain) |
| Hgene | ITCH | chr20:33080402 | chr20:35176435 | ENST00000262650 | + | 24 | 26 | 1_115 | 846 | 904.0 | Domain | C2 |
| Hgene | ITCH | chr20:33080402 | chr20:35176435 | ENST00000262650 | + | 24 | 26 | 326_359 | 846 | 904.0 | Domain | WW 1 |
| Hgene | ITCH | chr20:33080402 | chr20:35176435 | ENST00000262650 | + | 24 | 26 | 358_391 | 846 | 904.0 | Domain | WW 2 |
| Hgene | ITCH | chr20:33080402 | chr20:35176435 | ENST00000262650 | + | 24 | 26 | 438_471 | 846 | 904.0 | Domain | WW 3 |
| Hgene | ITCH | chr20:33080402 | chr20:35176435 | ENST00000262650 | + | 24 | 26 | 478_511 | 846 | 904.0 | Domain | WW 4 |
| Hgene | ITCH | chr20:33080402 | chr20:35176435 | ENST00000374864 | + | 23 | 25 | 1_115 | 805 | 863.0 | Domain | C2 |
| Hgene | ITCH | chr20:33080402 | chr20:35176435 | ENST00000374864 | + | 23 | 25 | 326_359 | 805 | 863.0 | Domain | WW 1 |
| Hgene | ITCH | chr20:33080402 | chr20:35176435 | ENST00000374864 | + | 23 | 25 | 358_391 | 805 | 863.0 | Domain | WW 2 |
| Hgene | ITCH | chr20:33080402 | chr20:35176435 | ENST00000374864 | + | 23 | 25 | 438_471 | 805 | 863.0 | Domain | WW 3 |
| Hgene | ITCH | chr20:33080402 | chr20:35176435 | ENST00000374864 | + | 23 | 25 | 478_511 | 805 | 863.0 | Domain | WW 4 |
| Hgene | ITCH | chr20:33080402 | chr20:35176435 | ENST00000535650 | + | 22 | 24 | 1_115 | 695 | 753.0 | Domain | C2 |
| Hgene | ITCH | chr20:33080402 | chr20:35176435 | ENST00000535650 | + | 22 | 24 | 326_359 | 695 | 753.0 | Domain | WW 1 |
| Hgene | ITCH | chr20:33080402 | chr20:35176435 | ENST00000535650 | + | 22 | 24 | 358_391 | 695 | 753.0 | Domain | WW 2 |
| Hgene | ITCH | chr20:33080402 | chr20:35176435 | ENST00000535650 | + | 22 | 24 | 438_471 | 695 | 753.0 | Domain | WW 3 |
| Hgene | ITCH | chr20:33080402 | chr20:35176435 | ENST00000535650 | + | 22 | 24 | 478_511 | 695 | 753.0 | Domain | WW 4 |
| Hgene | ITCH | chr20:33080402 | chr20:35176435 | ENST00000262650 | + | 24 | 26 | 574_583 | 846 | 904.0 | Region | MAP kinase docking site |
| Hgene | ITCH | chr20:33080402 | chr20:35176435 | ENST00000374864 | + | 23 | 25 | 574_583 | 805 | 863.0 | Region | MAP kinase docking site |
| Hgene | ITCH | chr20:33080402 | chr20:35176435 | ENST00000535650 | + | 22 | 24 | 574_583 | 695 | 753.0 | Region | MAP kinase docking site |
| Tgene | MYL9 | chr20:33080402 | chr20:35176435 | ENST00000346786 | 0 | 3 | 42_53 | 0 | 119.0 | Calcium binding | . | |
| Tgene | MYL9 | chr20:33080402 | chr20:35176435 | ENST00000279022 | 1 | 4 | 134_169 | 61 | 173.0 | Domain | EF-hand 3 | |
| Tgene | MYL9 | chr20:33080402 | chr20:35176435 | ENST00000279022 | 1 | 4 | 98_133 | 61 | 173.0 | Domain | EF-hand 2 | |
| Tgene | MYL9 | chr20:33080402 | chr20:35176435 | ENST00000346786 | 0 | 3 | 134_169 | 0 | 119.0 | Domain | EF-hand 3 | |
| Tgene | MYL9 | chr20:33080402 | chr20:35176435 | ENST00000346786 | 0 | 3 | 29_64 | 0 | 119.0 | Domain | EF-hand 1 | |
| Tgene | MYL9 | chr20:33080402 | chr20:35176435 | ENST00000346786 | 0 | 3 | 98_133 | 0 | 119.0 | Domain | EF-hand 2 |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | ITCH | chr20:33080402 | chr20:35176435 | ENST00000262650 | + | 24 | 26 | 569_903 | 846 | 904.0 | Domain | HECT |
| Hgene | ITCH | chr20:33080402 | chr20:35176435 | ENST00000374864 | + | 23 | 25 | 569_903 | 805 | 863.0 | Domain | HECT |
| Hgene | ITCH | chr20:33080402 | chr20:35176435 | ENST00000535650 | + | 22 | 24 | 569_903 | 695 | 753.0 | Domain | HECT |
| Tgene | MYL9 | chr20:33080402 | chr20:35176435 | ENST00000279022 | 1 | 4 | 42_53 | 61 | 173.0 | Calcium binding | . | |
| Tgene | MYL9 | chr20:33080402 | chr20:35176435 | ENST00000279022 | 1 | 4 | 29_64 | 61 | 173.0 | Domain | EF-hand 1 |
Top |
Fusion Gene Sequence for ITCH-MYL9 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >40358_40358_1_ITCH-MYL9_ITCH_chr20_33080402_ENST00000262650_MYL9_chr20_35176435_ENST00000279022_length(transcript)=3586nt_BP=2675nt GTCTAGGGACTGAGGAGTCGCCGCCGCCCCGAGTCCCGGTACCATGCATTTCACGGTGGCCTTGTGGAGACAACGCCTTAACCCAAGGAA GTGACTCAAACTGTGAGAACTTCAGGTTTTCCAACCTATTGGTGGTATGTCTGACAGTGGATCACAACTTGGTTCAATGGGTAGCCTCAC CATGAAATCACAGCTTCAGATCACTGTCATCTCAGCAAAACTTAAGGAAAATAAGAAGAATTGGTTTGGACCAAGTCCTTACGTAGAGGT CACAGTAGATGGACAGTCAAAGAAGACAGAAAAATGCAACAACACAAACAGTCCCAAGTGGAAGCAACCCCTTACAGTTATCGTTACCCC TGTGAGTAAATTACATTTTCGTGTGTGGAGTCACCAGACACTGAAATCTGATGTTTTGTTGGGAACTGCTGCATTAGATATTTATGAAAC ATTAAAGTCAAACAATATGAAACTTGAAGAAGTAGTTGTGACTTTGCAGCTTGGAGGTGACAAAGAGCCAACAGAGACAATAGGAGACTT GTCAATTTGTCTTGATGGGCTACAGTTAGAGTCTGAAGTTGTTACCAATGGTGAAACTACATGTTCAGAAAATGGGGTTTCGCTCTGTTT GCCCAGGCTGGAGTGCAATAGTGCGATCTCGGCTCACTGCAACCTCTGCCTCCCGGGTTTAAGTGATTCTCCTATCTCAGCCTCCCGAGT AGCTGGGTTTACAGGTGCTTCTCAGAATGATGATGGCTCCAGATCCAAGGATGAAACAAGAGTGAGCACAAATGGATCAGATGACCCTGA AGATGCAGGAGCTGGTGAAAATAGGAGAGTCAGTGGGAATAATTCTCCATCACTCTCAAATGGTGGTTTTAAACCTTCTAGACCTCCAAG ACCTTCACGACCACCACCACCCACCCCACGTAGACCAGCATCTGTCAATGGTTCACCATCTGCCACTTCTGAAAGTGATGGGTCTAGTAC AGGCTCTCTGCCGCCGACAAATACAAATACAAATACATCTGAAGGAGCAACATCTGGATTAATAATTCCTCTTACTATATCTGGAGGCTC AGGCCCTAGGCCATTAAATCCTGTAACTCAAGCTCCCTTGCCACCTGGTTGGGAGCAGAGAGTGGACCAGCACGGGCGAGTTTACTATGT AGATCATGTTGAGAAAAGAACAACATGGGATAGACCAGAACCTCTACCTCCTGGCTGGGAACGGCGGGTTGACAACATGGGACGTATTTA TTATGTTGACCATTTCACAAGAACAACAACGTGGCAGAGGCCAACACTGGAATCCGTCCGGAACTATGAACAATGGCAGCTACAGCGTAG TCAGCTTCAAGGAGCAATGCAGCAGTTTAACCAGAGATTCATTTATGGGAATCAAGATTTATTTGCTACATCACAAAGTAAAGAATTTGA TCCTCTTGGTCCATTGCCACCTGGATGGGAGAAGAGAACAGACAGCAATGGCAGAGTATATTTCGTCAACCACAACACACGAATTACACA ATGGGAAGACCCCAGAAGTCAAGGTCAATTAAATGAAAAGCCCTTACCTGAAGGTTGGGAAATGAGATTCACAGTGGATGGAATTCCATA TTTTGTGGACCACAATAGAAGAACTACCACCTATATAGATCCCCGCACAGGAAAATCTGCCCTAGACAATGGACCTCAGATAGCCTATGT TCGGGACTTCAAAGCAAAGGTTCAGTATTTCCGGTTCTGGTGTCAGCAACTGGCCATGCCACAGCACATAAAGATTACAGTGACAAGAAA AACATTGTTTGAGGATTCCTTTCAACAGATAATGAGCTTCAGTCCCCAAGATCTGCGAAGACGTTTGTGGGTGATTTTTCCAGGAGAAGA AGGTTTAGATTATGGAGGTGTAGCAAGAGAATGGTTCTTTCTTTTGTCACATGAAGTGTTGAACCCAATGTATTGCCTGTTTGAATATGC AGGGAAGGATAACTACTGCTTGCAGATAAACCCCGCTTCTTACATCAATCCAGATCACCTGAAATATTTTCGTTTTATTGGCAGATTTAT TGCCATGGCTCTGTTCCATGGGAAATTCATAGACACGGGTTTTTCTTTACCATTCTATAAGCGTATCTTGAACAAACCAGTTGGACTCAA GGATTTAGAATCTATTGATCCAGAATTTTACAATTCTCTCATCTGGGTTAAGGAAAACAATATTGAGGAATGTGATTTGGAAATGTACTT CTCCGTTGACAAAGAAATTCTAGGTGAAATTAAGAGTCATGATCTGAAACCTAATGGTGGCAATATTCTTGTAACAGAAGAAAATAAAGA GGAATACATCAGAATGGTAGCTGAGTGGAGGTTGTCTCGAGGTGTTGAAGAACAGACACAAGCTTTCTTTGAAGGCTTTAATGAAATTCT TCCCCAGCAATATTTGCAATACTTTGATGCAAAGGAATTAGAGGTCCTTTTATGTGGAATGCAAGAGATTGATTTGAATGACTGGCAAAG ACATGCCATCTACCGTCATTATGCAAGGACCAGCAAACAAATCATGTGGTTTTGGCAGTTTGTTAAAGAAATTGATAATGAGAAGAGAAT GAGACTTCTGCAGTTTGTTACTGGAACCTGCCGATTGCCAGTAGGAGGATTTGCTGATCTCATGGGGAAGAACCCCACAGACGAATACCT GGAGGGCATGATGAGCGAGGCCCCGGGGCCCATCAACTTCACCATGTTCCTCACCATGTTTGGGGAGAAGCTGAACGGCACGGACCCCGA GGATGTGATTCGCAACGCCTTTGCCTGCTTCGACGAGGAAGCCTCAGGTTTCATCCATGAGGACCACCTCCGGGAGCTGCTCACCACCAT GGGTGACCGCTTCACAGATGAGGAAGTGGACGAGATGTACCGGGAGGCACCCATTGATAAGAAAGGCAACTTCAACTACGTGGAGTTCAC CCGCATCCTCAAACATGGCGCCAAGGATAAAGACGACTAGGCCACCCCAGCCCCCTGACACCCCAGCCCCCGCCAGTCACCCCTCCCCGC ACACACCCGTCCATACCAGCTCCCTGCCCATGACCCTCGCTCAGGGATCCCCCTTTGAGGGGTTAGGGTCCCAGTTCCCAGTGGAAGAAA CAGGCCAGGAGAAGTGCGTGCCGAGCTGAGGCAGATGTTCCCACAGTGACCCCAGAGCCCTGGGCTATAGTCTCTGACCCCTCCAAGGAA AGACCACCTTCTGGGGACATGGGCTGGAGGGCAGGACCTAGAGGCACCAAGGGAAGGCCCCATTCCGGGGCTGTTCCCCGAGGAGGAAGG GAAGGGGCTCTGTGTGCCCCCCAGGAGGAAGAGGCCCTGAGTCCTGGGATCAGACACCCCTTCACGTGTATCCCCACACAAATGCAAGCT CACCAAGGTCCCCTCTCAGTCCCCTTCCCTACACCCTGACCGGCCACTGCCGCACACCCACCCAGAGCACGCCACCCGCCATGGGAGTGT >40358_40358_1_ITCH-MYL9_ITCH_chr20_33080402_ENST00000262650_MYL9_chr20_35176435_ENST00000279022_length(amino acids)=957AA_BP=159 MSDSGSQLGSMGSLTMKSQLQITVISAKLKENKKNWFGPSPYVEVTVDGQSKKTEKCNNTNSPKWKQPLTVIVTPVSKLHFRVWSHQTLK SDVLLGTAALDIYETLKSNNMKLEEVVVTLQLGGDKEPTETIGDLSICLDGLQLESEVVTNGETTCSENGVSLCLPRLECNSAISAHCNL CLPGLSDSPISASRVAGFTGASQNDDGSRSKDETRVSTNGSDDPEDAGAGENRRVSGNNSPSLSNGGFKPSRPPRPSRPPPPTPRRPASV NGSPSATSESDGSSTGSLPPTNTNTNTSEGATSGLIIPLTISGGSGPRPLNPVTQAPLPPGWEQRVDQHGRVYYVDHVEKRTTWDRPEPL PPGWERRVDNMGRIYYVDHFTRTTTWQRPTLESVRNYEQWQLQRSQLQGAMQQFNQRFIYGNQDLFATSQSKEFDPLGPLPPGWEKRTDS NGRVYFVNHNTRITQWEDPRSQGQLNEKPLPEGWEMRFTVDGIPYFVDHNRRTTTYIDPRTGKSALDNGPQIAYVRDFKAKVQYFRFWCQ QLAMPQHIKITVTRKTLFEDSFQQIMSFSPQDLRRRLWVIFPGEEGLDYGGVAREWFFLLSHEVLNPMYCLFEYAGKDNYCLQINPASYI NPDHLKYFRFIGRFIAMALFHGKFIDTGFSLPFYKRILNKPVGLKDLESIDPEFYNSLIWVKENNIEECDLEMYFSVDKEILGEIKSHDL KPNGGNILVTEENKEEYIRMVAEWRLSRGVEEQTQAFFEGFNEILPQQYLQYFDAKELEVLLCGMQEIDLNDWQRHAIYRHYARTSKQIM WFWQFVKEIDNEKRMRLLQFVTGTCRLPVGGFADLMGKNPTDEYLEGMMSEAPGPINFTMFLTMFGEKLNGTDPEDVIRNAFACFDEEAS -------------------------------------------------------------- >40358_40358_2_ITCH-MYL9_ITCH_chr20_33080402_ENST00000374864_MYL9_chr20_35176435_ENST00000279022_length(transcript)=3540nt_BP=2629nt GCGCTCAGGCGCCTGCGCTTTAGCTGCTAACTCGCTCTAGTAGCTGTGGTCGGGGCTCGGGACCGGCTGCCATCTTAGTCTAGGGACTGA GGAGTCGCCGCCGCCCCGAGTCCCGGTACCATGCATTTCACGGTGGCCTTGTGGAGACAACGCCTTAACCCAAGGAAGTGACTCAAACTG TGAGAACTTCAGGTTTTCCAACCTATTGGTGGTATGTCTGACAGTGGATCACAACTTGGTTCAATGGGTAGCCTCACCATGAAATCACAG CTTCAGATCACTGTCATCTCAGCAAAACTTAAGGAAAATAAGAAGAATTGGTTTGGACCAAGTCCTTACGTAGAGGTCACAGTAGATGGA CAGTCAAAGAAGACAGAAAAATGCAACAACACAAACAGTCCCAAGTGGAAGCAACCCCTTACAGTTATCGTTACCCCTGTGAGTAAATTA CATTTTCGTGTGTGGAGTCACCAGACACTGAAATCTGATGTTTTGTTGGGAACTGCTGCATTAGATATTTATGAAACATTAAAGTCAAAC AATATGAAACTTGAAGAAGTAGTTGTGACTTTGCAGCTTGGAGGTGACAAAGAGCCAACAGAGACAATAGGAGACTTGTCAATTTGTCTT GATGGGCTACAGTTAGAGTCTGAAGTTGTTACCAATGGTGAAACTACATGTTCAGAAAGTGCTTCTCAGAATGATGATGGCTCCAGATCC AAGGATGAAACAAGAGTGAGCACAAATGGATCAGATGACCCTGAAGATGCAGGAGCTGGTGAAAATAGGAGAGTCAGTGGGAATAATTCT CCATCACTCTCAAATGGTGGTTTTAAACCTTCTAGACCTCCAAGACCTTCACGACCACCACCACCCACCCCACGTAGACCAGCATCTGTC AATGGTTCACCATCTGCCACTTCTGAAAGTGATGGGTCTAGTACAGGCTCTCTGCCGCCGACAAATACAAATACAAATACATCTGAAGGA GCAACATCTGGATTAATAATTCCTCTTACTATATCTGGAGGCTCAGGCCCTAGGCCATTAAATCCTGTAACTCAAGCTCCCTTGCCACCT GGTTGGGAGCAGAGAGTGGACCAGCACGGGCGAGTTTACTATGTAGATCATGTTGAGAAAAGAACAACATGGGATAGACCAGAACCTCTA CCTCCTGGCTGGGAACGGCGGGTTGACAACATGGGACGTATTTATTATGTTGACCATTTCACAAGAACAACAACGTGGCAGAGGCCAACA CTGGAATCCGTCCGGAACTATGAACAATGGCAGCTACAGCGTAGTCAGCTTCAAGGAGCAATGCAGCAGTTTAACCAGAGATTCATTTAT GGGAATCAAGATTTATTTGCTACATCACAAAGTAAAGAATTTGATCCTCTTGGTCCATTGCCACCTGGATGGGAGAAGAGAACAGACAGC AATGGCAGAGTATATTTCGTCAACCACAACACACGAATTACACAATGGGAAGACCCCAGAAGTCAAGGTCAATTAAATGAAAAGCCCTTA CCTGAAGGTTGGGAAATGAGATTCACAGTGGATGGAATTCCATATTTTGTGGACCACAATAGAAGAACTACCACCTATATAGATCCCCGC ACAGGAAAATCTGCCCTAGACAATGGACCTCAGATAGCCTATGTTCGGGACTTCAAAGCAAAGGTTCAGTATTTCCGGTTCTGGTGTCAG CAACTGGCCATGCCACAGCACATAAAGATTACAGTGACAAGAAAAACATTGTTTGAGGATTCCTTTCAACAGATAATGAGCTTCAGTCCC CAAGATCTGCGAAGACGTTTGTGGGTGATTTTTCCAGGAGAAGAAGGTTTAGATTATGGAGGTGTAGCAAGAGAATGGTTCTTTCTTTTG TCACATGAAGTGTTGAACCCAATGTATTGCCTGTTTGAATATGCAGGGAAGGATAACTACTGCTTGCAGATAAACCCCGCTTCTTACATC AATCCAGATCACCTGAAATATTTTCGTTTTATTGGCAGATTTATTGCCATGGCTCTGTTCCATGGGAAATTCATAGACACGGGTTTTTCT TTACCATTCTATAAGCGTATCTTGAACAAACCAGTTGGACTCAAGGATTTAGAATCTATTGATCCAGAATTTTACAATTCTCTCATCTGG GTTAAGGAAAACAATATTGAGGAATGTGATTTGGAAATGTACTTCTCCGTTGACAAAGAAATTCTAGGTGAAATTAAGAGTCATGATCTG AAACCTAATGGTGGCAATATTCTTGTAACAGAAGAAAATAAAGAGGAATACATCAGAATGGTAGCTGAGTGGAGGTTGTCTCGAGGTGTT GAAGAACAGACACAAGCTTTCTTTGAAGGCTTTAATGAAATTCTTCCCCAGCAATATTTGCAATACTTTGATGCAAAGGAATTAGAGGTC CTTTTATGTGGAATGCAAGAGATTGATTTGAATGACTGGCAAAGACATGCCATCTACCGTCATTATGCAAGGACCAGCAAACAAATCATG TGGTTTTGGCAGTTTGTTAAAGAAATTGATAATGAGAAGAGAATGAGACTTCTGCAGTTTGTTACTGGAACCTGCCGATTGCCAGTAGGA GGATTTGCTGATCTCATGGGGAAGAACCCCACAGACGAATACCTGGAGGGCATGATGAGCGAGGCCCCGGGGCCCATCAACTTCACCATG TTCCTCACCATGTTTGGGGAGAAGCTGAACGGCACGGACCCCGAGGATGTGATTCGCAACGCCTTTGCCTGCTTCGACGAGGAAGCCTCA GGTTTCATCCATGAGGACCACCTCCGGGAGCTGCTCACCACCATGGGTGACCGCTTCACAGATGAGGAAGTGGACGAGATGTACCGGGAG GCACCCATTGATAAGAAAGGCAACTTCAACTACGTGGAGTTCACCCGCATCCTCAAACATGGCGCCAAGGATAAAGACGACTAGGCCACC CCAGCCCCCTGACACCCCAGCCCCCGCCAGTCACCCCTCCCCGCACACACCCGTCCATACCAGCTCCCTGCCCATGACCCTCGCTCAGGG ATCCCCCTTTGAGGGGTTAGGGTCCCAGTTCCCAGTGGAAGAAACAGGCCAGGAGAAGTGCGTGCCGAGCTGAGGCAGATGTTCCCACAG TGACCCCAGAGCCCTGGGCTATAGTCTCTGACCCCTCCAAGGAAAGACCACCTTCTGGGGACATGGGCTGGAGGGCAGGACCTAGAGGCA CCAAGGGAAGGCCCCATTCCGGGGCTGTTCCCCGAGGAGGAAGGGAAGGGGCTCTGTGTGCCCCCCAGGAGGAAGAGGCCCTGAGTCCTG GGATCAGACACCCCTTCACGTGTATCCCCACACAAATGCAAGCTCACCAAGGTCCCCTCTCAGTCCCCTTCCCTACACCCTGACCGGCCA CTGCCGCACACCCACCCAGAGCACGCCACCCGCCATGGGAGTGTGCTCAGGAGTCGCGGGCAGCGTGGACATCTGTCCCAGAGGGGGCAG >40358_40358_2_ITCH-MYL9_ITCH_chr20_33080402_ENST00000374864_MYL9_chr20_35176435_ENST00000279022_length(amino acids)=916AA_BP=805 MSDSGSQLGSMGSLTMKSQLQITVISAKLKENKKNWFGPSPYVEVTVDGQSKKTEKCNNTNSPKWKQPLTVIVTPVSKLHFRVWSHQTLK SDVLLGTAALDIYETLKSNNMKLEEVVVTLQLGGDKEPTETIGDLSICLDGLQLESEVVTNGETTCSESASQNDDGSRSKDETRVSTNGS DDPEDAGAGENRRVSGNNSPSLSNGGFKPSRPPRPSRPPPPTPRRPASVNGSPSATSESDGSSTGSLPPTNTNTNTSEGATSGLIIPLTI SGGSGPRPLNPVTQAPLPPGWEQRVDQHGRVYYVDHVEKRTTWDRPEPLPPGWERRVDNMGRIYYVDHFTRTTTWQRPTLESVRNYEQWQ LQRSQLQGAMQQFNQRFIYGNQDLFATSQSKEFDPLGPLPPGWEKRTDSNGRVYFVNHNTRITQWEDPRSQGQLNEKPLPEGWEMRFTVD GIPYFVDHNRRTTTYIDPRTGKSALDNGPQIAYVRDFKAKVQYFRFWCQQLAMPQHIKITVTRKTLFEDSFQQIMSFSPQDLRRRLWVIF PGEEGLDYGGVAREWFFLLSHEVLNPMYCLFEYAGKDNYCLQINPASYINPDHLKYFRFIGRFIAMALFHGKFIDTGFSLPFYKRILNKP VGLKDLESIDPEFYNSLIWVKENNIEECDLEMYFSVDKEILGEIKSHDLKPNGGNILVTEENKEEYIRMVAEWRLSRGVEEQTQAFFEGF NEILPQQYLQYFDAKELEVLLCGMQEIDLNDWQRHAIYRHYARTSKQIMWFWQFVKEIDNEKRMRLLQFVTGTCRLPVGGFADLMGKNPT DEYLEGMMSEAPGPINFTMFLTMFGEKLNGTDPEDVIRNAFACFDEEASGFIHEDHLRELLTTMGDRFTDEEVDEMYREAPIDKKGNFNY -------------------------------------------------------------- >40358_40358_3_ITCH-MYL9_ITCH_chr20_33080402_ENST00000535650_MYL9_chr20_35176435_ENST00000279022_length(transcript)=3357nt_BP=2446nt AGCTGTGGTCGGGGCTCGGGACCGGCTGCCATCTTAGTCTAGGGACTGAGGAGTCGCCGCCGCCCCGAGTCCCGGTACCATGCATTTCAC GGTGGCCTTGTGGAGACAACGCCTTAACCCAAGGAAGTGACTCAAACTGTGAGAACTTCAGGTTTTCCAACCTATTGGTGGTATGTCTGA CAGTGGATCACAACTTGGTTCAATGGGTAGCCTCACCATGAAATCACAGCTTCAGATCACTGTATCGTTACCCCTGTGAGTAAATTACAT TTTCGTGTGTGGAGTCACCAGACACTGAAATCTGATGTTTTGTTGGGAACTGCTGCATTAGATATTTATGAAACATTAAAGTCAAACAAT ATGAAACTTGAAGAAGTAGTTGTGACTTTGCAGCTTGGAGGTGACAAAGAGCCAACAGAGACAATAGGAGACTTGTCAATTTGTCTTGAT GGGCTACAGTTAGAGTCTGAAGTTGTTACCAATGGTGAAACTACATGTTCAGAAAGTGCTTCTCAGAATGATGATGGCTCCAGATCCAAG GATGAAACAAGAGTGAGCACAAATGGATCAGATGACCCTGAAGATGCAGGAGCTGGTGAAAATAGGAGAGTCAGTGGGAATAATTCTCCA TCACTCTCAAATGGTGGTTTTAAACCTTCTAGACCTCCAAGACCTTCACGACCACCACCACCCACCCCACGTAGACCAGCATCTGTCAAT GGTTCACCATCTGCCACTTCTGAAAGTGATGGGTCTAGTACAGGCTCTCTGCCGCCGACAAATACAAATACAAATACATCTGAAGGAGCA ACATCTGGATTAATAATTCCTCTTACTATATCTGGAGGCTCAGGCCCTAGGCCATTAAATCCTGTAACTCAAGCTCCCTTGCCACCTGGT TGGGAGCAGAGAGTGGACCAGCACGGGCGAGTTTACTATGTAGATCATGTTGAGAAAAGAACAACATGGGATAGACCAGAACCTCTACCT CCTGGCTGGGAACGGCGGGTTGACAACATGGGACGTATTTATTATGTTGACCATTTCACAAGAACAACAACGTGGCAGAGGCCAACACTG GAATCCGTCCGGAACTATGAACAATGGCAGCTACAGCGTAGTCAGCTTCAAGGAGCAATGCAGCAGTTTAACCAGAGATTCATTTATGGG AATCAAGATTTATTTGCTACATCACAAAGTAAAGAATTTGATCCTCTTGGTCCATTGCCACCTGGATGGGAGAAGAGAACAGACAGCAAT GGCAGAGTATATTTCGTCAACCACAACACACGAATTACACAATGGGAAGACCCCAGAAGTCAAGGTCAATTAAATGAAAAGCCCTTACCT GAAGGTTGGGAAATGAGATTCACAGTGGATGGAATTCCATATTTTGTGGACCACAATAGAAGAACTACCACCTATATAGATCCCCGCACA GGAAAATCTGCCCTAGACAATGGACCTCAGATAGCCTATGTTCGGGACTTCAAAGCAAAGGTTCAGTATTTCCGGTTCTGGTGTCAGCAA CTGGCCATGCCACAGCACATAAAGATTACAGTGACAAGAAAAACATTGTTTGAGGATTCCTTTCAACAGATAATGAGCTTCAGTCCCCAA GATCTGCGAAGACGTTTGTGGGTGATTTTTCCAGGAGAAGAAGGTTTAGATTATGGAGGTGTAGCAAGAGAATGGTTCTTTCTTTTGTCA CATGAAGTGTTGAACCCAATGTATTGCCTGTTTGAATATGCAGGGAAGGATAACTACTGCTTGCAGATAAACCCCGCTTCTTACATCAAT CCAGATCACCTGAAATATTTTCGTTTTATTGGCAGATTTATTGCCATGGCTCTGTTCCATGGGAAATTCATAGACACGGGTTTTTCTTTA CCATTCTATAAGCGTATCTTGAACAAACCAGTTGGACTCAAGGATTTAGAATCTATTGATCCAGAATTTTACAATTCTCTCATCTGGGTT AAGGAAAACAATATTGAGGAATGTGATTTGGAAATGTACTTCTCCGTTGACAAAGAAATTCTAGGTGAAATTAAGAGTCATGATCTGAAA CCTAATGGTGGCAATATTCTTGTAACAGAAGAAAATAAAGAGGAATACATCAGAATGGTAGCTGAGTGGAGGTTGTCTCGAGGTGTTGAA GAACAGACACAAGCTTTCTTTGAAGGCTTTAATGAAATTCTTCCCCAGCAATATTTGCAATACTTTGATGCAAAGGAATTAGAGGTCCTT TTATGTGGAATGCAAGAGATTGATTTGAATGACTGGCAAAGACATGCCATCTACCGTCATTATGCAAGGACCAGCAAACAAATCATGTGG TTTTGGCAGTTTGTTAAAGAAATTGATAATGAGAAGAGAATGAGACTTCTGCAGTTTGTTACTGGAACCTGCCGATTGCCAGTAGGAGGA TTTGCTGATCTCATGGGGAAGAACCCCACAGACGAATACCTGGAGGGCATGATGAGCGAGGCCCCGGGGCCCATCAACTTCACCATGTTC CTCACCATGTTTGGGGAGAAGCTGAACGGCACGGACCCCGAGGATGTGATTCGCAACGCCTTTGCCTGCTTCGACGAGGAAGCCTCAGGT TTCATCCATGAGGACCACCTCCGGGAGCTGCTCACCACCATGGGTGACCGCTTCACAGATGAGGAAGTGGACGAGATGTACCGGGAGGCA CCCATTGATAAGAAAGGCAACTTCAACTACGTGGAGTTCACCCGCATCCTCAAACATGGCGCCAAGGATAAAGACGACTAGGCCACCCCA GCCCCCTGACACCCCAGCCCCCGCCAGTCACCCCTCCCCGCACACACCCGTCCATACCAGCTCCCTGCCCATGACCCTCGCTCAGGGATC CCCCTTTGAGGGGTTAGGGTCCCAGTTCCCAGTGGAAGAAACAGGCCAGGAGAAGTGCGTGCCGAGCTGAGGCAGATGTTCCCACAGTGA CCCCAGAGCCCTGGGCTATAGTCTCTGACCCCTCCAAGGAAAGACCACCTTCTGGGGACATGGGCTGGAGGGCAGGACCTAGAGGCACCA AGGGAAGGCCCCATTCCGGGGCTGTTCCCCGAGGAGGAAGGGAAGGGGCTCTGTGTGCCCCCCAGGAGGAAGAGGCCCTGAGTCCTGGGA TCAGACACCCCTTCACGTGTATCCCCACACAAATGCAAGCTCACCAAGGTCCCCTCTCAGTCCCCTTCCCTACACCCTGACCGGCCACTG CCGCACACCCACCCAGAGCACGCCACCCGCCATGGGAGTGTGCTCAGGAGTCGCGGGCAGCGTGGACATCTGTCCCAGAGGGGGCAGAAT >40358_40358_3_ITCH-MYL9_ITCH_chr20_33080402_ENST00000535650_MYL9_chr20_35176435_ENST00000279022_length(amino acids)=828AA_BP=717 MKSDVLLGTAALDIYETLKSNNMKLEEVVVTLQLGGDKEPTETIGDLSICLDGLQLESEVVTNGETTCSESASQNDDGSRSKDETRVSTN GSDDPEDAGAGENRRVSGNNSPSLSNGGFKPSRPPRPSRPPPPTPRRPASVNGSPSATSESDGSSTGSLPPTNTNTNTSEGATSGLIIPL TISGGSGPRPLNPVTQAPLPPGWEQRVDQHGRVYYVDHVEKRTTWDRPEPLPPGWERRVDNMGRIYYVDHFTRTTTWQRPTLESVRNYEQ WQLQRSQLQGAMQQFNQRFIYGNQDLFATSQSKEFDPLGPLPPGWEKRTDSNGRVYFVNHNTRITQWEDPRSQGQLNEKPLPEGWEMRFT VDGIPYFVDHNRRTTTYIDPRTGKSALDNGPQIAYVRDFKAKVQYFRFWCQQLAMPQHIKITVTRKTLFEDSFQQIMSFSPQDLRRRLWV IFPGEEGLDYGGVAREWFFLLSHEVLNPMYCLFEYAGKDNYCLQINPASYINPDHLKYFRFIGRFIAMALFHGKFIDTGFSLPFYKRILN KPVGLKDLESIDPEFYNSLIWVKENNIEECDLEMYFSVDKEILGEIKSHDLKPNGGNILVTEENKEEYIRMVAEWRLSRGVEEQTQAFFE GFNEILPQQYLQYFDAKELEVLLCGMQEIDLNDWQRHAIYRHYARTSKQIMWFWQFVKEIDNEKRMRLLQFVTGTCRLPVGGFADLMGKN PTDEYLEGMMSEAPGPINFTMFLTMFGEKLNGTDPEDVIRNAFACFDEEASGFIHEDHLRELLTTMGDRFTDEEVDEMYREAPIDKKGNF -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for ITCH-MYL9 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for ITCH-MYL9 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for ITCH-MYL9 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
