
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:KAT6A-SPPL2A (FusionGDB2 ID:41217) |
Fusion Gene Summary for KAT6A-SPPL2A |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: KAT6A-SPPL2A | Fusion gene ID: 41217 | Hgene | Tgene | Gene symbol | KAT6A | SPPL2A | Gene ID | 7994 | 84888 |
| Gene name | lysine acetyltransferase 6A | signal peptide peptidase like 2A | |
| Synonyms | ARTHS|MOZ|MRD32|MYST-3|MYST3|RUNXBP2|ZC2HC6A|ZNF220 | IMP3|PSL2 | |
| Cytomap | 8p11.21 | 15q21.2 | |
| Type of gene | protein-coding | protein-coding | |
| Description | histone acetyltransferase KAT6AK(lysine) acetyltransferase 6AMOZ, YBF2/SAS3, SAS2 and TIP60 protein 3MYST histone acetyltransferase (monocytic leukemia) 3histone acetyltransferase MYST3monocytic leukemia zinc finger proteinrunt-related transcription | signal peptide peptidase-like 2AIMP-3SPP-like 2Aintramembrane cleaving proteaseintramembrane protease 3presenilin-like protein 2 | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | Q92794 | Q8TCT8 | |
| Ensembl transtripts involved in fusion gene | ENST00000265713, ENST00000396930, ENST00000406337, ENST00000485568, | ENST00000559293, ENST00000261854, | |
| Fusion gene scores | * DoF score | 26 X 32 X 13=10816 | 11 X 9 X 7=693 |
| # samples | 42 | 11 | |
| ** MAII score | log2(42/10816*10)=-4.68663391861606 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(11/693*10)=-2.65535182861255 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: KAT6A [Title/Abstract] AND SPPL2A [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | KAT6A(41832221)-SPPL2A(51041943), # samples:2 | ||
| Anticipated loss of major functional domain due to fusion event. | KAT6A-SPPL2A seems lost the major protein functional domain in Hgene partner, which is a CGC due to the frame-shifted ORF. KAT6A-SPPL2A seems lost the major protein functional domain in Hgene partner, which is a epigenetic factor due to the frame-shifted ORF. KAT6A-SPPL2A seems lost the major protein functional domain in Hgene partner, which is a essential gene due to the frame-shifted ORF. KAT6A-SPPL2A seems lost the major protein functional domain in Hgene partner, which is a IUPHAR drug target due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | KAT6A | GO:0006473 | protein acetylation | 23431171 |
| Hgene | KAT6A | GO:0016573 | histone acetylation | 11742995|17925393 |
| Hgene | KAT6A | GO:0030099 | myeloid cell differentiation | 11742995 |
| Hgene | KAT6A | GO:0043966 | histone H3 acetylation | 16387653 |
| Hgene | KAT6A | GO:0045892 | negative regulation of transcription, DNA-templated | 11742995 |
| Hgene | KAT6A | GO:0045893 | positive regulation of transcription, DNA-templated | 11742995|11965546|18794358 |
| Tgene | SPPL2A | GO:0006509 | membrane protein ectodomain proteolysis | 16829951|17965014 |
| Tgene | SPPL2A | GO:0031293 | membrane protein intracellular domain proteolysis | 16829951|17557115 |
| Tgene | SPPL2A | GO:0033619 | membrane protein proteolysis | 2313285 |
 Fusion gene breakpoints across KAT6A (5'-gene) Fusion gene breakpoints across KAT6A (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
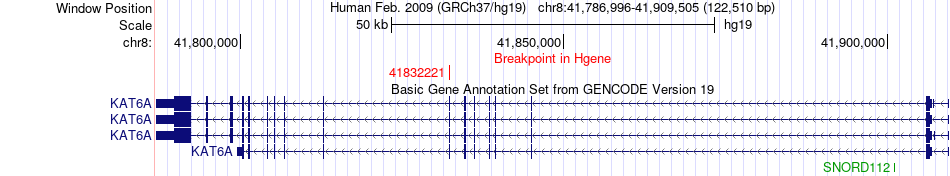 |
 Fusion gene breakpoints across SPPL2A (3'-gene) Fusion gene breakpoints across SPPL2A (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
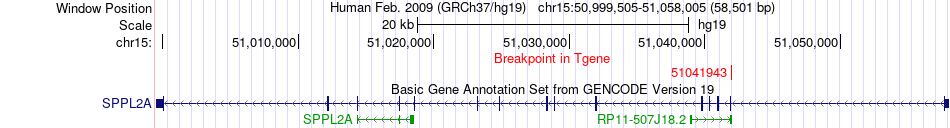 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | ESCA | TCGA-VR-A8EW | KAT6A | chr8 | 41832221 | - | SPPL2A | chr15 | 51041943 | - |
Top |
Fusion Gene ORF analysis for KAT6A-SPPL2A |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-intron | ENST00000265713 | ENST00000559293 | KAT6A | chr8 | 41832221 | - | SPPL2A | chr15 | 51041943 | - |
| 5CDS-intron | ENST00000396930 | ENST00000559293 | KAT6A | chr8 | 41832221 | - | SPPL2A | chr15 | 51041943 | - |
| 5CDS-intron | ENST00000406337 | ENST00000559293 | KAT6A | chr8 | 41832221 | - | SPPL2A | chr15 | 51041943 | - |
| 5CDS-intron | ENST00000485568 | ENST00000559293 | KAT6A | chr8 | 41832221 | - | SPPL2A | chr15 | 51041943 | - |
| Frame-shift | ENST00000406337 | ENST00000261854 | KAT6A | chr8 | 41832221 | - | SPPL2A | chr15 | 51041943 | - |
| In-frame | ENST00000265713 | ENST00000261854 | KAT6A | chr8 | 41832221 | - | SPPL2A | chr15 | 51041943 | - |
| In-frame | ENST00000396930 | ENST00000261854 | KAT6A | chr8 | 41832221 | - | SPPL2A | chr15 | 51041943 | - |
| In-frame | ENST00000485568 | ENST00000261854 | KAT6A | chr8 | 41832221 | - | SPPL2A | chr15 | 51041943 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000265713 | KAT6A | chr8 | 41832221 | - | ENST00000261854 | SPPL2A | chr15 | 51041943 | - | 3882 | 1894 | 412 | 3390 | 992 |
| ENST00000396930 | KAT6A | chr8 | 41832221 | - | ENST00000261854 | SPPL2A | chr15 | 51041943 | - | 4014 | 2026 | 544 | 3522 | 992 |
| ENST00000485568 | KAT6A | chr8 | 41832221 | - | ENST00000261854 | SPPL2A | chr15 | 51041943 | - | 3882 | 1894 | 412 | 3390 | 992 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000265713 | ENST00000261854 | KAT6A | chr8 | 41832221 | - | SPPL2A | chr15 | 51041943 | - | 0.000160429 | 0.99983954 |
| ENST00000396930 | ENST00000261854 | KAT6A | chr8 | 41832221 | - | SPPL2A | chr15 | 51041943 | - | 0.000190752 | 0.9998093 |
| ENST00000485568 | ENST00000261854 | KAT6A | chr8 | 41832221 | - | SPPL2A | chr15 | 51041943 | - | 0.000160429 | 0.99983954 |
Top |
Fusion Genomic Features for KAT6A-SPPL2A |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
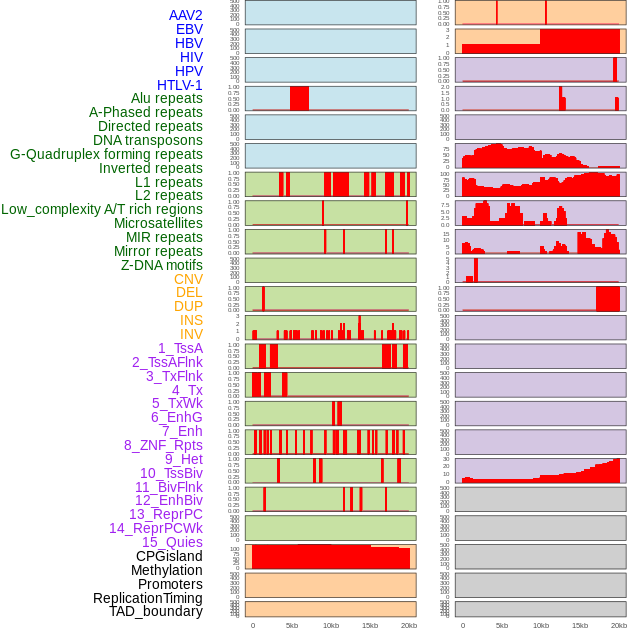 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for KAT6A-SPPL2A |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr8:41832221/chr15:51041943) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| KAT6A | SPPL2A |
| FUNCTION: Histone acetyltransferase that acetylates lysine residues in histone H3 and histone H4 (in vitro). Component of the MOZ/MORF complex which has a histone H3 acetyltransferase activity. May act as a transcriptional coactivator for RUNX1 and RUNX2. Acetylates p53/TP53 at 'Lys-120' and 'Lys-382' and controls its transcriptional activity via association with PML. {ECO:0000269|PubMed:11742995, ECO:0000269|PubMed:11965546, ECO:0000269|PubMed:12771199, ECO:0000269|PubMed:16387653, ECO:0000269|PubMed:17925393, ECO:0000269|PubMed:23431171}. | FUNCTION: Intramembrane-cleaving aspartic protease (I-CLiP) that cleaves type II membrane signal peptides in the hydrophobic plane of the membrane. Functions in FASLG, ITM2B and TNF processing (PubMed:16829952, PubMed:16829951, PubMed:17557115, PubMed:17965014). Catalyzes the intramembrane cleavage of the anchored fragment of shed TNF-alpha (TNF), which promotes the release of the intracellular domain (ICD) for signaling to the nucleus (PubMed:16829952). Also responsible for the intramembrane cleavage of Fas antigen ligand FASLG, which promotes the release of the intracellular FasL domain (FasL ICD) (PubMed:17557115). May play a role in the regulation of innate and adaptive immunity (PubMed:16829952). Catalyzes the intramembrane cleavage of the simian foamy virus envelope glycoprotein gp130 independently of prior ectodomain shedding by furin or furin-like proprotein convertase (PC)-mediated cleavage proteolysis (PubMed:23132852). {ECO:0000269|PubMed:16829951, ECO:0000269|PubMed:16829952, ECO:0000269|PubMed:17557115, ECO:0000269|PubMed:17965014, ECO:0000269|PubMed:23132852}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | KAT6A | chr8:41832221 | chr15:51041943 | ENST00000265713 | - | 8 | 17 | 371_379 | 494 | 2005.0 | Compositional bias | Note=Poly-Ser |
| Hgene | KAT6A | chr8:41832221 | chr15:51041943 | ENST00000396930 | - | 9 | 18 | 371_379 | 494 | 2005.0 | Compositional bias | Note=Poly-Ser |
| Hgene | KAT6A | chr8:41832221 | chr15:51041943 | ENST00000406337 | - | 9 | 18 | 371_379 | 494 | 2005.0 | Compositional bias | Note=Poly-Ser |
| Hgene | KAT6A | chr8:41832221 | chr15:51041943 | ENST00000265713 | - | 8 | 17 | 95_171 | 494 | 2005.0 | Domain | H15 |
| Hgene | KAT6A | chr8:41832221 | chr15:51041943 | ENST00000396930 | - | 9 | 18 | 95_171 | 494 | 2005.0 | Domain | H15 |
| Hgene | KAT6A | chr8:41832221 | chr15:51041943 | ENST00000406337 | - | 9 | 18 | 95_171 | 494 | 2005.0 | Domain | H15 |
| Hgene | KAT6A | chr8:41832221 | chr15:51041943 | ENST00000265713 | - | 8 | 17 | 1_144 | 494 | 2005.0 | Region | Note=Required for activation of RUNX1-1 |
| Hgene | KAT6A | chr8:41832221 | chr15:51041943 | ENST00000265713 | - | 8 | 17 | 52_166 | 494 | 2005.0 | Region | Note=Required for nuclear localization |
| Hgene | KAT6A | chr8:41832221 | chr15:51041943 | ENST00000396930 | - | 9 | 18 | 1_144 | 494 | 2005.0 | Region | Note=Required for activation of RUNX1-1 |
| Hgene | KAT6A | chr8:41832221 | chr15:51041943 | ENST00000396930 | - | 9 | 18 | 52_166 | 494 | 2005.0 | Region | Note=Required for nuclear localization |
| Hgene | KAT6A | chr8:41832221 | chr15:51041943 | ENST00000406337 | - | 9 | 18 | 1_144 | 494 | 2005.0 | Region | Note=Required for activation of RUNX1-1 |
| Hgene | KAT6A | chr8:41832221 | chr15:51041943 | ENST00000406337 | - | 9 | 18 | 52_166 | 494 | 2005.0 | Region | Note=Required for nuclear localization |
| Hgene | KAT6A | chr8:41832221 | chr15:51041943 | ENST00000265713 | - | 8 | 17 | 206_265 | 494 | 2005.0 | Zinc finger | PHD-type 1 |
| Hgene | KAT6A | chr8:41832221 | chr15:51041943 | ENST00000265713 | - | 8 | 17 | 259_313 | 494 | 2005.0 | Zinc finger | PHD-type 2 |
| Hgene | KAT6A | chr8:41832221 | chr15:51041943 | ENST00000396930 | - | 9 | 18 | 206_265 | 494 | 2005.0 | Zinc finger | PHD-type 1 |
| Hgene | KAT6A | chr8:41832221 | chr15:51041943 | ENST00000396930 | - | 9 | 18 | 259_313 | 494 | 2005.0 | Zinc finger | PHD-type 2 |
| Hgene | KAT6A | chr8:41832221 | chr15:51041943 | ENST00000406337 | - | 9 | 18 | 206_265 | 494 | 2005.0 | Zinc finger | PHD-type 1 |
| Hgene | KAT6A | chr8:41832221 | chr15:51041943 | ENST00000406337 | - | 9 | 18 | 259_313 | 494 | 2005.0 | Zinc finger | PHD-type 2 |
| Tgene | SPPL2A | chr8:41832221 | chr15:51041943 | ENST00000261854 | 0 | 15 | 63_151 | 22 | 521.0 | Domain | Note=PA | |
| Tgene | SPPL2A | chr8:41832221 | chr15:51041943 | ENST00000261854 | 0 | 15 | 463_465 | 22 | 521.0 | Motif | Note=PAL | |
| Tgene | SPPL2A | chr8:41832221 | chr15:51041943 | ENST00000261854 | 0 | 15 | 495_498 | 22 | 521.0 | Motif | Note=YXXo lysosomal targeting motif | |
| Tgene | SPPL2A | chr8:41832221 | chr15:51041943 | ENST00000261854 | 0 | 15 | 194_220 | 22 | 521.0 | Topological domain | Cytoplasmic | |
| Tgene | SPPL2A | chr8:41832221 | chr15:51041943 | ENST00000261854 | 0 | 15 | 242_247 | 22 | 521.0 | Topological domain | Lumenal | |
| Tgene | SPPL2A | chr8:41832221 | chr15:51041943 | ENST00000261854 | 0 | 15 | 269_285 | 22 | 521.0 | Topological domain | Cytoplasmic | |
| Tgene | SPPL2A | chr8:41832221 | chr15:51041943 | ENST00000261854 | 0 | 15 | 26_172 | 22 | 521.0 | Topological domain | Lumenal | |
| Tgene | SPPL2A | chr8:41832221 | chr15:51041943 | ENST00000261854 | 0 | 15 | 307_311 | 22 | 521.0 | Topological domain | Lumenal | |
| Tgene | SPPL2A | chr8:41832221 | chr15:51041943 | ENST00000261854 | 0 | 15 | 333_340 | 22 | 521.0 | Topological domain | Cytoplasmic | |
| Tgene | SPPL2A | chr8:41832221 | chr15:51041943 | ENST00000261854 | 0 | 15 | 362_399 | 22 | 521.0 | Topological domain | Lumenal | |
| Tgene | SPPL2A | chr8:41832221 | chr15:51041943 | ENST00000261854 | 0 | 15 | 421_437 | 22 | 521.0 | Topological domain | Cytoplasmic | |
| Tgene | SPPL2A | chr8:41832221 | chr15:51041943 | ENST00000261854 | 0 | 15 | 459_460 | 22 | 521.0 | Topological domain | Lumenal | |
| Tgene | SPPL2A | chr8:41832221 | chr15:51041943 | ENST00000261854 | 0 | 15 | 482_520 | 22 | 521.0 | Topological domain | Cytoplasmic | |
| Tgene | SPPL2A | chr8:41832221 | chr15:51041943 | ENST00000261854 | 0 | 15 | 173_193 | 22 | 521.0 | Transmembrane | Helical | |
| Tgene | SPPL2A | chr8:41832221 | chr15:51041943 | ENST00000261854 | 0 | 15 | 221_241 | 22 | 521.0 | Transmembrane | Helical | |
| Tgene | SPPL2A | chr8:41832221 | chr15:51041943 | ENST00000261854 | 0 | 15 | 248_268 | 22 | 521.0 | Transmembrane | Helical | |
| Tgene | SPPL2A | chr8:41832221 | chr15:51041943 | ENST00000261854 | 0 | 15 | 286_306 | 22 | 521.0 | Transmembrane | Helical | |
| Tgene | SPPL2A | chr8:41832221 | chr15:51041943 | ENST00000261854 | 0 | 15 | 312_332 | 22 | 521.0 | Transmembrane | Helical | |
| Tgene | SPPL2A | chr8:41832221 | chr15:51041943 | ENST00000261854 | 0 | 15 | 341_361 | 22 | 521.0 | Transmembrane | Helical | |
| Tgene | SPPL2A | chr8:41832221 | chr15:51041943 | ENST00000261854 | 0 | 15 | 400_420 | 22 | 521.0 | Transmembrane | Helical | |
| Tgene | SPPL2A | chr8:41832221 | chr15:51041943 | ENST00000261854 | 0 | 15 | 438_458 | 22 | 521.0 | Transmembrane | Helical | |
| Tgene | SPPL2A | chr8:41832221 | chr15:51041943 | ENST00000261854 | 0 | 15 | 461_481 | 22 | 521.0 | Transmembrane | Helical |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | KAT6A | chr8:41832221 | chr15:51041943 | ENST00000265713 | - | 8 | 17 | 1019_1026 | 494 | 2005.0 | Compositional bias | Note=Poly-Arg |
| Hgene | KAT6A | chr8:41832221 | chr15:51041943 | ENST00000265713 | - | 8 | 17 | 1069_1078 | 494 | 2005.0 | Compositional bias | Note=Poly-Glu |
| Hgene | KAT6A | chr8:41832221 | chr15:51041943 | ENST00000265713 | - | 8 | 17 | 1147_1150 | 494 | 2005.0 | Compositional bias | Note=Poly-Lys |
| Hgene | KAT6A | chr8:41832221 | chr15:51041943 | ENST00000265713 | - | 8 | 17 | 1221_1242 | 494 | 2005.0 | Compositional bias | Note=Glu-rich |
| Hgene | KAT6A | chr8:41832221 | chr15:51041943 | ENST00000265713 | - | 8 | 17 | 1267_1302 | 494 | 2005.0 | Compositional bias | Note=Glu-rich |
| Hgene | KAT6A | chr8:41832221 | chr15:51041943 | ENST00000265713 | - | 8 | 17 | 1411_1414 | 494 | 2005.0 | Compositional bias | Note=Poly-Glu |
| Hgene | KAT6A | chr8:41832221 | chr15:51041943 | ENST00000265713 | - | 8 | 17 | 1593_1597 | 494 | 2005.0 | Compositional bias | Note=Poly-Ser |
| Hgene | KAT6A | chr8:41832221 | chr15:51041943 | ENST00000265713 | - | 8 | 17 | 1643_1704 | 494 | 2005.0 | Compositional bias | Note=Gln/Pro-rich |
| Hgene | KAT6A | chr8:41832221 | chr15:51041943 | ENST00000265713 | - | 8 | 17 | 1897_1977 | 494 | 2005.0 | Compositional bias | Note=Met-rich |
| Hgene | KAT6A | chr8:41832221 | chr15:51041943 | ENST00000265713 | - | 8 | 17 | 788_801 | 494 | 2005.0 | Compositional bias | Note=Poly-Glu |
| Hgene | KAT6A | chr8:41832221 | chr15:51041943 | ENST00000265713 | - | 8 | 17 | 989_995 | 494 | 2005.0 | Compositional bias | Note=Poly-Glu |
| Hgene | KAT6A | chr8:41832221 | chr15:51041943 | ENST00000396930 | - | 9 | 18 | 1019_1026 | 494 | 2005.0 | Compositional bias | Note=Poly-Arg |
| Hgene | KAT6A | chr8:41832221 | chr15:51041943 | ENST00000396930 | - | 9 | 18 | 1069_1078 | 494 | 2005.0 | Compositional bias | Note=Poly-Glu |
| Hgene | KAT6A | chr8:41832221 | chr15:51041943 | ENST00000396930 | - | 9 | 18 | 1147_1150 | 494 | 2005.0 | Compositional bias | Note=Poly-Lys |
| Hgene | KAT6A | chr8:41832221 | chr15:51041943 | ENST00000396930 | - | 9 | 18 | 1221_1242 | 494 | 2005.0 | Compositional bias | Note=Glu-rich |
| Hgene | KAT6A | chr8:41832221 | chr15:51041943 | ENST00000396930 | - | 9 | 18 | 1267_1302 | 494 | 2005.0 | Compositional bias | Note=Glu-rich |
| Hgene | KAT6A | chr8:41832221 | chr15:51041943 | ENST00000396930 | - | 9 | 18 | 1411_1414 | 494 | 2005.0 | Compositional bias | Note=Poly-Glu |
| Hgene | KAT6A | chr8:41832221 | chr15:51041943 | ENST00000396930 | - | 9 | 18 | 1593_1597 | 494 | 2005.0 | Compositional bias | Note=Poly-Ser |
| Hgene | KAT6A | chr8:41832221 | chr15:51041943 | ENST00000396930 | - | 9 | 18 | 1643_1704 | 494 | 2005.0 | Compositional bias | Note=Gln/Pro-rich |
| Hgene | KAT6A | chr8:41832221 | chr15:51041943 | ENST00000396930 | - | 9 | 18 | 1897_1977 | 494 | 2005.0 | Compositional bias | Note=Met-rich |
| Hgene | KAT6A | chr8:41832221 | chr15:51041943 | ENST00000396930 | - | 9 | 18 | 788_801 | 494 | 2005.0 | Compositional bias | Note=Poly-Glu |
| Hgene | KAT6A | chr8:41832221 | chr15:51041943 | ENST00000396930 | - | 9 | 18 | 989_995 | 494 | 2005.0 | Compositional bias | Note=Poly-Glu |
| Hgene | KAT6A | chr8:41832221 | chr15:51041943 | ENST00000406337 | - | 9 | 18 | 1019_1026 | 494 | 2005.0 | Compositional bias | Note=Poly-Arg |
| Hgene | KAT6A | chr8:41832221 | chr15:51041943 | ENST00000406337 | - | 9 | 18 | 1069_1078 | 494 | 2005.0 | Compositional bias | Note=Poly-Glu |
| Hgene | KAT6A | chr8:41832221 | chr15:51041943 | ENST00000406337 | - | 9 | 18 | 1147_1150 | 494 | 2005.0 | Compositional bias | Note=Poly-Lys |
| Hgene | KAT6A | chr8:41832221 | chr15:51041943 | ENST00000406337 | - | 9 | 18 | 1221_1242 | 494 | 2005.0 | Compositional bias | Note=Glu-rich |
| Hgene | KAT6A | chr8:41832221 | chr15:51041943 | ENST00000406337 | - | 9 | 18 | 1267_1302 | 494 | 2005.0 | Compositional bias | Note=Glu-rich |
| Hgene | KAT6A | chr8:41832221 | chr15:51041943 | ENST00000406337 | - | 9 | 18 | 1411_1414 | 494 | 2005.0 | Compositional bias | Note=Poly-Glu |
| Hgene | KAT6A | chr8:41832221 | chr15:51041943 | ENST00000406337 | - | 9 | 18 | 1593_1597 | 494 | 2005.0 | Compositional bias | Note=Poly-Ser |
| Hgene | KAT6A | chr8:41832221 | chr15:51041943 | ENST00000406337 | - | 9 | 18 | 1643_1704 | 494 | 2005.0 | Compositional bias | Note=Gln/Pro-rich |
| Hgene | KAT6A | chr8:41832221 | chr15:51041943 | ENST00000406337 | - | 9 | 18 | 1897_1977 | 494 | 2005.0 | Compositional bias | Note=Met-rich |
| Hgene | KAT6A | chr8:41832221 | chr15:51041943 | ENST00000406337 | - | 9 | 18 | 788_801 | 494 | 2005.0 | Compositional bias | Note=Poly-Glu |
| Hgene | KAT6A | chr8:41832221 | chr15:51041943 | ENST00000406337 | - | 9 | 18 | 989_995 | 494 | 2005.0 | Compositional bias | Note=Poly-Glu |
| Hgene | KAT6A | chr8:41832221 | chr15:51041943 | ENST00000265713 | - | 8 | 17 | 504_778 | 494 | 2005.0 | Domain | MYST-type HAT |
| Hgene | KAT6A | chr8:41832221 | chr15:51041943 | ENST00000396930 | - | 9 | 18 | 504_778 | 494 | 2005.0 | Domain | MYST-type HAT |
| Hgene | KAT6A | chr8:41832221 | chr15:51041943 | ENST00000406337 | - | 9 | 18 | 504_778 | 494 | 2005.0 | Domain | MYST-type HAT |
| Hgene | KAT6A | chr8:41832221 | chr15:51041943 | ENST00000265713 | - | 8 | 17 | 1913_1948 | 494 | 2005.0 | Region | Note=Required for activation of RUNX1-2 |
| Hgene | KAT6A | chr8:41832221 | chr15:51041943 | ENST00000265713 | - | 8 | 17 | 488_778 | 494 | 2005.0 | Region | Note=Catalytic |
| Hgene | KAT6A | chr8:41832221 | chr15:51041943 | ENST00000265713 | - | 8 | 17 | 645_649 | 494 | 2005.0 | Region | Acetyl-CoA binding |
| Hgene | KAT6A | chr8:41832221 | chr15:51041943 | ENST00000265713 | - | 8 | 17 | 654_660 | 494 | 2005.0 | Region | Acetyl-CoA binding |
| Hgene | KAT6A | chr8:41832221 | chr15:51041943 | ENST00000396930 | - | 9 | 18 | 1913_1948 | 494 | 2005.0 | Region | Note=Required for activation of RUNX1-2 |
| Hgene | KAT6A | chr8:41832221 | chr15:51041943 | ENST00000396930 | - | 9 | 18 | 488_778 | 494 | 2005.0 | Region | Note=Catalytic |
| Hgene | KAT6A | chr8:41832221 | chr15:51041943 | ENST00000396930 | - | 9 | 18 | 645_649 | 494 | 2005.0 | Region | Acetyl-CoA binding |
| Hgene | KAT6A | chr8:41832221 | chr15:51041943 | ENST00000396930 | - | 9 | 18 | 654_660 | 494 | 2005.0 | Region | Acetyl-CoA binding |
| Hgene | KAT6A | chr8:41832221 | chr15:51041943 | ENST00000406337 | - | 9 | 18 | 1913_1948 | 494 | 2005.0 | Region | Note=Required for activation of RUNX1-2 |
| Hgene | KAT6A | chr8:41832221 | chr15:51041943 | ENST00000406337 | - | 9 | 18 | 488_778 | 494 | 2005.0 | Region | Note=Catalytic |
| Hgene | KAT6A | chr8:41832221 | chr15:51041943 | ENST00000406337 | - | 9 | 18 | 645_649 | 494 | 2005.0 | Region | Acetyl-CoA binding |
| Hgene | KAT6A | chr8:41832221 | chr15:51041943 | ENST00000406337 | - | 9 | 18 | 654_660 | 494 | 2005.0 | Region | Acetyl-CoA binding |
| Hgene | KAT6A | chr8:41832221 | chr15:51041943 | ENST00000265713 | - | 8 | 17 | 537_562 | 494 | 2005.0 | Zinc finger | C2HC MYST-type |
| Hgene | KAT6A | chr8:41832221 | chr15:51041943 | ENST00000396930 | - | 9 | 18 | 537_562 | 494 | 2005.0 | Zinc finger | C2HC MYST-type |
| Hgene | KAT6A | chr8:41832221 | chr15:51041943 | ENST00000406337 | - | 9 | 18 | 537_562 | 494 | 2005.0 | Zinc finger | C2HC MYST-type |
Top |
Fusion Gene Sequence for KAT6A-SPPL2A |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >41217_41217_1_KAT6A-SPPL2A_KAT6A_chr8_41832221_ENST00000265713_SPPL2A_chr15_51041943_ENST00000261854_length(transcript)=3882nt_BP=1894nt ACTGTCTCCAAGATGGCGGCCGTGTCAGTTTGGGGCATCTCCGCGGTCCGGCCCGGGGCCCCGGGATCTCGGCTGTCCTTCCTCCCGGCA TAAGATGCACATTTTTCTGCTCTGGAGCCGGGAATGAAATATTCTTGAGTTCTTACAACTTTATGACGAGACCCATGTGTGGTGCTATTG AGAAATTCATTGGGAAGTTGGAAGACATTTCAATCAACAGGTTGTTTTGGTTTCTATAGTACAATTGGGGTGGCATTCTGTTTTGTGAAA GGAGGAAGGACTTAGGCCAGAAAACTCATATGCTATGGTTAACTGGTTCCCAGCCTCCGAGAATCTTGTTTTCCATGGTGTAAAACTTAC TCAGCATCAGGATAAGGGATAACGACTCTATGGATATACAGAATCCTTCACCATGGTAAAACTCGCAAACCCGCTTTATACTGAGTGGAT TTTGGAGGCCATCAAAAAAGTGAAAAAGCAGAAACAGCGTCCTTCAGAAGAAAGGATATGCAATGCTGTGTCTTCATCCCATGGCTTGGA TCGTAAAACTGTTTTAGAACAATTGGAGTTGAGTGTTAAAGATGGAACAATTTTAAAAGTCTCAAATAAAGGACTCAATTCCTATAAAGA TCCTGATAATCCTGGGCGAATAGCACTTCCTAAGCCTCGGAACCATGGAAAATTGGATAATAAACAAAATGTGGATTGGAATAAACTGAT AAAGCGGGCAGTTGAGGGCTTGGCAGAGTCTGGTGGCTCAACTTTGAAAAGCATTGAACGTTTTTTGAAAGGTCAGAAGGATGTGTCTGC ATTATTCGGAGGCAGTGCTGCCTCTGGCTTTCACCAGCAGTTACGATTGGCTATCAAACGTGCCATTGGCCACGGCAGACTCCTTAAAGA TGGACCTCTTTATCGGCTCAACACTAAAGCAACCAACGTGGATGGGAAAGAGAGTTGTGAGTCTCTTTCCTGTTTACCTCCAGTGTCCCT TCTTCCACATGAAAAGGATAAGCCGGTTGCTGAACCAATCCCCATCTGTAGTTTCTGTCTTGGTACAAAAGAACAAAACCGAGAAAAGAA GCCAGAGGAACTCATCTCCTGTGCCGACTGTGGCAACAGTGGCCATCCATCCTGTTTAAAGTTTTCCCCTGAACTAACGGTTCGAGTGAA GGCCTTACGGTGGCAGTGCATCGAGTGTAAAACATGCAGCTCCTGTCGAGATCAAGGCAAAAATGCGGATAACATGCTCTTTTGTGATTC ATGTGACCGAGGTTTTCACATGGAGTGTTGTGATCCGCCACTCACCCGTATGCCAAAAGGCATGTGGATATGTCAAATATGTCGACCTAG GAAAAAAGGACGAAAACTTCTACAAAAGAAGGCAGCACAGATAAAACGGCGCTATACTAATCCAATAGGACGTCCAAAAAACAGGTTAAA GAAACAAAACACGGTATCAAAAGGTCCCTTCAGCAAAGTTCGAACTGGCCCTGGAAGGGGTAGGAAACGAAAAATCACTCTTTCCAGCCA ATCAGCATCATCATCATCAGAAGAAGGATATTTAGAGCGGATAGATGGCTTGGACTTCTGCAGAGATAGCAATGTCTCCTTGAAGTTCAA CAAGAAAACCAAAGGGCTCATTGATGGCCTTACCAAATTTTTTACCCCTTCCCCTGATGGGCGGAAAGCTCGGGGGGAAGTGGTGGACTA CTCTGAGCAATATCGAATCAGAAAGAGGGGCAACAGGAAATCAAGCACTTCAGATTGGCCCACAGACAATCAGGATGGCTGGGATGGCAA ACAAGAAAATGAGGAGCGACTTTTTGGGAGCCAGGAAATCATGACTGAGAAAGATATGGAATTATTTCGTGATATCCAAGAACAAGCACT GCAGACAGCCGCTCAGGAAGCAATCTTGCATGCGTCTGGAAATGGCACAACCAAGGACTACTGCATGCTTTATAACCCTTATTGGACAGC TCTTCCAAGTACCCTAGAAAATGCAACTTCCATTAGTTTGATGAATCTGACTTCCACACCACTATGCAACCTTTCTGATATTCCTCCTGT TGGCATAAAGAGCAAAGCAGTTGTGGTTCCATGGGGAAGCTGCCATTTTCTTGAAAAAGCCAGAATTGCACAGAAAGGAGGTGCTGAAGC AATGTTAGTTGTCAATAACAGTGTCCTATTTCCTCCCTCAGGTAACAGATCTGAATTTCCTGATGTGAAAATACTGATTGCATTTATAAG CTACAAAGACTTTAGAGATATGAACCAGACTCTAGGAGATAACATTACTGTGAAAATGTATTCTCCATCGTGGCCTAACTTTGATTATAC TATGGTGGTTATTTTTGTAATTGCGGTGTTCACTGTGGCATTAGGTGGATACTGGAGTGGACTAGTTGAATTGGAAAACTTGAAAGCAGT GACAACTGAAGATAGAGAAATGAGGAAAAAGAAGGAAGAATATTTAACTTTTAGTCCTCTTACAGTTGTAATATTTGTGGTCATCTGCTG TGTTATGATGGTCTTACTTTATTTCTTCTACAAATGGTTGGTTTATGTTATGATAGCAATTTTCTGCATAGCATCAGCAATGAGTCTGTA CAACTGTCTTGCTGCACTAATTCATAAGATACCATATGGACAATGCACGATTGCATGTCGTGGCAAAAACATGGAAGTGAGACTTATTTT TCTCTCTGGACTGTGCATAGCAGTAGCTGTTGTTTGGGCTGTGTTTCGAAATGAAGACAGGTGGGCTTGGATTTTACAGGATATCTTGGG GATTGCTTTCTGTCTGAATTTAATTAAAACACTGAAGTTGCCCAACTTCAAGTCATGTGTGATACTTCTAGGCCTTCTCCTCCTCTATGA TGTATTTTTTGTTTTCATAACACCATTCATCACAAAGAATGGTGAGAGTATCATGGTTGAACTCGCAGCTGGACCTTTTGGAAATAATGA AAAGTTGCCAGTAGTCATCAGAGTACCAAAACTGATCTATTTCTCAGTAATGAGTGTGTGCCTCATGCCTGTTTCAATATTGGGTTTTGG AGACATTATTGTACCAGGCCTGTTGATTGCATACTGTAGAAGATTTGATGTTCAGACTGGTTCTTCTTACATATACTATGTTTCGTCTAC AGTTGCCTATGCTATTGGCATGATACTTACATTTGTTGTTCTGGTGCTGATGAAAAAGGGGCAACCTGCTCTCCTCTATTTAGTACCTTG CACACTTATTACTGCCTCAGTTGTTGCCTGGAGACGTAAGGAAATGAAAAAGTTCTGGAAAGGTAACAGCTATCAGATGATGGACCATTT GGATTGTGCAACAAATGAAGAAAACCCTGTGATATCTGGTGAACAGATTGTCCAGCAATAATATTATGTGGAACTGCTATAATGTGTCAT TGATTTTCTACAAATAGACTTCGACTTTTTAAATTGACTTTTGAATTGACAATCTGAAAGAGTCTTCAATGATATGCTTGCAAAAATATA TTTTTATGAGCTGGTACTGACAGTTACATCATAAATAACTAAAACGCTTTGCTTTTAATGTTAAAGTTGTGCCTTCACATTAAATAAAAC ATATGGTCTGTGTAGTTTCCGAGATGTACTATATACAGTATATTTTTCTAAAAAAAAATGCCTTGACCCACCCCTATACTTTTTTTTAAC GAAATCTGTTTCTTTCAGCTCTGATGGTAGATACAAGTAGTTACTATTTCTGAATGGGTTTTACTGTAAAGAATTATGCAAATTGGCCAG GCGCGGTGGCTCACGCCTGTAATCCCAACACTTTGGGAGGCCAAGGCGGGCGGATCACGAGGTCAGGAGATCGAGACCATCCTGGCTAAC >41217_41217_1_KAT6A-SPPL2A_KAT6A_chr8_41832221_ENST00000265713_SPPL2A_chr15_51041943_ENST00000261854_length(amino acids)=992AA_BP=494 MVKLANPLYTEWILEAIKKVKKQKQRPSEERICNAVSSSHGLDRKTVLEQLELSVKDGTILKVSNKGLNSYKDPDNPGRIALPKPRNHGK LDNKQNVDWNKLIKRAVEGLAESGGSTLKSIERFLKGQKDVSALFGGSAASGFHQQLRLAIKRAIGHGRLLKDGPLYRLNTKATNVDGKE SCESLSCLPPVSLLPHEKDKPVAEPIPICSFCLGTKEQNREKKPEELISCADCGNSGHPSCLKFSPELTVRVKALRWQCIECKTCSSCRD QGKNADNMLFCDSCDRGFHMECCDPPLTRMPKGMWICQICRPRKKGRKLLQKKAAQIKRRYTNPIGRPKNRLKKQNTVSKGPFSKVRTGP GRGRKRKITLSSQSASSSSEEGYLERIDGLDFCRDSNVSLKFNKKTKGLIDGLTKFFTPSPDGRKARGEVVDYSEQYRIRKRGNRKSSTS DWPTDNQDGWDGKQENEERLFGSQEIMTEKDMELFRDIQEQALQTAAQEAILHASGNGTTKDYCMLYNPYWTALPSTLENATSISLMNLT STPLCNLSDIPPVGIKSKAVVVPWGSCHFLEKARIAQKGGAEAMLVVNNSVLFPPSGNRSEFPDVKILIAFISYKDFRDMNQTLGDNITV KMYSPSWPNFDYTMVVIFVIAVFTVALGGYWSGLVELENLKAVTTEDREMRKKKEEYLTFSPLTVVIFVVICCVMMVLLYFFYKWLVYVM IAIFCIASAMSLYNCLAALIHKIPYGQCTIACRGKNMEVRLIFLSGLCIAVAVVWAVFRNEDRWAWILQDILGIAFCLNLIKTLKLPNFK SCVILLGLLLLYDVFFVFITPFITKNGESIMVELAAGPFGNNEKLPVVIRVPKLIYFSVMSVCLMPVSILGFGDIIVPGLLIAYCRRFDV QTGSSYIYYVSSTVAYAIGMILTFVVLVLMKKGQPALLYLVPCTLITASVVAWRRKEMKKFWKGNSYQMMDHLDCATNEENPVISGEQIV -------------------------------------------------------------- >41217_41217_2_KAT6A-SPPL2A_KAT6A_chr8_41832221_ENST00000396930_SPPL2A_chr15_51041943_ENST00000261854_length(transcript)=4014nt_BP=2026nt ACTGTCTCCAAGATGGCGGCCGTGTCAGTTTGGGGCATCTCCGCGGTCCGGCCCGGGGCCCCGGGATCTCGGCTGTCCTTCCTCCCGGAT TCTTTCTACTAATCCAGATACTTGTTGAAGTGCTGACTAGTTTCTTGGGGAAAGATGTCAAGGAAGAGTTGAAATTCCTAGACTGGTGAG CAGAGATTGCGGAGTACAGAATAGCCATCACCCACTCTGGCATAAGATGCACATTTTTCTGCTCTGGAGCCGGGAATGAAATATTCTTGA GTTCTTACAACTTTATGACGAGACCCATGTGTGGTGCTATTGAGAAATTCATTGGGAAGTTGGAAGACATTTCAATCAACAGGTTGTTTT GGTTTCTATAGTACAATTGGGGTGGCATTCTGTTTTGTGAAAGGAGGAAGGACTTAGGCCAGAAAACTCATATGCTATGGTTAACTGGTT CCCAGCCTCCGAGAATCTTGTTTTCCATGGTGTAAAACTTACTCAGCATCAGGATAAGGGATAACGACTCTATGGATATACAGAATCCTT CACCATGGTAAAACTCGCAAACCCGCTTTATACTGAGTGGATTTTGGAGGCCATCAAAAAAGTGAAAAAGCAGAAACAGCGTCCTTCAGA AGAAAGGATATGCAATGCTGTGTCTTCATCCCATGGCTTGGATCGTAAAACTGTTTTAGAACAATTGGAGTTGAGTGTTAAAGATGGAAC AATTTTAAAAGTCTCAAATAAAGGACTCAATTCCTATAAAGATCCTGATAATCCTGGGCGAATAGCACTTCCTAAGCCTCGGAACCATGG AAAATTGGATAATAAACAAAATGTGGATTGGAATAAACTGATAAAGCGGGCAGTTGAGGGCTTGGCAGAGTCTGGTGGCTCAACTTTGAA AAGCATTGAACGTTTTTTGAAAGGTCAGAAGGATGTGTCTGCATTATTCGGAGGCAGTGCTGCCTCTGGCTTTCACCAGCAGTTACGATT GGCTATCAAACGTGCCATTGGCCACGGCAGACTCCTTAAAGATGGACCTCTTTATCGGCTCAACACTAAAGCAACCAACGTGGATGGGAA AGAGAGTTGTGAGTCTCTTTCCTGTTTACCTCCAGTGTCCCTTCTTCCACATGAAAAGGATAAGCCGGTTGCTGAACCAATCCCCATCTG TAGTTTCTGTCTTGGTACAAAAGAACAAAACCGAGAAAAGAAGCCAGAGGAACTCATCTCCTGTGCCGACTGTGGCAACAGTGGCCATCC ATCCTGTTTAAAGTTTTCCCCTGAACTAACGGTTCGAGTGAAGGCCTTACGGTGGCAGTGCATCGAGTGTAAAACATGCAGCTCCTGTCG AGATCAAGGCAAAAATGCGGATAACATGCTCTTTTGTGATTCATGTGACCGAGGTTTTCACATGGAGTGTTGTGATCCGCCACTCACCCG TATGCCAAAAGGCATGTGGATATGTCAAATATGTCGACCTAGGAAAAAAGGACGAAAACTTCTACAAAAGAAGGCAGCACAGATAAAACG GCGCTATACTAATCCAATAGGACGTCCAAAAAACAGGTTAAAGAAACAAAACACGGTATCAAAAGGTCCCTTCAGCAAAGTTCGAACTGG CCCTGGAAGGGGTAGGAAACGAAAAATCACTCTTTCCAGCCAATCAGCATCATCATCATCAGAAGAAGGATATTTAGAGCGGATAGATGG CTTGGACTTCTGCAGAGATAGCAATGTCTCCTTGAAGTTCAACAAGAAAACCAAAGGGCTCATTGATGGCCTTACCAAATTTTTTACCCC TTCCCCTGATGGGCGGAAAGCTCGGGGGGAAGTGGTGGACTACTCTGAGCAATATCGAATCAGAAAGAGGGGCAACAGGAAATCAAGCAC TTCAGATTGGCCCACAGACAATCAGGATGGCTGGGATGGCAAACAAGAAAATGAGGAGCGACTTTTTGGGAGCCAGGAAATCATGACTGA GAAAGATATGGAATTATTTCGTGATATCCAAGAACAAGCACTGCAGACAGCCGCTCAGGAAGCAATCTTGCATGCGTCTGGAAATGGCAC AACCAAGGACTACTGCATGCTTTATAACCCTTATTGGACAGCTCTTCCAAGTACCCTAGAAAATGCAACTTCCATTAGTTTGATGAATCT GACTTCCACACCACTATGCAACCTTTCTGATATTCCTCCTGTTGGCATAAAGAGCAAAGCAGTTGTGGTTCCATGGGGAAGCTGCCATTT TCTTGAAAAAGCCAGAATTGCACAGAAAGGAGGTGCTGAAGCAATGTTAGTTGTCAATAACAGTGTCCTATTTCCTCCCTCAGGTAACAG ATCTGAATTTCCTGATGTGAAAATACTGATTGCATTTATAAGCTACAAAGACTTTAGAGATATGAACCAGACTCTAGGAGATAACATTAC TGTGAAAATGTATTCTCCATCGTGGCCTAACTTTGATTATACTATGGTGGTTATTTTTGTAATTGCGGTGTTCACTGTGGCATTAGGTGG ATACTGGAGTGGACTAGTTGAATTGGAAAACTTGAAAGCAGTGACAACTGAAGATAGAGAAATGAGGAAAAAGAAGGAAGAATATTTAAC TTTTAGTCCTCTTACAGTTGTAATATTTGTGGTCATCTGCTGTGTTATGATGGTCTTACTTTATTTCTTCTACAAATGGTTGGTTTATGT TATGATAGCAATTTTCTGCATAGCATCAGCAATGAGTCTGTACAACTGTCTTGCTGCACTAATTCATAAGATACCATATGGACAATGCAC GATTGCATGTCGTGGCAAAAACATGGAAGTGAGACTTATTTTTCTCTCTGGACTGTGCATAGCAGTAGCTGTTGTTTGGGCTGTGTTTCG AAATGAAGACAGGTGGGCTTGGATTTTACAGGATATCTTGGGGATTGCTTTCTGTCTGAATTTAATTAAAACACTGAAGTTGCCCAACTT CAAGTCATGTGTGATACTTCTAGGCCTTCTCCTCCTCTATGATGTATTTTTTGTTTTCATAACACCATTCATCACAAAGAATGGTGAGAG TATCATGGTTGAACTCGCAGCTGGACCTTTTGGAAATAATGAAAAGTTGCCAGTAGTCATCAGAGTACCAAAACTGATCTATTTCTCAGT AATGAGTGTGTGCCTCATGCCTGTTTCAATATTGGGTTTTGGAGACATTATTGTACCAGGCCTGTTGATTGCATACTGTAGAAGATTTGA TGTTCAGACTGGTTCTTCTTACATATACTATGTTTCGTCTACAGTTGCCTATGCTATTGGCATGATACTTACATTTGTTGTTCTGGTGCT GATGAAAAAGGGGCAACCTGCTCTCCTCTATTTAGTACCTTGCACACTTATTACTGCCTCAGTTGTTGCCTGGAGACGTAAGGAAATGAA AAAGTTCTGGAAAGGTAACAGCTATCAGATGATGGACCATTTGGATTGTGCAACAAATGAAGAAAACCCTGTGATATCTGGTGAACAGAT TGTCCAGCAATAATATTATGTGGAACTGCTATAATGTGTCATTGATTTTCTACAAATAGACTTCGACTTTTTAAATTGACTTTTGAATTG ACAATCTGAAAGAGTCTTCAATGATATGCTTGCAAAAATATATTTTTATGAGCTGGTACTGACAGTTACATCATAAATAACTAAAACGCT TTGCTTTTAATGTTAAAGTTGTGCCTTCACATTAAATAAAACATATGGTCTGTGTAGTTTCCGAGATGTACTATATACAGTATATTTTTC TAAAAAAAAATGCCTTGACCCACCCCTATACTTTTTTTTAACGAAATCTGTTTCTTTCAGCTCTGATGGTAGATACAAGTAGTTACTATT TCTGAATGGGTTTTACTGTAAAGAATTATGCAAATTGGCCAGGCGCGGTGGCTCACGCCTGTAATCCCAACACTTTGGGAGGCCAAGGCG >41217_41217_2_KAT6A-SPPL2A_KAT6A_chr8_41832221_ENST00000396930_SPPL2A_chr15_51041943_ENST00000261854_length(amino acids)=992AA_BP=494 MVKLANPLYTEWILEAIKKVKKQKQRPSEERICNAVSSSHGLDRKTVLEQLELSVKDGTILKVSNKGLNSYKDPDNPGRIALPKPRNHGK LDNKQNVDWNKLIKRAVEGLAESGGSTLKSIERFLKGQKDVSALFGGSAASGFHQQLRLAIKRAIGHGRLLKDGPLYRLNTKATNVDGKE SCESLSCLPPVSLLPHEKDKPVAEPIPICSFCLGTKEQNREKKPEELISCADCGNSGHPSCLKFSPELTVRVKALRWQCIECKTCSSCRD QGKNADNMLFCDSCDRGFHMECCDPPLTRMPKGMWICQICRPRKKGRKLLQKKAAQIKRRYTNPIGRPKNRLKKQNTVSKGPFSKVRTGP GRGRKRKITLSSQSASSSSEEGYLERIDGLDFCRDSNVSLKFNKKTKGLIDGLTKFFTPSPDGRKARGEVVDYSEQYRIRKRGNRKSSTS DWPTDNQDGWDGKQENEERLFGSQEIMTEKDMELFRDIQEQALQTAAQEAILHASGNGTTKDYCMLYNPYWTALPSTLENATSISLMNLT STPLCNLSDIPPVGIKSKAVVVPWGSCHFLEKARIAQKGGAEAMLVVNNSVLFPPSGNRSEFPDVKILIAFISYKDFRDMNQTLGDNITV KMYSPSWPNFDYTMVVIFVIAVFTVALGGYWSGLVELENLKAVTTEDREMRKKKEEYLTFSPLTVVIFVVICCVMMVLLYFFYKWLVYVM IAIFCIASAMSLYNCLAALIHKIPYGQCTIACRGKNMEVRLIFLSGLCIAVAVVWAVFRNEDRWAWILQDILGIAFCLNLIKTLKLPNFK SCVILLGLLLLYDVFFVFITPFITKNGESIMVELAAGPFGNNEKLPVVIRVPKLIYFSVMSVCLMPVSILGFGDIIVPGLLIAYCRRFDV QTGSSYIYYVSSTVAYAIGMILTFVVLVLMKKGQPALLYLVPCTLITASVVAWRRKEMKKFWKGNSYQMMDHLDCATNEENPVISGEQIV -------------------------------------------------------------- >41217_41217_3_KAT6A-SPPL2A_KAT6A_chr8_41832221_ENST00000485568_SPPL2A_chr15_51041943_ENST00000261854_length(transcript)=3882nt_BP=1894nt ACTGTCTCCAAGATGGCGGCCGTGTCAGTTTGGGGCATCTCCGCGGTCCGGCCCGGGGCCCCGGGATCTCGGCTGTCCTTCCTCCCGGCA TAAGATGCACATTTTTCTGCTCTGGAGCCGGGAATGAAATATTCTTGAGTTCTTACAACTTTATGACGAGACCCATGTGTGGTGCTATTG AGAAATTCATTGGGAAGTTGGAAGACATTTCAATCAACAGGTTGTTTTGGTTTCTATAGTACAATTGGGGTGGCATTCTGTTTTGTGAAA GGAGGAAGGACTTAGGCCAGAAAACTCATATGCTATGGTTAACTGGTTCCCAGCCTCCGAGAATCTTGTTTTCCATGGTGTAAAACTTAC TCAGCATCAGGATAAGGGATAACGACTCTATGGATATACAGAATCCTTCACCATGGTAAAACTCGCAAACCCGCTTTATACTGAGTGGAT TTTGGAGGCCATCAAAAAAGTGAAAAAGCAGAAACAGCGTCCTTCAGAAGAAAGGATATGCAATGCTGTGTCTTCATCCCATGGCTTGGA TCGTAAAACTGTTTTAGAACAATTGGAGTTGAGTGTTAAAGATGGAACAATTTTAAAAGTCTCAAATAAAGGACTCAATTCCTATAAAGA TCCTGATAATCCTGGGCGAATAGCACTTCCTAAGCCTCGGAACCATGGAAAATTGGATAATAAACAAAATGTGGATTGGAATAAACTGAT AAAGCGGGCAGTTGAGGGCTTGGCAGAGTCTGGTGGCTCAACTTTGAAAAGCATTGAACGTTTTTTGAAAGGTCAGAAGGATGTGTCTGC ATTATTCGGAGGCAGTGCTGCCTCTGGCTTTCACCAGCAGTTACGATTGGCTATCAAACGTGCCATTGGCCACGGCAGACTCCTTAAAGA TGGACCTCTTTATCGGCTCAACACTAAAGCAACCAACGTGGATGGGAAAGAGAGTTGTGAGTCTCTTTCCTGTTTACCTCCAGTGTCCCT TCTTCCACATGAAAAGGATAAGCCGGTTGCTGAACCAATCCCCATCTGTAGTTTCTGTCTTGGTACAAAAGAACAAAACCGAGAAAAGAA GCCAGAGGAACTCATCTCCTGTGCCGACTGTGGCAACAGTGGCCATCCATCCTGTTTAAAGTTTTCCCCTGAACTAACGGTTCGAGTGAA GGCCTTACGGTGGCAGTGCATCGAGTGTAAAACATGCAGCTCCTGTCGAGATCAAGGCAAAAATGCGGATAACATGCTCTTTTGTGATTC ATGTGACCGAGGTTTTCACATGGAGTGTTGTGATCCGCCACTCACCCGTATGCCAAAAGGCATGTGGATATGTCAAATATGTCGACCTAG GAAAAAAGGACGAAAACTTCTACAAAAGAAGGCAGCACAGATAAAACGGCGCTATACTAATCCAATAGGACGTCCAAAAAACAGGTTAAA GAAACAAAACACGGTATCAAAAGGTCCCTTCAGCAAAGTTCGAACTGGCCCTGGAAGGGGTAGGAAACGAAAAATCACTCTTTCCAGCCA ATCAGCATCATCATCATCAGAAGAAGGATATTTAGAGCGGATAGATGGCTTGGACTTCTGCAGAGATAGCAATGTCTCCTTGAAGTTCAA CAAGAAAACCAAAGGGCTCATTGATGGCCTTACCAAATTTTTTACCCCTTCCCCTGATGGGCGGAAAGCTCGGGGGGAAGTGGTGGACTA CTCTGAGCAATATCGAATCAGAAAGAGGGGCAACAGGAAATCAAGCACTTCAGATTGGCCCACAGACAATCAGGATGGCTGGGATGGCAA ACAAGAAAATGAGGAGCGACTTTTTGGGAGCCAGGAAATCATGACTGAGAAAGATATGGAATTATTTCGTGATATCCAAGAACAAGCACT GCAGACAGCCGCTCAGGAAGCAATCTTGCATGCGTCTGGAAATGGCACAACCAAGGACTACTGCATGCTTTATAACCCTTATTGGACAGC TCTTCCAAGTACCCTAGAAAATGCAACTTCCATTAGTTTGATGAATCTGACTTCCACACCACTATGCAACCTTTCTGATATTCCTCCTGT TGGCATAAAGAGCAAAGCAGTTGTGGTTCCATGGGGAAGCTGCCATTTTCTTGAAAAAGCCAGAATTGCACAGAAAGGAGGTGCTGAAGC AATGTTAGTTGTCAATAACAGTGTCCTATTTCCTCCCTCAGGTAACAGATCTGAATTTCCTGATGTGAAAATACTGATTGCATTTATAAG CTACAAAGACTTTAGAGATATGAACCAGACTCTAGGAGATAACATTACTGTGAAAATGTATTCTCCATCGTGGCCTAACTTTGATTATAC TATGGTGGTTATTTTTGTAATTGCGGTGTTCACTGTGGCATTAGGTGGATACTGGAGTGGACTAGTTGAATTGGAAAACTTGAAAGCAGT GACAACTGAAGATAGAGAAATGAGGAAAAAGAAGGAAGAATATTTAACTTTTAGTCCTCTTACAGTTGTAATATTTGTGGTCATCTGCTG TGTTATGATGGTCTTACTTTATTTCTTCTACAAATGGTTGGTTTATGTTATGATAGCAATTTTCTGCATAGCATCAGCAATGAGTCTGTA CAACTGTCTTGCTGCACTAATTCATAAGATACCATATGGACAATGCACGATTGCATGTCGTGGCAAAAACATGGAAGTGAGACTTATTTT TCTCTCTGGACTGTGCATAGCAGTAGCTGTTGTTTGGGCTGTGTTTCGAAATGAAGACAGGTGGGCTTGGATTTTACAGGATATCTTGGG GATTGCTTTCTGTCTGAATTTAATTAAAACACTGAAGTTGCCCAACTTCAAGTCATGTGTGATACTTCTAGGCCTTCTCCTCCTCTATGA TGTATTTTTTGTTTTCATAACACCATTCATCACAAAGAATGGTGAGAGTATCATGGTTGAACTCGCAGCTGGACCTTTTGGAAATAATGA AAAGTTGCCAGTAGTCATCAGAGTACCAAAACTGATCTATTTCTCAGTAATGAGTGTGTGCCTCATGCCTGTTTCAATATTGGGTTTTGG AGACATTATTGTACCAGGCCTGTTGATTGCATACTGTAGAAGATTTGATGTTCAGACTGGTTCTTCTTACATATACTATGTTTCGTCTAC AGTTGCCTATGCTATTGGCATGATACTTACATTTGTTGTTCTGGTGCTGATGAAAAAGGGGCAACCTGCTCTCCTCTATTTAGTACCTTG CACACTTATTACTGCCTCAGTTGTTGCCTGGAGACGTAAGGAAATGAAAAAGTTCTGGAAAGGTAACAGCTATCAGATGATGGACCATTT GGATTGTGCAACAAATGAAGAAAACCCTGTGATATCTGGTGAACAGATTGTCCAGCAATAATATTATGTGGAACTGCTATAATGTGTCAT TGATTTTCTACAAATAGACTTCGACTTTTTAAATTGACTTTTGAATTGACAATCTGAAAGAGTCTTCAATGATATGCTTGCAAAAATATA TTTTTATGAGCTGGTACTGACAGTTACATCATAAATAACTAAAACGCTTTGCTTTTAATGTTAAAGTTGTGCCTTCACATTAAATAAAAC ATATGGTCTGTGTAGTTTCCGAGATGTACTATATACAGTATATTTTTCTAAAAAAAAATGCCTTGACCCACCCCTATACTTTTTTTTAAC GAAATCTGTTTCTTTCAGCTCTGATGGTAGATACAAGTAGTTACTATTTCTGAATGGGTTTTACTGTAAAGAATTATGCAAATTGGCCAG GCGCGGTGGCTCACGCCTGTAATCCCAACACTTTGGGAGGCCAAGGCGGGCGGATCACGAGGTCAGGAGATCGAGACCATCCTGGCTAAC >41217_41217_3_KAT6A-SPPL2A_KAT6A_chr8_41832221_ENST00000485568_SPPL2A_chr15_51041943_ENST00000261854_length(amino acids)=992AA_BP=494 MVKLANPLYTEWILEAIKKVKKQKQRPSEERICNAVSSSHGLDRKTVLEQLELSVKDGTILKVSNKGLNSYKDPDNPGRIALPKPRNHGK LDNKQNVDWNKLIKRAVEGLAESGGSTLKSIERFLKGQKDVSALFGGSAASGFHQQLRLAIKRAIGHGRLLKDGPLYRLNTKATNVDGKE SCESLSCLPPVSLLPHEKDKPVAEPIPICSFCLGTKEQNREKKPEELISCADCGNSGHPSCLKFSPELTVRVKALRWQCIECKTCSSCRD QGKNADNMLFCDSCDRGFHMECCDPPLTRMPKGMWICQICRPRKKGRKLLQKKAAQIKRRYTNPIGRPKNRLKKQNTVSKGPFSKVRTGP GRGRKRKITLSSQSASSSSEEGYLERIDGLDFCRDSNVSLKFNKKTKGLIDGLTKFFTPSPDGRKARGEVVDYSEQYRIRKRGNRKSSTS DWPTDNQDGWDGKQENEERLFGSQEIMTEKDMELFRDIQEQALQTAAQEAILHASGNGTTKDYCMLYNPYWTALPSTLENATSISLMNLT STPLCNLSDIPPVGIKSKAVVVPWGSCHFLEKARIAQKGGAEAMLVVNNSVLFPPSGNRSEFPDVKILIAFISYKDFRDMNQTLGDNITV KMYSPSWPNFDYTMVVIFVIAVFTVALGGYWSGLVELENLKAVTTEDREMRKKKEEYLTFSPLTVVIFVVICCVMMVLLYFFYKWLVYVM IAIFCIASAMSLYNCLAALIHKIPYGQCTIACRGKNMEVRLIFLSGLCIAVAVVWAVFRNEDRWAWILQDILGIAFCLNLIKTLKLPNFK SCVILLGLLLLYDVFFVFITPFITKNGESIMVELAAGPFGNNEKLPVVIRVPKLIYFSVMSVCLMPVSILGFGDIIVPGLLIAYCRRFDV QTGSSYIYYVSSTVAYAIGMILTFVVLVLMKKGQPALLYLVPCTLITASVVAWRRKEMKKFWKGNSYQMMDHLDCATNEENPVISGEQIV -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for KAT6A-SPPL2A |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
| Hgene | KAT6A | chr8:41832221 | chr15:51041943 | ENST00000265713 | - | 8 | 17 | 144_664 | 494.0 | 2005.0 | PML |
| Hgene | KAT6A | chr8:41832221 | chr15:51041943 | ENST00000265713 | - | 8 | 17 | 1517_1741 | 494.0 | 2005.0 | PML |
| Hgene | KAT6A | chr8:41832221 | chr15:51041943 | ENST00000396930 | - | 9 | 18 | 144_664 | 494.0 | 2005.0 | PML |
| Hgene | KAT6A | chr8:41832221 | chr15:51041943 | ENST00000396930 | - | 9 | 18 | 1517_1741 | 494.0 | 2005.0 | PML |
| Hgene | KAT6A | chr8:41832221 | chr15:51041943 | ENST00000406337 | - | 9 | 18 | 144_664 | 494.0 | 2005.0 | PML |
| Hgene | KAT6A | chr8:41832221 | chr15:51041943 | ENST00000406337 | - | 9 | 18 | 1517_1741 | 494.0 | 2005.0 | PML |
| Hgene | KAT6A | chr8:41832221 | chr15:51041943 | ENST00000265713 | - | 8 | 17 | 312_664 | 494.0 | 2005.0 | RUNX1-1 |
| Hgene | KAT6A | chr8:41832221 | chr15:51041943 | ENST00000396930 | - | 9 | 18 | 312_664 | 494.0 | 2005.0 | RUNX1-1 |
| Hgene | KAT6A | chr8:41832221 | chr15:51041943 | ENST00000406337 | - | 9 | 18 | 312_664 | 494.0 | 2005.0 | RUNX1-1 |
| Hgene | KAT6A | chr8:41832221 | chr15:51041943 | ENST00000265713 | - | 8 | 17 | 1517_1642 | 494.0 | 2005.0 | RUNX1-2 |
| Hgene | KAT6A | chr8:41832221 | chr15:51041943 | ENST00000396930 | - | 9 | 18 | 1517_1642 | 494.0 | 2005.0 | RUNX1-2 |
| Hgene | KAT6A | chr8:41832221 | chr15:51041943 | ENST00000406337 | - | 9 | 18 | 1517_1642 | 494.0 | 2005.0 | RUNX1-2 |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for KAT6A-SPPL2A |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for KAT6A-SPPL2A |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
