
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:ITLN1-DOCK11 (FusionGDB2 ID:40606) |
Fusion Gene Summary for ITLN1-DOCK11 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: ITLN1-DOCK11 | Fusion gene ID: 40606 | Hgene | Tgene | Gene symbol | ITLN1 | DOCK11 | Gene ID | 55600 | 139818 |
| Gene name | intelectin 1 | dedicator of cytokinesis 11 | |
| Synonyms | HL-1|HL1|INTL|ITLN|LFR|hIntL|omentin | ACG|ZIZ2|bB128O4.1 | |
| Cytomap | 1q23.3 | Xq24 | |
| Type of gene | protein-coding | protein-coding | |
| Description | intelectin-1ITLN-1endothelial lectin HL-1galactofuranose-binding lectinintelectin 1 (galactofuranose binding)intestinal lactoferrin receptor | dedicator of cytokinesis protein 11Activated Cdc42-associated guanine nucleotide exchange factor, ACGbB128O4.1.1 (novel protein)zizimin-2zizimin2 | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | Q8WWA0 | Q5JSL3 | |
| Ensembl transtripts involved in fusion gene | ENST00000326245, ENST00000487531, | ENST00000276202, ENST00000276204, | |
| Fusion gene scores | * DoF score | 6 X 5 X 2=60 | 11 X 12 X 7=924 |
| # samples | 6 | 13 | |
| ** MAII score | log2(6/60*10)=0 | log2(13/924*10)=-2.8293812283876 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: ITLN1 [Title/Abstract] AND DOCK11 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | ITLN1(160851748)-DOCK11(117742246), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | ITLN1 | GO:0001934 | positive regulation of protein phosphorylation | 16531507 |
| Hgene | ITLN1 | GO:0046326 | positive regulation of glucose import | 16531507 |
| Hgene | ITLN1 | GO:0070207 | protein homotrimerization | 26148048 |
 Fusion gene breakpoints across ITLN1 (5'-gene) Fusion gene breakpoints across ITLN1 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
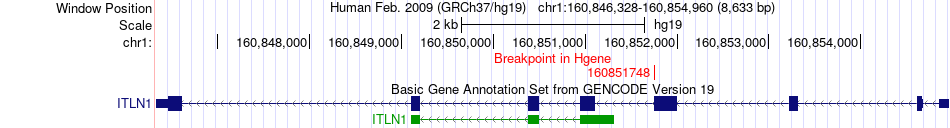 |
 Fusion gene breakpoints across DOCK11 (3'-gene) Fusion gene breakpoints across DOCK11 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
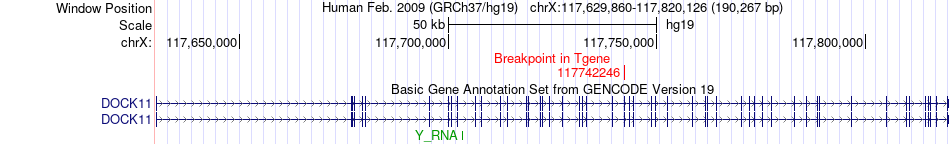 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | MESO | TCGA-MQ-A4LC-01A | ITLN1 | chr1 | 160851748 | - | DOCK11 | chrX | 117742246 | + |
Top |
Fusion Gene ORF analysis for ITLN1-DOCK11 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| In-frame | ENST00000326245 | ENST00000276202 | ITLN1 | chr1 | 160851748 | - | DOCK11 | chrX | 117742246 | + |
| In-frame | ENST00000326245 | ENST00000276204 | ITLN1 | chr1 | 160851748 | - | DOCK11 | chrX | 117742246 | + |
| intron-3CDS | ENST00000487531 | ENST00000276202 | ITLN1 | chr1 | 160851748 | - | DOCK11 | chrX | 117742246 | + |
| intron-3CDS | ENST00000487531 | ENST00000276204 | ITLN1 | chr1 | 160851748 | - | DOCK11 | chrX | 117742246 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000326245 | ITLN1 | chr1 | 160851748 | - | ENST00000276204 | DOCK11 | chrX | 117742246 | + | 4307 | 521 | 336 | 3950 | 1204 |
| ENST00000326245 | ITLN1 | chr1 | 160851748 | - | ENST00000276202 | DOCK11 | chrX | 117742246 | + | 4279 | 521 | 336 | 3938 | 1200 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000326245 | ENST00000276204 | ITLN1 | chr1 | 160851748 | - | DOCK11 | chrX | 117742246 | + | 0.000438433 | 0.9995616 |
| ENST00000326245 | ENST00000276202 | ITLN1 | chr1 | 160851748 | - | DOCK11 | chrX | 117742246 | + | 0.000758088 | 0.99924195 |
Top |
Fusion Genomic Features for ITLN1-DOCK11 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
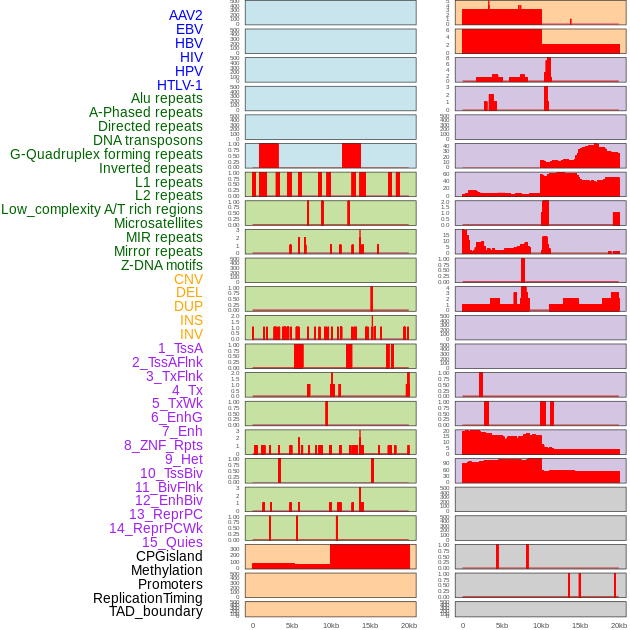 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for ITLN1-DOCK11 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr1:160851748/chrX:117742246) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| ITLN1 | DOCK11 |
| FUNCTION: Lectin that specifically recognizes microbial carbohydrate chains in a calcium-dependent manner (PubMed:11313366, PubMed:26148048). Binds to microbial glycans that contain a terminal acyclic 1,2-diol moiety, including beta-linked D-galactofuranose (beta-Galf), D-phosphoglycerol-modified glycans, D-glycero-D-talo-oct-2-ulosonic acid (KO) and 3-deoxy-D-manno-oct-2-ulosonic acid (KDO) (PubMed:26148048). Binds to glycans from Gram-positive and Gram-negative bacteria, including K.pneumoniae, S.pneumoniae, Y.pestis, P.mirabilis and P.vulgaris (PubMed:26148048). Does not bind human glycans (PubMed:26148048). Probably plays a role in the defense system against microorganisms (Probable). May function as adipokine that has no effect on basal glucose uptake but enhances insulin-stimulated glucose uptake in adipocytes (PubMed:16531507). Increases AKT phosphorylation in the absence and presence of insulin (PubMed:16531507). May interact with lactoferrin/LTF and increase its uptake, and may thereby play a role in iron absorption (PubMed:11747454, PubMed:23921499). {ECO:0000269|PubMed:11313366, ECO:0000269|PubMed:16531507, ECO:0000269|PubMed:23921499, ECO:0000269|PubMed:26148048, ECO:0000305, ECO:0000305|PubMed:11747454}. | FUNCTION: Guanine nucleotide-exchange factor (GEF) that activates CDC42 by exchanging bound GDP for free GTP. Required for marginal zone (MZ) B-cell development, is associated with early bone marrow B-cell development, MZ B-cell formation, MZ B-cell number and marginal metallophilic macrophages morphology. Facilitates filopodia formation through the activation of CDC42. {ECO:0000250|UniProtKB:A2AF47}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | DOCK11 | chr1:160851748 | chrX:117742246 | ENST00000276202 | 0 | 53 | 1609_2036 | 0 | 2074.0 | Domain | DOCKER | |
| Tgene | DOCK11 | chr1:160851748 | chrX:117742246 | ENST00000276202 | 0 | 53 | 165_272 | 0 | 2074.0 | Domain | PH | |
| Tgene | DOCK11 | chr1:160851748 | chrX:117742246 | ENST00000276202 | 0 | 53 | 640_818 | 0 | 2074.0 | Domain | C2 DOCK-type |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | ITLN1 | chr1:160851748 | chrX:117742246 | ENST00000326245 | - | 4 | 8 | 32_255 | 135 | 314.0 | Domain | Fibrinogen C-terminal |
| Hgene | ITLN1 | chr1:160851748 | chrX:117742246 | ENST00000326245 | - | 4 | 8 | 262_263 | 135 | 314.0 | Region | Carbohydrate binding |
Top |
Fusion Gene Sequence for ITLN1-DOCK11 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >40606_40606_1_ITLN1-DOCK11_ITLN1_chr1_160851748_ENST00000326245_DOCK11_chrX_117742246_ENST00000276202_length(transcript)=4279nt_BP=521nt AGGAGCGTTTTTGGAGAAAGCTGCACTCTGTTGAGCTCCAGGGCGCAGTGGAGGGAGGGAGTGAAGGAGCTCTCTGTACCCAAGGAAAGT GCAGCTGAGACTCAGACAAGATTACAATGAACCAACTCAGCTTCCTGCTGTTTCTCATAGCGACCACCAGAGGATGGAGTACAGATGAGG CTAATACTTACTTCAAGGAATGGACCTGTTCTTCGTCTCCATCTCTGCCCAGAAGCTGCAAGGAAATCAAAGACGAATGTCCTAGTGCAT TTGATGGCCTGTATTTTCTCCGCACTGAGAATGGTGTTATCTACCAGACCTTCTGTGACATGACCTCTGGGGGTGGCGGCTGGACCCTGG TGGCCAGCGTGCACGAGAATGACATGCGTGGGAAGTGCACGGTGGGCGATCGCTGGTCCAGTCAGCAGGGCAGCAAAGCAGTCTACCCAG AGGGGGACGGCAACTGGGCCAACTACAACACCTTTGGATCTGCAGAGGCGGCCACGAGCGATGACTACAAGAACCCTGGCTACTACGATG ATAGCAATATTGAAACAGTCTGCAGATTTTTTATCAATAAACAAATTGCTAAAGTACTCATGGTTTTTCTTTGAAATAATTGCAAAGTCA ATGGCCACATACTTGTTGGAAGAGAATAAGATTAAGCTTCCCCGAGGCCAGAGATTTCCCGAGACATATCATCATGTCTTACATTCACTG CTTCTTGCAATAATTCCCCATGTGACTATTCGGTATGCGGAGATTCCCGATGAGTCCAGAAATGTGAACTATAGTTTGGCTAGCTTCCTG AAGCGCTGTTTGACACTAATGGATAGAGGATTTATTTTCAATTTAATAAATGACTATATATCTGGATTCAGCCCCAAAGATCCTAAGGTT CTGGCTGAATACAAGTTTGAATTTCTGCAAACAATTTGCAATCACGAACATTACATTCCTCTGAACTTGCCAATGGCATTTGCAAAACCT AAACTGCAGCGGGTTCAAGATTCAAATCTTGAATACAGTTTATCAGATGAGTATTGCAAGCATCACTTCTTGGTTGGTCTACTTCTGAGG GAAACTTCCATTGCTCTTCAGGACAATTATGAGATCAGATATACAGCTATCTCTGTTATAAAGAATCTTTTGATAAAACATGCATTTGAC ACAAGATACCAGCACAAGAACCAACAAGCCAAAATAGCACAATTGTACCTCCCCTTTGTTGGACTACTTTTGGAAAATATACAGCGATTA GCAGGTCGAGATACCTTGTATTCTTGTGCAGCCATGCCTAATTCTGCATCCAGAGATGAGTTTCCATGTGGCTTTACTTCACCTGCCAAT AGAGGGAGTCTGAGCACTGACAAAGACACCGCTTATGGGTCTTTTCAAAATGGACATGGAATTAAGAGAGAAGATTCAAGAGGTTCCCTC ATCCCAGAAGGAGCAACAGGATTTCCAGATCAGGGCAACACTGGTGAAAATACCCGACAGAGTTCTACAAGGAGTAGTGTATCCCAGTAT AACCGCCTGGATCAGTATGAAATCAGAAGCCTCCTGATGTGCTACCTGTATATAGTAAAAATGATTTCAGAAGATACTCTCTTAACTTAC TGGAATAAAGTATCACCTCAGGAGCTCATAAACATTCTTATACTTTTAGAAGTATGCTTGTTTCACTTTAGATATATGGGGAAAAGAAAC ATAGCAAGGGTGCATGATGCCTGGCTGTCAAAACACTTCGGAATAGACCGAAAATCGCAAACCATGCCTGCTCTTCGAAACAGATCAGGA GTAATGCAGGCCCGGCTTCAGCATCTTAGTAGCCTAGAAAGTTCATTTACACTTAATCACAGTTCTACAACAACTGAAGCAGACATTTTC CACCAGGCACTTCTTGAAGGCAATACAGCTACTGAAGTTTCCCTAACAGTACTAGACACCATATCATTTTTCACTCAGTGCTTCAAGACC CAACTTTTAAATAATGATGGCCATAACCCATTAATGAAAAAAGTGTTTGATATACATCTTGCTTTTCTTAAAAATGGACAATCTGAAGTG TCGCTGAAACATGTATTTGCCTCACTGAGAGCTTTCATCAGTAAGTTTCCTTCAGCATTTTTCAAAGGAAGAGTAAACATGTGTGCTGCA TTTTGCTATGAGGTTTTAAAGTGCTGCACATCGAAGATTAGCTCAACCAGGAATGAAGCATCTGCACTTTTGTATCTTTTGATGAGAAAC AACTTTGAGTATACCAAAAGGAAAACCTTTTTGAGGACACATCTACAGATAATAATTGCTGTAAGCCAACTGATAGCTGATGTAGCACTA AGCGGAGGATCAAGATTTCAGGAGTCTTTATTCATTATCAATAATTTTGCAAATAGTGACAGACCTATGAAGGCAACTGCCTTTCCCGCA GAAGTCAAAGACTTGACCAAGAGAATCCGCACTGTTCTTATGGCCACTGCCCAAATGAAGGAGCATGAGAAAGACCCTGAAATGCTAATT GATCTCCAGTATAGCTTAGCCAAGTCCTATGCAAGCACCCCAGAGCTCAGGAAAACCTGGCTTGATAGCATGGCCAAGATTCATGTAAAA AATGGAGATTTTTCAGAGGCTGCGATGTGTTATGTCCATGTAGCAGCTCTAGTTGCAGAGTTTCTTCATCGAAAAAAATTATTTCCTAAC GGATGTTCAGCGTTCAAGAAAATTACTCCCAATATAGATGAAGAAGGAGCAATGAAAGAAGATGCTGGGATGATGGATGTCCATTATAGT GAAGAGGTCTTGCTGGAGTTGCTAGAACAATGTGTGGATGGCTTATGGAAGGCAGAACGTTATGAAATAATTTCTGAGATTTCCAAGTTG ATCGTTCCAATTTATGAGAAACGTCGTGAGTTTGAGAAACTTACTCAAGTTTATAGAACTCTTCATGGAGCTTACACAAAAATTCTGGAA GTTATGCATACAAAAAAGAGACTTTTAGGCACTTTCTTCAGAGTTGCCTTTTATGGCCAATCTTTTTTTGAAGAAGAAGATGGAAAGGAG TACATCTATAAAGAACCAAAGCTCACTGGCCTCTCAGAAATTTCCTTGAGACTTGTTAAACTTTATGGTGAAAAGTTTGGTACGGAGAAT GTCAAAATAATTCAGGATTCAGACAAGGTAAATGCCAAAGAGCTTGATCCAAAATATGCTCATATACAAGTTACTTATGTGAAGCCTTAC TTTGATGACAAAGAACTCACAGAAAGGAAGACCGAGTTTGAAAGAAATCATAATATCAGCAGATTTGTTTTTGAGGCCCCTTACACTTTA TCAGGCAAAAAACAGGGCTGTATAGAAGAACAGTGCAAACGCCGTACAATCTTGACAACTTCAAACTCGTTTCCTTACGTGAAGAAGAGG ATTCCTATTAACTGTGAACAGCAGATTAATTTAAAACCAATTGATGTTGCCACTGATGAAATAAAAGATAAAACTGCAGAGCTGCAAAAG CTTTGCTCCTCTACTGACGTGGACATGATTCAGCTCCAACTTAAATTGCAGGGCTGTGTTTCTGTGCAGGTCAATGCTGGTCCATTAGCA TATGCAAGAGCTTTCTTAAATGACAGCCAAGCTAGCAAGTATCCACCTAAGAAAGTGAGTGAGTTGAAAGACATGTTTAGGAAATTTATA CAAGCATGCAGCATTGCACTTGAACTAAATGAGCGGCTAATTAAAGAAGATCAAGTTGAGTACCATGAAGGGCTAAAGTCAAATTTCAGA GACATGGTAAAAGAATTATCTGACATTATCCATGAGCAGATATTACAAGAAGACACAATGCATTCTCCCTGGATGAGCAACACATTACAT GTATTTTGTGCAATTAGTGGTACATCAAGTGACCGAGGTTATGGTTCCCCAAGATACGCTGAAGTGTGAGGAAATGCAGATGTACGTGAC AATGAGACTGACCTTTCTCAGGAATATTTGGAGCTGTGCAAATGTTAAAATTTAAAGATTTGATATACATGGAGTGTTTCTTCTCGACAC CAAAATTTTCATGTGTTCCAACAGGGTGCTTACATATTTGTAAATAAGCAACTTGAAAGTGCCTGGAAAATTGCACCACTGTGCTTGGTT TGTACTTTTTTAGGTAAATCTATATGCTGAAAAGTAGAGCTCAAAAACAGTAGTTCAATTTGCTTAATTATTGCTTAAAATAATGGTACT >40606_40606_1_ITLN1-DOCK11_ITLN1_chr1_160851748_ENST00000326245_DOCK11_chrX_117742246_ENST00000276202_length(amino acids)=1200AA_BP=62 MGVAAGPWWPACTRMTCVGSARWAIAGPVSRAAKQSTQRGTATGPTTTPLDLQRRPRAMTTRTLATTMIAILKQSADFLSINKLLKYSWF FFEIIAKSMATYLLEENKIKLPRGQRFPETYHHVLHSLLLAIIPHVTIRYAEIPDESRNVNYSLASFLKRCLTLMDRGFIFNLINDYISG FSPKDPKVLAEYKFEFLQTICNHEHYIPLNLPMAFAKPKLQRVQDSNLEYSLSDEYCKHHFLVGLLLRETSIALQDNYEIRYTAISVIKN LLIKHAFDTRYQHKNQQAKIAQLYLPFVGLLLENIQRLAGRDTLYSCAAMPNSASRDEFPCGFTSPANRGSLSTDKDTAYGSFQNGHGIK REDSRGSLIPEGATGFPDQGNTGENTRQSSTRSSVSQYNRLDQYEIRSLLMCYLYIVKMISEDTLLTYWNKVSPQELINILILLEVCLFH FRYMGKRNIARVHDAWLSKHFGIDRKSQTMPALRNRSGVMQARLQHLSSLESSFTLNHSSTTTEADIFHQALLEGNTATEVSLTVLDTIS FFTQCFKTQLLNNDGHNPLMKKVFDIHLAFLKNGQSEVSLKHVFASLRAFISKFPSAFFKGRVNMCAAFCYEVLKCCTSKISSTRNEASA LLYLLMRNNFEYTKRKTFLRTHLQIIIAVSQLIADVALSGGSRFQESLFIINNFANSDRPMKATAFPAEVKDLTKRIRTVLMATAQMKEH EKDPEMLIDLQYSLAKSYASTPELRKTWLDSMAKIHVKNGDFSEAAMCYVHVAALVAEFLHRKKLFPNGCSAFKKITPNIDEEGAMKEDA GMMDVHYSEEVLLELLEQCVDGLWKAERYEIISEISKLIVPIYEKRREFEKLTQVYRTLHGAYTKILEVMHTKKRLLGTFFRVAFYGQSF FEEEDGKEYIYKEPKLTGLSEISLRLVKLYGEKFGTENVKIIQDSDKVNAKELDPKYAHIQVTYVKPYFDDKELTERKTEFERNHNISRF VFEAPYTLSGKKQGCIEEQCKRRTILTTSNSFPYVKKRIPINCEQQINLKPIDVATDEIKDKTAELQKLCSSTDVDMIQLQLKLQGCVSV QVNAGPLAYARAFLNDSQASKYPPKKVSELKDMFRKFIQACSIALELNERLIKEDQVEYHEGLKSNFRDMVKELSDIIHEQILQEDTMHS -------------------------------------------------------------- >40606_40606_2_ITLN1-DOCK11_ITLN1_chr1_160851748_ENST00000326245_DOCK11_chrX_117742246_ENST00000276204_length(transcript)=4307nt_BP=521nt AGGAGCGTTTTTGGAGAAAGCTGCACTCTGTTGAGCTCCAGGGCGCAGTGGAGGGAGGGAGTGAAGGAGCTCTCTGTACCCAAGGAAAGT GCAGCTGAGACTCAGACAAGATTACAATGAACCAACTCAGCTTCCTGCTGTTTCTCATAGCGACCACCAGAGGATGGAGTACAGATGAGG CTAATACTTACTTCAAGGAATGGACCTGTTCTTCGTCTCCATCTCTGCCCAGAAGCTGCAAGGAAATCAAAGACGAATGTCCTAGTGCAT TTGATGGCCTGTATTTTCTCCGCACTGAGAATGGTGTTATCTACCAGACCTTCTGTGACATGACCTCTGGGGGTGGCGGCTGGACCCTGG TGGCCAGCGTGCACGAGAATGACATGCGTGGGAAGTGCACGGTGGGCGATCGCTGGTCCAGTCAGCAGGGCAGCAAAGCAGTCTACCCAG AGGGGGACGGCAACTGGGCCAACTACAACACCTTTGGATCTGCAGAGGCGGCCACGAGCGATGACTACAAGAACCCTGGCTACTACGATG ATAGCAATATTGAAACAGTCTGCAGATTTTTTATCAATAAACAAATTGCTAAAGTACTCATGGTTTTTCTTTGAAATAATTGCAAAGTCA ATGGCCACATACTTGTTGGAAGAGAATAAGATTAAGCTTCCCCGAGGCCAGAGATTTCCCGAGACATATCATCATGTCTTACATTCACTG CTTCTTGCAATAATTCCCCATGTGACTATTCGGTATGCGGAGATTCCCGATGAGTCCAGAAATGTGAACTATAGTTTGGCTAGCTTCCTG AAGCGCTGTTTGACACTAATGGATAGAGGATTTATTTTCAATTTAATAAATGACTATATATCTGGATTCAGCCCCAAAGATCCTAAGGTT CTGGCTGAATACAAGTTTGAATTTCTGCAAACAATTTGCAATCACGAACATTACATTCCTCTGAACTTGCCAATGGCATTTGCAAAACCT AAACTGCAGCGGGTTCAAGATTCAAATCTTGAATACAGTTTATCAGATGAGTATTGCAAGCATCACTTCTTGGTTGGTCTACTTCTGAGG GAAACTTCCATTGCTCTTCAGGACAATTATGAGATCAGATATACAGCTATCTCTGTTATAAAGAATCTTTTGATAAAACATGCATTTGAC ACAAGATACCAGCACAAGAACCAACAAGCCAAAATAGCACAATTGTACCTCCCCTTTGTTGGACTACTTTTGGAAAATATACAGCGATTA GCAGGTCGAGATACCTTGTATTCTTGTGCAGCCATGCCTAATTCTGCATCCAGAGATGAGTTTCCATGTGGCTTTACTTCACCTGCCAAT AGAGGGAGTCTGAGCACTGACAAAGACACCGCTTATGGGTCTTTTCAAAATGGACATGGAATTAAGAGAGAAGATTCAAGAGGTTCCCTC ATCCCAGAAGGAGCAACAGGATTTCCAGATCAGGGCAACACTGGTGAAAATACCCGACAGAGTTCTACAAGGAGTAGTGTATCCCAGTAT AACCGCCTGGATCAGTATGAAATCAGAAGCCTCCTGATGTGCTACCTGTATATAGTAAAAATGATTTCAGAAGATACTCTCTTAACTTAC TGGAATAAAGTATCACCTCAGGAGCTCATAAACATTCTTATACTTTTAGAAGTATGCTTGTTTCACTTTAGATATATGGGGAAAAGAAAC ATAGCAAGGGTGCATGATGCCTGGCTGTCAAAACACTTCGGAATAGACCGAAAATCGCAAACCATGCCTGCTCTTCGAAACAGATCAGGA GTAATGCAGGCCCGGCTTCAGCATCTTAGTAGCCTAGAAAGTTCATTTACACTTAATCACAGTTCTACAACAACTGAAGCAGACATTTTC CACCAGGCACTTCTTGAAGGCAATACAGCTACTGAAGTTTCCCTAACAGTACTAGACACCATATCATTTTTCACTCAGTGCTTCAAGACC CAACTTTTAAATAATGATGGCCATAACCCATTAATGAAAAAAGTGTTTGATATACATCTTGCTTTTCTTAAAAATGGACAATCTGAAGTG TCGCTGAAACATGTATTTGCCTCACTGAGAGCTTTCATCAGTAAGTTTCCTTCAGCATTTTTCAAAGGAAGAGTAAACATGTGTGCTGCA TTTTGCTATGAGGTTTTAAAGTGCTGCACATCGAAGATTAGCTCAACCAGGAATGAAGCATCTGCACTTTTGTATCTTTTGATGAGAAAC AACTTTGAGTATACCAAAAGGAAAACCTTTTTGAGGACACATCTACAGATAATAATTGCTGTAAGCCAACTGATAGCTGATGTAGCACTA AGCGGAGGATCAAGATTTCAGGAGTCTTTATTCATTATCAATAATTTTGCAAATAGTGACAGACCTATGAAGGCAACTGCCTTTCCCGCA GAAGTCAAAGACTTGACCAAGAGAATCCGCACTGTTCTTATGGCCACTGCCCAAATGAAGGAGCATGAGAAAGACCCTGAAATGCTAATT GATCTCCAGTATAGCTTAGCCAAGTCCTATGCAAGCACCCCAGAGCTCAGGAAAACCTGGCTTGATAGCATGGCCAAGATTCATGTAAAA AATGGAGATTTTTCAGAGGCTGCGATGTGTTATGTCCATGTAGCAGCTCTAGTTGCAGAGTTTCTTCATCGAAAAAAATTATTTCCTAAC GGATGTTCAGCGTTCAAGAAAATTACTCCCAATATAGATGAAGAAGGAGCAATGAAAGAAGATGCTGGGATGATGGATGTCCATTATAGT GAAGAGGTCTTGCTGGAGTTGCTAGAACAATGTGTGGATGGCTTATGGAAGGCAGAACGTTATGAAATAATTTCTGAGATTTCCAAGTTG ATCGTTCCAATTTATGAGAAACGTCGTGAGTTTGAGAAACTTACTCAAGTTTATAGAACTCTTCATGGAGCTTACACAAAAATTCTGGAA GTTATGCATACAAAAAAGAGACTTTTAGGCACTTTCTTCAGAGTTGCCTTTTATGGCCAATCTTTTTTTGAAGAAGAAGATGGAAAGGAG TACATCTATAAAGAACCAAAGCTCACTGGCCTCTCAGAAATTTCCTTGAGACTTGTTAAACTTTATGGTGAAAAGTTTGGTACGGAGAAT GTCAAAATAATTCAGGATTCAGACAAGGTAAATGCCAAAGAGCTTGATCCAAAATATGCTCATATACAAGTTACTTATGTGAAGCCTTAC TTTGATGACAAAGAACTCACAGAAAGGAAGACCGAGTTTGAAAGAAATCATAATATCAGCAGATTTGTTTTTGAGGCCCCTTACACTTTA TCAGGCAAAAAACAGGGCTGTATAGAAGAACAGTGCAAACGCCGTACAATCTTGACAACTTCAAACTCGTTTCCTTACGTGAAGAAGAGG ATTCCTATTAACTGTGAACAGCAGATTAATTTAAAACCAATTGATGTTGCCACTGATGAAATAAAAGATAAAACTGCAGAGCTGCAAAAG CTTTGCTCCTCTACTGACGTGGACATGATTCAGCTCCAACTTAAATTGCAGGGCTGTGTTTCTGTGCAGGTCAATGCTGGTCCATTAGCA TATGCAAGAGCTTTCTTAAATGACAGCCAAGCTAGCAAGTATCCACCTAAGAAAGTGAGTGAGTTGAAAGACATGTTTAGGAAATTTATA CAAGCATGCAGCATTGCACTTGAACTAAATGAGCGGCTAATTAAAGAAGATCAAGTTGAGTACCATGAAGGGCTAAAGTCAAATTTCAGA GACATGGTAAAAGAATTATCTGACATTATCCATGAGCAGGCAAGTATTAGGATATTACAAGAAGACACAATGCATTCTCCCTGGATGAGC AACACATTACATGTATTTTGTGCAATTAGTGGTACATCAAGTGACCGAGGTTATGGTTCCCCAAGATACGCTGAAGTGTGAGGAAATGCA GATGTACGTGACAATGAGACTGACCTTTCTCAGGAATATTTGGAGCTGTGCAAATGTTAAAATTTAAAGATTTGATATACATGGAGTGTT TCTTCTCGACACCAAAATTTTCATGTGTTCCAACAGGGTGCTTACATATTTGTAAATAAGCAACTTGAAAGTGCCTGGAAAATTGCACCA CTGTGCTTGGTTTGTACTTTTTTAGGTAAATCTATATGCTGAAAAGTAGAGCTCAAAAACAGTAGTTCAATTTGCTTAATTATTGCTTAA >40606_40606_2_ITLN1-DOCK11_ITLN1_chr1_160851748_ENST00000326245_DOCK11_chrX_117742246_ENST00000276204_length(amino acids)=1204AA_BP=62 MGVAAGPWWPACTRMTCVGSARWAIAGPVSRAAKQSTQRGTATGPTTTPLDLQRRPRAMTTRTLATTMIAILKQSADFLSINKLLKYSWF FFEIIAKSMATYLLEENKIKLPRGQRFPETYHHVLHSLLLAIIPHVTIRYAEIPDESRNVNYSLASFLKRCLTLMDRGFIFNLINDYISG FSPKDPKVLAEYKFEFLQTICNHEHYIPLNLPMAFAKPKLQRVQDSNLEYSLSDEYCKHHFLVGLLLRETSIALQDNYEIRYTAISVIKN LLIKHAFDTRYQHKNQQAKIAQLYLPFVGLLLENIQRLAGRDTLYSCAAMPNSASRDEFPCGFTSPANRGSLSTDKDTAYGSFQNGHGIK REDSRGSLIPEGATGFPDQGNTGENTRQSSTRSSVSQYNRLDQYEIRSLLMCYLYIVKMISEDTLLTYWNKVSPQELINILILLEVCLFH FRYMGKRNIARVHDAWLSKHFGIDRKSQTMPALRNRSGVMQARLQHLSSLESSFTLNHSSTTTEADIFHQALLEGNTATEVSLTVLDTIS FFTQCFKTQLLNNDGHNPLMKKVFDIHLAFLKNGQSEVSLKHVFASLRAFISKFPSAFFKGRVNMCAAFCYEVLKCCTSKISSTRNEASA LLYLLMRNNFEYTKRKTFLRTHLQIIIAVSQLIADVALSGGSRFQESLFIINNFANSDRPMKATAFPAEVKDLTKRIRTVLMATAQMKEH EKDPEMLIDLQYSLAKSYASTPELRKTWLDSMAKIHVKNGDFSEAAMCYVHVAALVAEFLHRKKLFPNGCSAFKKITPNIDEEGAMKEDA GMMDVHYSEEVLLELLEQCVDGLWKAERYEIISEISKLIVPIYEKRREFEKLTQVYRTLHGAYTKILEVMHTKKRLLGTFFRVAFYGQSF FEEEDGKEYIYKEPKLTGLSEISLRLVKLYGEKFGTENVKIIQDSDKVNAKELDPKYAHIQVTYVKPYFDDKELTERKTEFERNHNISRF VFEAPYTLSGKKQGCIEEQCKRRTILTTSNSFPYVKKRIPINCEQQINLKPIDVATDEIKDKTAELQKLCSSTDVDMIQLQLKLQGCVSV QVNAGPLAYARAFLNDSQASKYPPKKVSELKDMFRKFIQACSIALELNERLIKEDQVEYHEGLKSNFRDMVKELSDIIHEQASIRILQED -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for ITLN1-DOCK11 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for ITLN1-DOCK11 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for ITLN1-DOCK11 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
