
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:ITGB1-DYM (FusionGDB2 ID:40507) |
Fusion Gene Summary for ITGB1-DYM |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: ITGB1-DYM | Fusion gene ID: 40507 | Hgene | Tgene | Gene symbol | ITGB1 | DYM | Gene ID | 3688 | 54808 |
| Gene name | integrin subunit beta 1 | dymeclin | |
| Synonyms | CD29|FNRB|GPIIA|MDF2|MSK12|VLA-BETA|VLAB | DMC|SMC | |
| Cytomap | 10p11.22 | 18q21.1 | |
| Type of gene | protein-coding | protein-coding | |
| Description | integrin beta-1glycoprotein IIaintegrin VLA-4 beta subunitintegrin beta 1integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen CD29 includes MDF2, MSK12)very late activation protein, beta polypeptide | dymeclindyggve-Melchior-Clausen syndrome protein | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | P05556 | Q7RTS9 | |
| Ensembl transtripts involved in fusion gene | ENST00000302278, ENST00000374956, ENST00000396033, ENST00000423113, ENST00000484088, | ENST00000578396, ENST00000584977, ENST00000269445, ENST00000442713, | |
| Fusion gene scores | * DoF score | 10 X 7 X 8=560 | 19 X 11 X 13=2717 |
| # samples | 11 | 24 | |
| ** MAII score | log2(11/560*10)=-2.34792330342031 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(24/2717*10)=-3.50090825461346 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: ITGB1 [Title/Abstract] AND DYM [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | ITGB1(33214799)-DYM(46798673), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | ITGB1-DYM seems lost the major protein functional domain in Hgene partner, which is a IUPHAR drug target due to the frame-shifted ORF. ITGB1-DYM seems lost the major protein functional domain in Hgene partner, which is a tumor suppressor due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | ITGB1 | GO:0007155 | cell adhesion | 19703720 |
| Hgene | ITGB1 | GO:0007159 | leukocyte cell-cell adhesion | 1715889 |
| Hgene | ITGB1 | GO:0007229 | integrin-mediated signaling pathway | 31331973 |
| Hgene | ITGB1 | GO:0010710 | regulation of collagen catabolic process | 10455171 |
| Hgene | ITGB1 | GO:0010763 | positive regulation of fibroblast migration | 26763945 |
| Hgene | ITGB1 | GO:0023035 | CD40 signaling pathway | 31331973 |
| Hgene | ITGB1 | GO:0033627 | cell adhesion mediated by integrin | 12807887|17158881 |
| Hgene | ITGB1 | GO:0051897 | positive regulation of protein kinase B signaling | 24044949 |
| Hgene | ITGB1 | GO:0090303 | positive regulation of wound healing | 26763945 |
| Hgene | ITGB1 | GO:1903078 | positive regulation of protein localization to plasma membrane | 10455171 |
| Hgene | ITGB1 | GO:2000273 | positive regulation of signaling receptor activity | 24044949 |
 Fusion gene breakpoints across ITGB1 (5'-gene) Fusion gene breakpoints across ITGB1 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
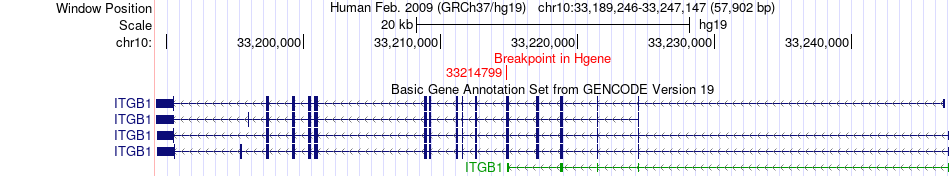 |
 Fusion gene breakpoints across DYM (3'-gene) Fusion gene breakpoints across DYM (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
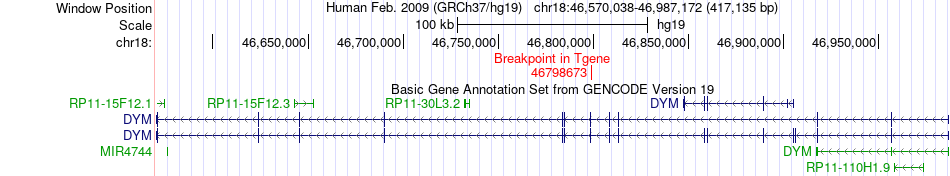 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | BRCA | TCGA-E2-A9RU-01A | ITGB1 | chr10 | 33214799 | - | DYM | chr18 | 46798673 | - |
Top |
Fusion Gene ORF analysis for ITGB1-DYM |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-intron | ENST00000302278 | ENST00000578396 | ITGB1 | chr10 | 33214799 | - | DYM | chr18 | 46798673 | - |
| 5CDS-intron | ENST00000302278 | ENST00000584977 | ITGB1 | chr10 | 33214799 | - | DYM | chr18 | 46798673 | - |
| 5CDS-intron | ENST00000374956 | ENST00000578396 | ITGB1 | chr10 | 33214799 | - | DYM | chr18 | 46798673 | - |
| 5CDS-intron | ENST00000374956 | ENST00000584977 | ITGB1 | chr10 | 33214799 | - | DYM | chr18 | 46798673 | - |
| 5CDS-intron | ENST00000396033 | ENST00000578396 | ITGB1 | chr10 | 33214799 | - | DYM | chr18 | 46798673 | - |
| 5CDS-intron | ENST00000396033 | ENST00000584977 | ITGB1 | chr10 | 33214799 | - | DYM | chr18 | 46798673 | - |
| 5CDS-intron | ENST00000423113 | ENST00000578396 | ITGB1 | chr10 | 33214799 | - | DYM | chr18 | 46798673 | - |
| 5CDS-intron | ENST00000423113 | ENST00000584977 | ITGB1 | chr10 | 33214799 | - | DYM | chr18 | 46798673 | - |
| Frame-shift | ENST00000302278 | ENST00000269445 | ITGB1 | chr10 | 33214799 | - | DYM | chr18 | 46798673 | - |
| Frame-shift | ENST00000302278 | ENST00000442713 | ITGB1 | chr10 | 33214799 | - | DYM | chr18 | 46798673 | - |
| Frame-shift | ENST00000374956 | ENST00000269445 | ITGB1 | chr10 | 33214799 | - | DYM | chr18 | 46798673 | - |
| Frame-shift | ENST00000374956 | ENST00000442713 | ITGB1 | chr10 | 33214799 | - | DYM | chr18 | 46798673 | - |
| Frame-shift | ENST00000423113 | ENST00000269445 | ITGB1 | chr10 | 33214799 | - | DYM | chr18 | 46798673 | - |
| Frame-shift | ENST00000423113 | ENST00000442713 | ITGB1 | chr10 | 33214799 | - | DYM | chr18 | 46798673 | - |
| In-frame | ENST00000396033 | ENST00000269445 | ITGB1 | chr10 | 33214799 | - | DYM | chr18 | 46798673 | - |
| In-frame | ENST00000396033 | ENST00000442713 | ITGB1 | chr10 | 33214799 | - | DYM | chr18 | 46798673 | - |
| intron-3CDS | ENST00000484088 | ENST00000269445 | ITGB1 | chr10 | 33214799 | - | DYM | chr18 | 46798673 | - |
| intron-3CDS | ENST00000484088 | ENST00000442713 | ITGB1 | chr10 | 33214799 | - | DYM | chr18 | 46798673 | - |
| intron-intron | ENST00000484088 | ENST00000578396 | ITGB1 | chr10 | 33214799 | - | DYM | chr18 | 46798673 | - |
| intron-intron | ENST00000484088 | ENST00000584977 | ITGB1 | chr10 | 33214799 | - | DYM | chr18 | 46798673 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000396033 | ITGB1 | chr10 | 33214799 | - | ENST00000442713 | DYM | chr18 | 46798673 | - | 2193 | 922 | 40 | 1806 | 588 |
| ENST00000396033 | ITGB1 | chr10 | 33214799 | - | ENST00000269445 | DYM | chr18 | 46798673 | - | 2042 | 922 | 40 | 1806 | 588 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000396033 | ENST00000442713 | ITGB1 | chr10 | 33214799 | - | DYM | chr18 | 46798673 | - | 0.000485087 | 0.99951494 |
| ENST00000396033 | ENST00000269445 | ITGB1 | chr10 | 33214799 | - | DYM | chr18 | 46798673 | - | 0.000398697 | 0.9996013 |
Top |
Fusion Genomic Features for ITGB1-DYM |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
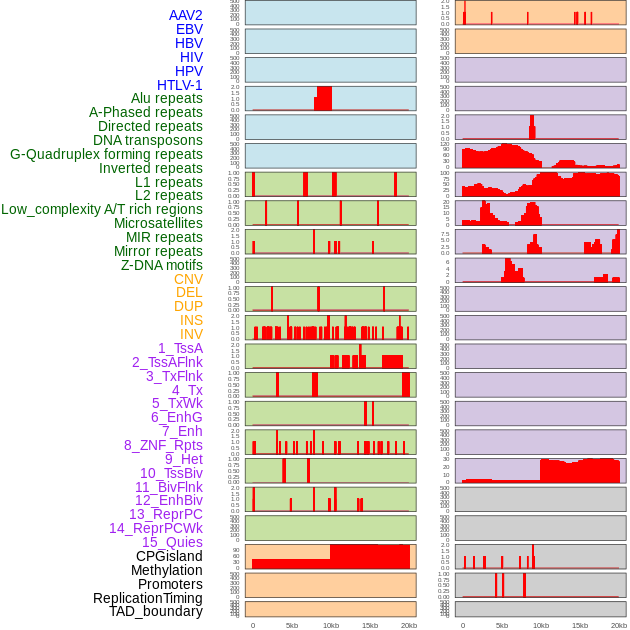 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for ITGB1-DYM |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr10:33214799/chr18:46798673) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| ITGB1 | DYM |
| FUNCTION: Integrins alpha-1/beta-1, alpha-2/beta-1, alpha-10/beta-1 and alpha-11/beta-1 are receptors for collagen. Integrins alpha-1/beta-1 and alpha-2/beta-2 recognize the proline-hydroxylated sequence G-F-P-G-E-R in collagen. Integrins alpha-2/beta-1, alpha-3/beta-1, alpha-4/beta-1, alpha-5/beta-1, alpha-8/beta-1, alpha-10/beta-1, alpha-11/beta-1 and alpha-V/beta-1 are receptors for fibronectin. Alpha-4/beta-1 recognizes one or more domains within the alternatively spliced CS-1 and CS-5 regions of fibronectin. Integrin alpha-5/beta-1 is a receptor for fibrinogen. Integrin alpha-1/beta-1, alpha-2/beta-1, alpha-6/beta-1 and alpha-7/beta-1 are receptors for lamimin. Integrin alpha-6/beta-1 (ITGA6:ITGB1) is present in oocytes and is involved in sperm-egg fusion (By similarity). Integrin alpha-4/beta-1 is a receptor for VCAM1. It recognizes the sequence Q-I-D-S in VCAM1. Integrin alpha-9/beta-1 is a receptor for VCAM1, cytotactin and osteopontin. It recognizes the sequence A-E-I-D-G-I-E-L in cytotactin. Integrin alpha-3/beta-1 is a receptor for epiligrin, thrombospondin and CSPG4. Alpha-3/beta-1 may mediate with LGALS3 the stimulation by CSPG4 of endothelial cells migration. Integrin alpha-V/beta-1 is a receptor for vitronectin. Beta-1 integrins recognize the sequence R-G-D in a wide array of ligands. When associated with alpha-7 integrin, regulates cell adhesion and laminin matrix deposition. Involved in promoting endothelial cell motility and angiogenesis. Involved in osteoblast compaction through the fibronectin fibrillogenesis cell-mediated matrix assembly process and the formation of mineralized bone nodules. May be involved in up-regulation of the activity of kinases such as PKC via binding to KRT1. Together with KRT1 and RACK1, serves as a platform for SRC activation or inactivation. Plays a mechanistic adhesive role during telophase, required for the successful completion of cytokinesis. Integrin alpha-3/beta-1 provides a docking site for FAP (seprase) at invadopodia plasma membranes in a collagen-dependent manner and hence may participate in the adhesion, formation of invadopodia and matrix degradation processes, promoting cell invasion. ITGA4:ITGB1 binds to fractalkine (CX3CL1) and may act as its coreceptor in CX3CR1-dependent fractalkine signaling (PubMed:23125415, PubMed:24789099). ITGA4:ITGB1 and ITGA5:ITGB1 bind to PLA2G2A via a site (site 2) which is distinct from the classical ligand-binding site (site 1) and this induces integrin conformational changes and enhanced ligand binding to site 1 (PubMed:18635536, PubMed:25398877). ITGA5:ITGB1 acts as a receptor for fibrillin-1 (FBN1) and mediates R-G-D-dependent cell adhesion to FBN1 (PubMed:12807887, PubMed:17158881). ITGA5:ITGB1 is a receptor for IL1B and binding is essential for IL1B signaling (PubMed:29030430). ITGA5:ITGB3 is a receptor for soluble CD40LG and is required for CD40/CD40LG signaling (PubMed:31331973). {ECO:0000250|UniProtKB:P09055, ECO:0000269|PubMed:10455171, ECO:0000269|PubMed:12473654, ECO:0000269|PubMed:12807887, ECO:0000269|PubMed:16256741, ECO:0000269|PubMed:17158881, ECO:0000269|PubMed:18635536, ECO:0000269|PubMed:18804435, ECO:0000269|PubMed:19064666, ECO:0000269|PubMed:21768292, ECO:0000269|PubMed:23125415, ECO:0000269|PubMed:24789099, ECO:0000269|PubMed:25398877, ECO:0000269|PubMed:29030430, ECO:0000269|PubMed:31331973, ECO:0000269|PubMed:7523423}.; FUNCTION: [Isoform 2]: Interferes with isoform 1 resulting in a dominant negative effect on cell adhesion and migration (in vitro). {ECO:0000305|PubMed:2249781}.; FUNCTION: [Isoform 5]: Isoform 5 displaces isoform 1 in striated muscles. {ECO:0000250|UniProtKB:P09055}.; FUNCTION: (Microbial infection) Integrin ITGA2:ITGB1 acts as a receptor for Human echoviruses 1 and 8. {ECO:0000269|PubMed:8411387}.; FUNCTION: (Microbial infection) Acts as a receptor for Cytomegalovirus/HHV-5. {ECO:0000269|PubMed:20660204}.; FUNCTION: (Microbial infection) Acts as a receptor for Epstein-Barr virus/HHV-4. {ECO:0000269|PubMed:17945327}.; FUNCTION: (Microbial infection) Integrin ITGA5:ITGB1 acts as a receptor for Human parvovirus B19. {ECO:0000269|PubMed:12907437}.; FUNCTION: (Microbial infection) Integrin ITGA2:ITGB1 acts as a receptor for Human rotavirus. {ECO:0000269|PubMed:12941907}.; FUNCTION: (Microbial infection) Acts as a receptor for Mammalian reovirus. {ECO:0000269|PubMed:16501085}.; FUNCTION: (Microbial infection) In case of HIV-1 infection, integrin ITGA5:ITGB1 binding to extracellular viral Tat protein seems to enhance angiogenesis in Kaposi's sarcoma lesions. {ECO:0000269|PubMed:10397733}. | FUNCTION: Necessary for correct organization of Golgi apparatus. Involved in bone development. {ECO:0000269|PubMed:21280149}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | ITGB1 | chr10:33214799 | chr18:46798673 | ENST00000302278 | - | 6 | 16 | 140_378 | 262 | 799.0 | Domain | Note=VWFA |
| Hgene | ITGB1 | chr10:33214799 | chr18:46798673 | ENST00000374956 | - | 6 | 17 | 140_378 | 262 | 826.0 | Domain | Note=VWFA |
| Hgene | ITGB1 | chr10:33214799 | chr18:46798673 | ENST00000396033 | - | 6 | 16 | 140_378 | 262 | 799.0 | Domain | Note=VWFA |
| Hgene | ITGB1 | chr10:33214799 | chr18:46798673 | ENST00000423113 | - | 5 | 16 | 140_378 | 262 | 1217.0 | Domain | Note=VWFA |
| Hgene | ITGB1 | chr10:33214799 | chr18:46798673 | ENST00000302278 | - | 6 | 16 | 295_314 | 262 | 799.0 | Region | CX3CL1-binding |
| Hgene | ITGB1 | chr10:33214799 | chr18:46798673 | ENST00000302278 | - | 6 | 16 | 466_635 | 262 | 799.0 | Region | Note=Cysteine-rich tandem repeats |
| Hgene | ITGB1 | chr10:33214799 | chr18:46798673 | ENST00000374956 | - | 6 | 17 | 295_314 | 262 | 826.0 | Region | CX3CL1-binding |
| Hgene | ITGB1 | chr10:33214799 | chr18:46798673 | ENST00000374956 | - | 6 | 17 | 466_635 | 262 | 826.0 | Region | Note=Cysteine-rich tandem repeats |
| Hgene | ITGB1 | chr10:33214799 | chr18:46798673 | ENST00000396033 | - | 6 | 16 | 295_314 | 262 | 799.0 | Region | CX3CL1-binding |
| Hgene | ITGB1 | chr10:33214799 | chr18:46798673 | ENST00000396033 | - | 6 | 16 | 466_635 | 262 | 799.0 | Region | Note=Cysteine-rich tandem repeats |
| Hgene | ITGB1 | chr10:33214799 | chr18:46798673 | ENST00000423113 | - | 5 | 16 | 295_314 | 262 | 1217.0 | Region | CX3CL1-binding |
| Hgene | ITGB1 | chr10:33214799 | chr18:46798673 | ENST00000423113 | - | 5 | 16 | 466_635 | 262 | 1217.0 | Region | Note=Cysteine-rich tandem repeats |
| Hgene | ITGB1 | chr10:33214799 | chr18:46798673 | ENST00000302278 | - | 6 | 16 | 466_515 | 262 | 799.0 | Repeat | Note=I |
| Hgene | ITGB1 | chr10:33214799 | chr18:46798673 | ENST00000302278 | - | 6 | 16 | 516_559 | 262 | 799.0 | Repeat | Note=II |
| Hgene | ITGB1 | chr10:33214799 | chr18:46798673 | ENST00000302278 | - | 6 | 16 | 560_598 | 262 | 799.0 | Repeat | Note=III |
| Hgene | ITGB1 | chr10:33214799 | chr18:46798673 | ENST00000302278 | - | 6 | 16 | 599_635 | 262 | 799.0 | Repeat | Note=IV |
| Hgene | ITGB1 | chr10:33214799 | chr18:46798673 | ENST00000374956 | - | 6 | 17 | 466_515 | 262 | 826.0 | Repeat | Note=I |
| Hgene | ITGB1 | chr10:33214799 | chr18:46798673 | ENST00000374956 | - | 6 | 17 | 516_559 | 262 | 826.0 | Repeat | Note=II |
| Hgene | ITGB1 | chr10:33214799 | chr18:46798673 | ENST00000374956 | - | 6 | 17 | 560_598 | 262 | 826.0 | Repeat | Note=III |
| Hgene | ITGB1 | chr10:33214799 | chr18:46798673 | ENST00000374956 | - | 6 | 17 | 599_635 | 262 | 826.0 | Repeat | Note=IV |
| Hgene | ITGB1 | chr10:33214799 | chr18:46798673 | ENST00000396033 | - | 6 | 16 | 466_515 | 262 | 799.0 | Repeat | Note=I |
| Hgene | ITGB1 | chr10:33214799 | chr18:46798673 | ENST00000396033 | - | 6 | 16 | 516_559 | 262 | 799.0 | Repeat | Note=II |
| Hgene | ITGB1 | chr10:33214799 | chr18:46798673 | ENST00000396033 | - | 6 | 16 | 560_598 | 262 | 799.0 | Repeat | Note=III |
| Hgene | ITGB1 | chr10:33214799 | chr18:46798673 | ENST00000396033 | - | 6 | 16 | 599_635 | 262 | 799.0 | Repeat | Note=IV |
| Hgene | ITGB1 | chr10:33214799 | chr18:46798673 | ENST00000423113 | - | 5 | 16 | 466_515 | 262 | 1217.0 | Repeat | Note=I |
| Hgene | ITGB1 | chr10:33214799 | chr18:46798673 | ENST00000423113 | - | 5 | 16 | 516_559 | 262 | 1217.0 | Repeat | Note=II |
| Hgene | ITGB1 | chr10:33214799 | chr18:46798673 | ENST00000423113 | - | 5 | 16 | 560_598 | 262 | 1217.0 | Repeat | Note=III |
| Hgene | ITGB1 | chr10:33214799 | chr18:46798673 | ENST00000423113 | - | 5 | 16 | 599_635 | 262 | 1217.0 | Repeat | Note=IV |
| Hgene | ITGB1 | chr10:33214799 | chr18:46798673 | ENST00000302278 | - | 6 | 16 | 21_728 | 262 | 799.0 | Topological domain | Extracellular |
| Hgene | ITGB1 | chr10:33214799 | chr18:46798673 | ENST00000302278 | - | 6 | 16 | 752_798 | 262 | 799.0 | Topological domain | Cytoplasmic |
| Hgene | ITGB1 | chr10:33214799 | chr18:46798673 | ENST00000374956 | - | 6 | 17 | 21_728 | 262 | 826.0 | Topological domain | Extracellular |
| Hgene | ITGB1 | chr10:33214799 | chr18:46798673 | ENST00000374956 | - | 6 | 17 | 752_798 | 262 | 826.0 | Topological domain | Cytoplasmic |
| Hgene | ITGB1 | chr10:33214799 | chr18:46798673 | ENST00000396033 | - | 6 | 16 | 21_728 | 262 | 799.0 | Topological domain | Extracellular |
| Hgene | ITGB1 | chr10:33214799 | chr18:46798673 | ENST00000396033 | - | 6 | 16 | 752_798 | 262 | 799.0 | Topological domain | Cytoplasmic |
| Hgene | ITGB1 | chr10:33214799 | chr18:46798673 | ENST00000423113 | - | 5 | 16 | 21_728 | 262 | 1217.0 | Topological domain | Extracellular |
| Hgene | ITGB1 | chr10:33214799 | chr18:46798673 | ENST00000423113 | - | 5 | 16 | 752_798 | 262 | 1217.0 | Topological domain | Cytoplasmic |
| Hgene | ITGB1 | chr10:33214799 | chr18:46798673 | ENST00000302278 | - | 6 | 16 | 729_751 | 262 | 799.0 | Transmembrane | Helical |
| Hgene | ITGB1 | chr10:33214799 | chr18:46798673 | ENST00000374956 | - | 6 | 17 | 729_751 | 262 | 826.0 | Transmembrane | Helical |
| Hgene | ITGB1 | chr10:33214799 | chr18:46798673 | ENST00000396033 | - | 6 | 16 | 729_751 | 262 | 799.0 | Transmembrane | Helical |
| Hgene | ITGB1 | chr10:33214799 | chr18:46798673 | ENST00000423113 | - | 5 | 16 | 729_751 | 262 | 1217.0 | Transmembrane | Helical |
Top |
Fusion Gene Sequence for ITGB1-DYM |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >40507_40507_1_ITGB1-DYM_ITGB1_chr10_33214799_ENST00000396033_DYM_chr18_46798673_ENST00000269445_length(transcript)=2042nt_BP=922nt GAGCCAGCCCAGCCGCGTTCCGAACGTGAGGGTCGCCGGCCTGGGCGCTGTCACGTCGGGGCTGCCGGAGCTGCGGGGGACCGGGCCCGA ACGGCCCCTGACACCTGCGGTCTCCCGCCGGGCTGGGCAAGCGCAGATGAATTTACAACCAATTTTCTGGATTGGACTGATCAGTTCAGT TTGCTGTGTGTTTGCTCAAACAGATGAAAATAGATGTTTAAAAGCAAATGCCAAATCATGTGGAGAATGTATACAAGCAGGGCCAAATTG TGGGTGGTGCACAAATTCAACATTTTTACAGGAAGGAATGCCTACTTCTGCACGATGTGATGATTTAGAAGCCTTAAAAAAGAAGGGTTG CCCTCCAGATGACATAGAAAATCCCAGAGGCTCCAAAGATATAAAGAAAAATAAAAATGTAACCAACCGTAGCAAAGGAACAGCAGAGAA GCTCAAGCCAGAGGATATTACTCAGATCCAACCACAGCAGTTGGTTTTGCGATTAAGATCAGGGGAGCCACAGACATTTACATTAAAATT CAAGAGAGCTGAAGACTATCCCATTGACCTCTACTACCTTATGGACCTGTCTTACTCAATGAAAGACGATTTGGAGAATGTAAAAAGTCT TGGAACAGATCTGATGAATGAAATGAGGAGGATTACTTCGGACTTCAGAATTGGATTTGGCTCATTTGTGGAAAAGACTGTGATGCCTTA CATTAGCACAACACCAGCTAAGCTCAGGAACCCTTGCACAAGTGAACAGAACTGCACCAGCCCATTTAGCTACAAAAATGTGCTCAGTCT TACTAATAAAGGAGAAGTATTTAATGAACTTGTTGGAAAACAGCGCATATCTGGAAATTTGGATTCTCCAGAAGGTGGTTTCGATGCCAT CATGCAAGTTGCAGTTTGTGGAGTTTTACCAATTCTTGAGATTCTGTATCATGTTGAAGAAAGGAATTCACACCATGTGTATATGGCCCT TATAATATTGTTGATCCTTACGGAAGATGATGGCTTCAACAGATCCATTCATGAAGTGATACTAAAAAATATTACTTGGTATTCAGAACG AGTTTTAACTGAAATCTCCTTGGGGAGTCTCCTGATCCTGGTGGTAATAAGAACCATTCAATACAACATGACTAGGACACGAGACAAGTA CCTTCACACAAATTGTTTGGCAGCTTTAGCAAATATGTCGGCACAGTTTCGTTCTCTCCATCAGTATGCTGCCCAGAGGATCATCAGTTT ATTTTCTTTGCTGTCTAAAAAACACAACAAAGTTCTGGAACAAGCCACACAGTCCTTGAGAGGTTCGCTGAGTTCTAATGATGTTCCTCT ACCAGATTATGCACAAGACCTAAATGTCATTGAAGAAGTGATTCGAATGATGTTAGAGATCATCAACTCCTGCCTGACAAATTCCCTTCA CCACAACCCAAACTTGGTATACGCCCTGCTTTACAAACGCGATCTCTTTGAACAATTTCGAACTCATCCTTCATTTCAGGATATAATGCA AAATATTGATCTGGTGATCTCCTTCTTTAGCTCAAGGTTGCTGCAAGCTGGAGCTGAGCTGTCAGTGGAACGGGTCCTGGAAATCATTAA GCAAGGCGTCGTTGCGCTGCCCAAAGACAGACTGAAGAAATTTCCAGAATTGAAATTCAAATATGTGGAAGAGGAGCAGCCCGAGGAGTT TTTTATCCCCTATGTCTGGTCTCTTGTCTACAACTCAGCAGTCGGCCTGTACTGGAATCCACAGGACATCCAGCTGTTCACCATGGATTC CGACTGAGGGCAGGATGCTCTCCCACCCGGACCCCTCCAGCCAAGCAGCCCTTCAAGTTCTTTTATTTCTGGGTAACAGAAGTAGACAGA CAGGTTACTTGGTGTATCTTCTGTTAAAGAGGATTGCACGAGTGTGTTTTCCTCACACACTTTGATTTGGAGAATTGGTGCTAGTTGGCA >40507_40507_1_ITGB1-DYM_ITGB1_chr10_33214799_ENST00000396033_DYM_chr18_46798673_ENST00000269445_length(amino acids)=588AA_BP=294 MGAVTSGLPELRGTGPERPLTPAVSRRAGQAQMNLQPIFWIGLISSVCCVFAQTDENRCLKANAKSCGECIQAGPNCGWCTNSTFLQEGM PTSARCDDLEALKKKGCPPDDIENPRGSKDIKKNKNVTNRSKGTAEKLKPEDITQIQPQQLVLRLRSGEPQTFTLKFKRAEDYPIDLYYL MDLSYSMKDDLENVKSLGTDLMNEMRRITSDFRIGFGSFVEKTVMPYISTTPAKLRNPCTSEQNCTSPFSYKNVLSLTNKGEVFNELVGK QRISGNLDSPEGGFDAIMQVAVCGVLPILEILYHVEERNSHHVYMALIILLILTEDDGFNRSIHEVILKNITWYSERVLTEISLGSLLIL VVIRTIQYNMTRTRDKYLHTNCLAALANMSAQFRSLHQYAAQRIISLFSLLSKKHNKVLEQATQSLRGSLSSNDVPLPDYAQDLNVIEEV IRMMLEIINSCLTNSLHHNPNLVYALLYKRDLFEQFRTHPSFQDIMQNIDLVISFFSSRLLQAGAELSVERVLEIIKQGVVALPKDRLKK -------------------------------------------------------------- >40507_40507_2_ITGB1-DYM_ITGB1_chr10_33214799_ENST00000396033_DYM_chr18_46798673_ENST00000442713_length(transcript)=2193nt_BP=922nt GAGCCAGCCCAGCCGCGTTCCGAACGTGAGGGTCGCCGGCCTGGGCGCTGTCACGTCGGGGCTGCCGGAGCTGCGGGGGACCGGGCCCGA ACGGCCCCTGACACCTGCGGTCTCCCGCCGGGCTGGGCAAGCGCAGATGAATTTACAACCAATTTTCTGGATTGGACTGATCAGTTCAGT TTGCTGTGTGTTTGCTCAAACAGATGAAAATAGATGTTTAAAAGCAAATGCCAAATCATGTGGAGAATGTATACAAGCAGGGCCAAATTG TGGGTGGTGCACAAATTCAACATTTTTACAGGAAGGAATGCCTACTTCTGCACGATGTGATGATTTAGAAGCCTTAAAAAAGAAGGGTTG CCCTCCAGATGACATAGAAAATCCCAGAGGCTCCAAAGATATAAAGAAAAATAAAAATGTAACCAACCGTAGCAAAGGAACAGCAGAGAA GCTCAAGCCAGAGGATATTACTCAGATCCAACCACAGCAGTTGGTTTTGCGATTAAGATCAGGGGAGCCACAGACATTTACATTAAAATT CAAGAGAGCTGAAGACTATCCCATTGACCTCTACTACCTTATGGACCTGTCTTACTCAATGAAAGACGATTTGGAGAATGTAAAAAGTCT TGGAACAGATCTGATGAATGAAATGAGGAGGATTACTTCGGACTTCAGAATTGGATTTGGCTCATTTGTGGAAAAGACTGTGATGCCTTA CATTAGCACAACACCAGCTAAGCTCAGGAACCCTTGCACAAGTGAACAGAACTGCACCAGCCCATTTAGCTACAAAAATGTGCTCAGTCT TACTAATAAAGGAGAAGTATTTAATGAACTTGTTGGAAAACAGCGCATATCTGGAAATTTGGATTCTCCAGAAGGTGGTTTCGATGCCAT CATGCAAGTTGCAGTTTGTGGAGTTTTACCAATTCTTGAGATTCTGTATCATGTTGAAGAAAGGAATTCACACCATGTGTATATGGCCCT TATAATATTGTTGATCCTTACGGAAGATGATGGCTTCAACAGATCCATTCATGAAGTGATACTAAAAAATATTACTTGGTATTCAGAACG AGTTTTAACTGAAATCTCCTTGGGGAGTCTCCTGATCCTGGTGGTAATAAGAACCATTCAATACAACATGACTAGGACACGAGACAAGTA CCTTCACACAAATTGTTTGGCAGCTTTAGCAAATATGTCGGCACAGTTTCGTTCTCTCCATCAGTATGCTGCCCAGAGGATCATCAGTTT ATTTTCTTTGCTGTCTAAAAAACACAACAAAGTTCTGGAACAAGCCACACAGTCCTTGAGAGGTTCGCTGAGTTCTAATGATGTTCCTCT ACCAGATTATGCACAAGACCTAAATGTCATTGAAGAAGTGATTCGAATGATGTTAGAGATCATCAACTCCTGCCTGACAAATTCCCTTCA CCACAACCCAAACTTGGTATACGCCCTGCTTTACAAACGCGATCTCTTTGAACAATTTCGAACTCATCCTTCATTTCAGGATATAATGCA AAATATTGATCTGGTGATCTCCTTCTTTAGCTCAAGGTTGCTGCAAGCTGGAGCTGAGCTGTCAGTGGAACGGGTCCTGGAAATCATTAA GCAAGGCGTCGTTGCGCTGCCCAAAGACAGACTGAAGAAATTTCCAGAATTGAAATTCAAATATGTGGAAGAGGAGCAGCCCGAGGAGTT TTTTATCCCCTATGTCTGGTCTCTTGTCTACAACTCAGCAGTCGGCCTGTACTGGAATCCACAGGACATCCAGCTGTTCACCATGGATTC CGACTGAGGGCAGGATGCTCTCCCACCCGGACCCCTCCAGCCAAGCAGCCCTTCAAGTTCTTTTATTTCTGGGTAACAGAAGTAGACAGA CAGGTTACTTGGTGTATCTTCTGTTAAAGAGGATTGCACGAGTGTGTTTTCCTCACACACTTTGATTTGGAGAATTGGTGCTAGTTGGCA ATAGATAACTCAGCGTAGATAGTATTGCAAAAAGGGGAGGAAATACACAACAATAATAAATGTAAAAACCTGCTATTCAACATGCAGTTT TATTTCGAAGCCAAAAATCTAGAGCTTTCCCAAGATCCTGTTGCCTTAGGCACATCACACTTCAACAGTGCACACTATCCAACAGTGCAC >40507_40507_2_ITGB1-DYM_ITGB1_chr10_33214799_ENST00000396033_DYM_chr18_46798673_ENST00000442713_length(amino acids)=588AA_BP=294 MGAVTSGLPELRGTGPERPLTPAVSRRAGQAQMNLQPIFWIGLISSVCCVFAQTDENRCLKANAKSCGECIQAGPNCGWCTNSTFLQEGM PTSARCDDLEALKKKGCPPDDIENPRGSKDIKKNKNVTNRSKGTAEKLKPEDITQIQPQQLVLRLRSGEPQTFTLKFKRAEDYPIDLYYL MDLSYSMKDDLENVKSLGTDLMNEMRRITSDFRIGFGSFVEKTVMPYISTTPAKLRNPCTSEQNCTSPFSYKNVLSLTNKGEVFNELVGK QRISGNLDSPEGGFDAIMQVAVCGVLPILEILYHVEERNSHHVYMALIILLILTEDDGFNRSIHEVILKNITWYSERVLTEISLGSLLIL VVIRTIQYNMTRTRDKYLHTNCLAALANMSAQFRSLHQYAAQRIISLFSLLSKKHNKVLEQATQSLRGSLSSNDVPLPDYAQDLNVIEEV IRMMLEIINSCLTNSLHHNPNLVYALLYKRDLFEQFRTHPSFQDIMQNIDLVISFFSSRLLQAGAELSVERVLEIIKQGVVALPKDRLKK -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for ITGB1-DYM |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
| Hgene | ITGB1 | chr10:33214799 | chr18:46798673 | ENST00000302278 | - | 6 | 16 | 785_792 | 262.0 | 799.0 | ITGB1BP1 |
| Hgene | ITGB1 | chr10:33214799 | chr18:46798673 | ENST00000374956 | - | 6 | 17 | 785_792 | 262.0 | 826.0 | ITGB1BP1 |
| Hgene | ITGB1 | chr10:33214799 | chr18:46798673 | ENST00000396033 | - | 6 | 16 | 785_792 | 262.0 | 799.0 | ITGB1BP1 |
| Hgene | ITGB1 | chr10:33214799 | chr18:46798673 | ENST00000423113 | - | 5 | 16 | 785_792 | 262.0 | 1217.0 | ITGB1BP1 |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for ITGB1-DYM |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for ITGB1-DYM |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
