
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:KAZN-PRDM2 (FusionGDB2 ID:41288) |
Fusion Gene Summary for KAZN-PRDM2 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: KAZN-PRDM2 | Fusion gene ID: 41288 | Hgene | Tgene | Gene symbol | KAZN | PRDM2 | Gene ID | 23254 | 7799 |
| Gene name | kazrin, periplakin interacting protein | PR/SET domain 2 | |
| Synonyms | C1orf196|KAZ | HUMHOXY1|KMT8|KMT8A|MTB-ZF|RIZ|RIZ1|RIZ2 | |
| Cytomap | 1p36.21 | 1p36.21 | |
| Type of gene | protein-coding | protein-coding | |
| Description | kazrin | PR domain zinc finger protein 2GATA-3 binding protein G3BMTE-binding proteinPR domain 2PR domain containing 2, with ZNF domainPR domain-containing protein 2lysine N-methyltransferase 8retinoblastoma protein-binding zinc finger proteinretinoblastom | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | Q674X7 | . | |
| Ensembl transtripts involved in fusion gene | ENST00000361144, ENST00000376030, ENST00000400797, ENST00000400798, ENST00000422387, ENST00000503743, | ENST00000235372, ENST00000311066, ENST00000343137, ENST00000413440, ENST00000502727, ENST00000503842, ENST00000505823, ENST00000376048, | |
| Fusion gene scores | * DoF score | 15 X 11 X 8=1320 | 10 X 8 X 4=320 |
| # samples | 18 | 11 | |
| ** MAII score | log2(18/1320*10)=-2.87446911791614 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(11/320*10)=-1.5405683813627 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: KAZN [Title/Abstract] AND PRDM2 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | KAZN(15287371)-PRDM2(14142921), # samples:2 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Tgene | PRDM2 | GO:0045944 | positive regulation of transcription by RNA polymerase II | 8654390 |
 Fusion gene breakpoints across KAZN (5'-gene) Fusion gene breakpoints across KAZN (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
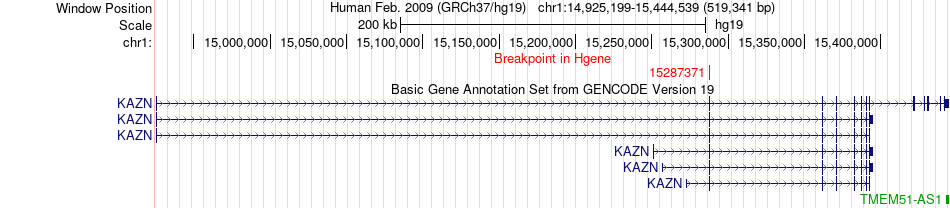 |
 Fusion gene breakpoints across PRDM2 (3'-gene) Fusion gene breakpoints across PRDM2 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
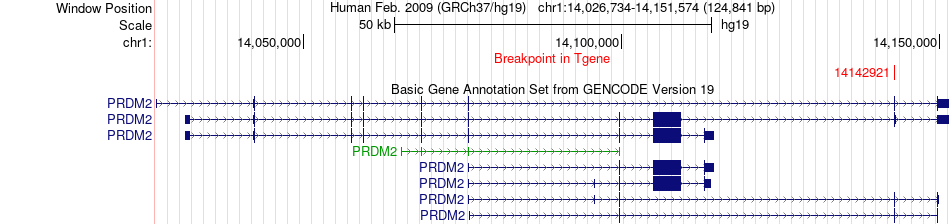 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | THCA | TCGA-EM-A4FU | KAZN | chr1 | 15287371 | + | PRDM2 | chr1 | 14142921 | + |
Top |
Fusion Gene ORF analysis for KAZN-PRDM2 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-intron | ENST00000361144 | ENST00000235372 | KAZN | chr1 | 15287371 | + | PRDM2 | chr1 | 14142921 | + |
| 5CDS-intron | ENST00000361144 | ENST00000311066 | KAZN | chr1 | 15287371 | + | PRDM2 | chr1 | 14142921 | + |
| 5CDS-intron | ENST00000361144 | ENST00000343137 | KAZN | chr1 | 15287371 | + | PRDM2 | chr1 | 14142921 | + |
| 5CDS-intron | ENST00000361144 | ENST00000413440 | KAZN | chr1 | 15287371 | + | PRDM2 | chr1 | 14142921 | + |
| 5CDS-intron | ENST00000361144 | ENST00000502727 | KAZN | chr1 | 15287371 | + | PRDM2 | chr1 | 14142921 | + |
| 5CDS-intron | ENST00000361144 | ENST00000503842 | KAZN | chr1 | 15287371 | + | PRDM2 | chr1 | 14142921 | + |
| 5CDS-intron | ENST00000361144 | ENST00000505823 | KAZN | chr1 | 15287371 | + | PRDM2 | chr1 | 14142921 | + |
| 5CDS-intron | ENST00000376030 | ENST00000235372 | KAZN | chr1 | 15287371 | + | PRDM2 | chr1 | 14142921 | + |
| 5CDS-intron | ENST00000376030 | ENST00000311066 | KAZN | chr1 | 15287371 | + | PRDM2 | chr1 | 14142921 | + |
| 5CDS-intron | ENST00000376030 | ENST00000343137 | KAZN | chr1 | 15287371 | + | PRDM2 | chr1 | 14142921 | + |
| 5CDS-intron | ENST00000376030 | ENST00000413440 | KAZN | chr1 | 15287371 | + | PRDM2 | chr1 | 14142921 | + |
| 5CDS-intron | ENST00000376030 | ENST00000502727 | KAZN | chr1 | 15287371 | + | PRDM2 | chr1 | 14142921 | + |
| 5CDS-intron | ENST00000376030 | ENST00000503842 | KAZN | chr1 | 15287371 | + | PRDM2 | chr1 | 14142921 | + |
| 5CDS-intron | ENST00000376030 | ENST00000505823 | KAZN | chr1 | 15287371 | + | PRDM2 | chr1 | 14142921 | + |
| 5CDS-intron | ENST00000400797 | ENST00000235372 | KAZN | chr1 | 15287371 | + | PRDM2 | chr1 | 14142921 | + |
| 5CDS-intron | ENST00000400797 | ENST00000311066 | KAZN | chr1 | 15287371 | + | PRDM2 | chr1 | 14142921 | + |
| 5CDS-intron | ENST00000400797 | ENST00000343137 | KAZN | chr1 | 15287371 | + | PRDM2 | chr1 | 14142921 | + |
| 5CDS-intron | ENST00000400797 | ENST00000413440 | KAZN | chr1 | 15287371 | + | PRDM2 | chr1 | 14142921 | + |
| 5CDS-intron | ENST00000400797 | ENST00000502727 | KAZN | chr1 | 15287371 | + | PRDM2 | chr1 | 14142921 | + |
| 5CDS-intron | ENST00000400797 | ENST00000503842 | KAZN | chr1 | 15287371 | + | PRDM2 | chr1 | 14142921 | + |
| 5CDS-intron | ENST00000400797 | ENST00000505823 | KAZN | chr1 | 15287371 | + | PRDM2 | chr1 | 14142921 | + |
| 5CDS-intron | ENST00000400798 | ENST00000235372 | KAZN | chr1 | 15287371 | + | PRDM2 | chr1 | 14142921 | + |
| 5CDS-intron | ENST00000400798 | ENST00000311066 | KAZN | chr1 | 15287371 | + | PRDM2 | chr1 | 14142921 | + |
| 5CDS-intron | ENST00000400798 | ENST00000343137 | KAZN | chr1 | 15287371 | + | PRDM2 | chr1 | 14142921 | + |
| 5CDS-intron | ENST00000400798 | ENST00000413440 | KAZN | chr1 | 15287371 | + | PRDM2 | chr1 | 14142921 | + |
| 5CDS-intron | ENST00000400798 | ENST00000502727 | KAZN | chr1 | 15287371 | + | PRDM2 | chr1 | 14142921 | + |
| 5CDS-intron | ENST00000400798 | ENST00000503842 | KAZN | chr1 | 15287371 | + | PRDM2 | chr1 | 14142921 | + |
| 5CDS-intron | ENST00000400798 | ENST00000505823 | KAZN | chr1 | 15287371 | + | PRDM2 | chr1 | 14142921 | + |
| 5CDS-intron | ENST00000422387 | ENST00000235372 | KAZN | chr1 | 15287371 | + | PRDM2 | chr1 | 14142921 | + |
| 5CDS-intron | ENST00000422387 | ENST00000311066 | KAZN | chr1 | 15287371 | + | PRDM2 | chr1 | 14142921 | + |
| 5CDS-intron | ENST00000422387 | ENST00000343137 | KAZN | chr1 | 15287371 | + | PRDM2 | chr1 | 14142921 | + |
| 5CDS-intron | ENST00000422387 | ENST00000413440 | KAZN | chr1 | 15287371 | + | PRDM2 | chr1 | 14142921 | + |
| 5CDS-intron | ENST00000422387 | ENST00000502727 | KAZN | chr1 | 15287371 | + | PRDM2 | chr1 | 14142921 | + |
| 5CDS-intron | ENST00000422387 | ENST00000503842 | KAZN | chr1 | 15287371 | + | PRDM2 | chr1 | 14142921 | + |
| 5CDS-intron | ENST00000422387 | ENST00000505823 | KAZN | chr1 | 15287371 | + | PRDM2 | chr1 | 14142921 | + |
| 5CDS-intron | ENST00000503743 | ENST00000235372 | KAZN | chr1 | 15287371 | + | PRDM2 | chr1 | 14142921 | + |
| 5CDS-intron | ENST00000503743 | ENST00000311066 | KAZN | chr1 | 15287371 | + | PRDM2 | chr1 | 14142921 | + |
| 5CDS-intron | ENST00000503743 | ENST00000343137 | KAZN | chr1 | 15287371 | + | PRDM2 | chr1 | 14142921 | + |
| 5CDS-intron | ENST00000503743 | ENST00000413440 | KAZN | chr1 | 15287371 | + | PRDM2 | chr1 | 14142921 | + |
| 5CDS-intron | ENST00000503743 | ENST00000502727 | KAZN | chr1 | 15287371 | + | PRDM2 | chr1 | 14142921 | + |
| 5CDS-intron | ENST00000503743 | ENST00000503842 | KAZN | chr1 | 15287371 | + | PRDM2 | chr1 | 14142921 | + |
| 5CDS-intron | ENST00000503743 | ENST00000505823 | KAZN | chr1 | 15287371 | + | PRDM2 | chr1 | 14142921 | + |
| In-frame | ENST00000361144 | ENST00000376048 | KAZN | chr1 | 15287371 | + | PRDM2 | chr1 | 14142921 | + |
| In-frame | ENST00000376030 | ENST00000376048 | KAZN | chr1 | 15287371 | + | PRDM2 | chr1 | 14142921 | + |
| In-frame | ENST00000400797 | ENST00000376048 | KAZN | chr1 | 15287371 | + | PRDM2 | chr1 | 14142921 | + |
| In-frame | ENST00000400798 | ENST00000376048 | KAZN | chr1 | 15287371 | + | PRDM2 | chr1 | 14142921 | + |
| In-frame | ENST00000422387 | ENST00000376048 | KAZN | chr1 | 15287371 | + | PRDM2 | chr1 | 14142921 | + |
| In-frame | ENST00000503743 | ENST00000376048 | KAZN | chr1 | 15287371 | + | PRDM2 | chr1 | 14142921 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000376030 | KAZN | chr1 | 15287371 | + | ENST00000376048 | PRDM2 | chr1 | 14142921 | + | 2763 | 712 | 150 | 881 | 243 |
| ENST00000503743 | KAZN | chr1 | 15287371 | + | ENST00000376048 | PRDM2 | chr1 | 14142921 | + | 2678 | 627 | 137 | 796 | 219 |
| ENST00000422387 | KAZN | chr1 | 15287371 | + | ENST00000376048 | PRDM2 | chr1 | 14142921 | + | 2750 | 699 | 137 | 868 | 243 |
| ENST00000361144 | KAZN | chr1 | 15287371 | + | ENST00000376048 | PRDM2 | chr1 | 14142921 | + | 2686 | 635 | 603 | 1 | 201 |
| ENST00000400798 | KAZN | chr1 | 15287371 | + | ENST00000376048 | PRDM2 | chr1 | 14142921 | + | 2488 | 437 | 19 | 606 | 195 |
| ENST00000400797 | KAZN | chr1 | 15287371 | + | ENST00000376048 | PRDM2 | chr1 | 14142921 | + | 2498 | 447 | 1654 | 1286 | 122 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000376030 | ENST00000376048 | KAZN | chr1 | 15287371 | + | PRDM2 | chr1 | 14142921 | + | 0.015859287 | 0.98414075 |
| ENST00000503743 | ENST00000376048 | KAZN | chr1 | 15287371 | + | PRDM2 | chr1 | 14142921 | + | 0.012687095 | 0.98731285 |
| ENST00000422387 | ENST00000376048 | KAZN | chr1 | 15287371 | + | PRDM2 | chr1 | 14142921 | + | 0.013555325 | 0.98644465 |
| ENST00000361144 | ENST00000376048 | KAZN | chr1 | 15287371 | + | PRDM2 | chr1 | 14142921 | + | 0.15692359 | 0.84307635 |
| ENST00000400798 | ENST00000376048 | KAZN | chr1 | 15287371 | + | PRDM2 | chr1 | 14142921 | + | 0.5898439 | 0.41015613 |
| ENST00000400797 | ENST00000376048 | KAZN | chr1 | 15287371 | + | PRDM2 | chr1 | 14142921 | + | 0.17502065 | 0.82497936 |
Top |
Fusion Genomic Features for KAZN-PRDM2 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| KAZN | chr1 | 15287371 | + | PRDM2 | chr1 | 14142921 | + | 5.20E-05 | 0.999948 |
| KAZN | chr1 | 15287371 | + | PRDM2 | chr1 | 14142921 | + | 5.20E-05 | 0.999948 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
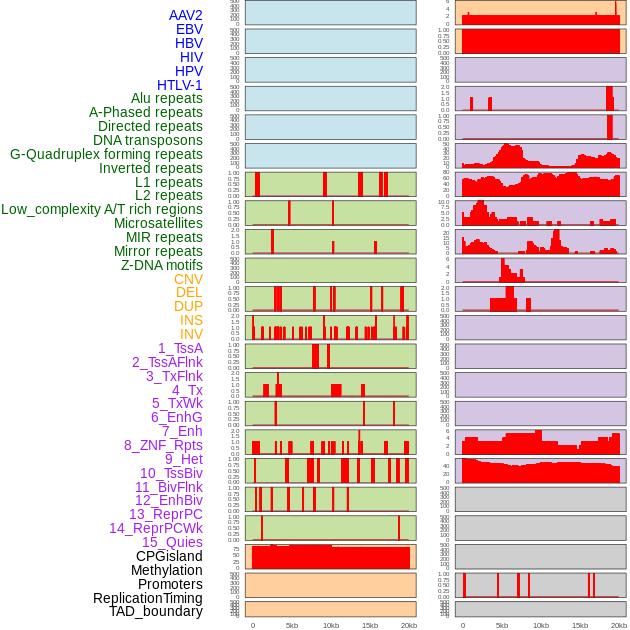 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
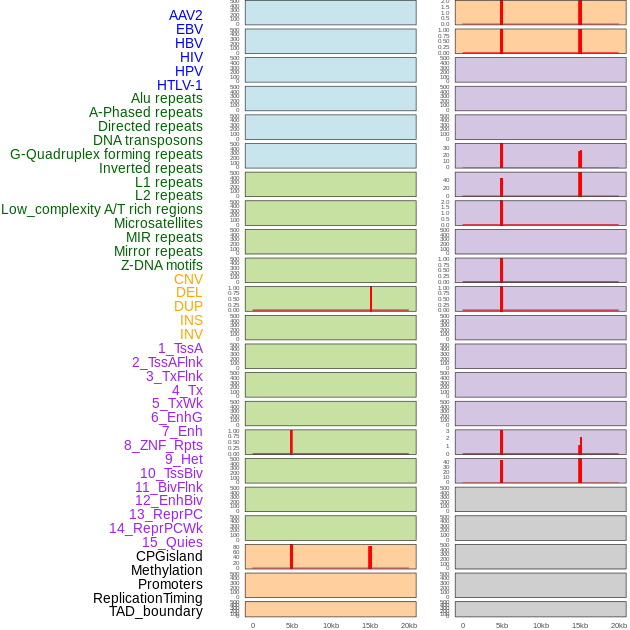 |
Top |
Fusion Protein Features for KAZN-PRDM2 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr1:15287371/chr1:14142921) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| KAZN | . |
| FUNCTION: Component of the cornified envelope of keratinocytes. May be involved in the interplay between adherens junctions and desmosomes. The function in the nucleus is not known. {ECO:0000269|PubMed:15337775}. | FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | PRDM2 | chr1:15287371 | chr1:14142921 | ENST00000311066 | 0 | 9 | 1052_1074 | 0 | 1683.0 | Compositional bias | Note=Poly-Ser | |
| Tgene | PRDM2 | chr1:15287371 | chr1:14142921 | ENST00000311066 | 0 | 9 | 1361_1447 | 0 | 1683.0 | Compositional bias | Note=Arg/Lys-rich (basic) | |
| Tgene | PRDM2 | chr1:15287371 | chr1:14142921 | ENST00000311066 | 0 | 9 | 268_296 | 0 | 1683.0 | Compositional bias | Note=Asp/Glu-rich (acidic) | |
| Tgene | PRDM2 | chr1:15287371 | chr1:14142921 | ENST00000311066 | 0 | 9 | 933_1049 | 0 | 1683.0 | Compositional bias | Note=Pro-rich | |
| Tgene | PRDM2 | chr1:15287371 | chr1:14142921 | ENST00000376048 | 5 | 8 | 1052_1074 | 170 | 227.0 | Compositional bias | Note=Poly-Ser | |
| Tgene | PRDM2 | chr1:15287371 | chr1:14142921 | ENST00000376048 | 5 | 8 | 1361_1447 | 170 | 227.0 | Compositional bias | Note=Arg/Lys-rich (basic) | |
| Tgene | PRDM2 | chr1:15287371 | chr1:14142921 | ENST00000376048 | 5 | 8 | 268_296 | 170 | 227.0 | Compositional bias | Note=Asp/Glu-rich (acidic) | |
| Tgene | PRDM2 | chr1:15287371 | chr1:14142921 | ENST00000376048 | 5 | 8 | 933_1049 | 170 | 227.0 | Compositional bias | Note=Pro-rich | |
| Tgene | PRDM2 | chr1:15287371 | chr1:14142921 | ENST00000311066 | 0 | 9 | 28_141 | 0 | 1683.0 | Domain | SET | |
| Tgene | PRDM2 | chr1:15287371 | chr1:14142921 | ENST00000311066 | 0 | 9 | 1028_1052 | 0 | 1683.0 | Motif | SH3-binding | |
| Tgene | PRDM2 | chr1:15287371 | chr1:14142921 | ENST00000311066 | 0 | 9 | 970_979 | 0 | 1683.0 | Motif | SH3-binding | |
| Tgene | PRDM2 | chr1:15287371 | chr1:14142921 | ENST00000311066 | 0 | 9 | 985_998 | 0 | 1683.0 | Motif | SH3-binding | |
| Tgene | PRDM2 | chr1:15287371 | chr1:14142921 | ENST00000376048 | 5 | 8 | 1028_1052 | 170 | 227.0 | Motif | SH3-binding | |
| Tgene | PRDM2 | chr1:15287371 | chr1:14142921 | ENST00000376048 | 5 | 8 | 970_979 | 170 | 227.0 | Motif | SH3-binding | |
| Tgene | PRDM2 | chr1:15287371 | chr1:14142921 | ENST00000376048 | 5 | 8 | 985_998 | 170 | 227.0 | Motif | SH3-binding | |
| Tgene | PRDM2 | chr1:15287371 | chr1:14142921 | ENST00000311066 | 0 | 9 | 294_316 | 0 | 1683.0 | Region | Note=Retinoblastoma protein binding | |
| Tgene | PRDM2 | chr1:15287371 | chr1:14142921 | ENST00000376048 | 5 | 8 | 294_316 | 170 | 227.0 | Region | Note=Retinoblastoma protein binding | |
| Tgene | PRDM2 | chr1:15287371 | chr1:14142921 | ENST00000311066 | 0 | 9 | 1134_1156 | 0 | 1683.0 | Zinc finger | C2H2-type 4 | |
| Tgene | PRDM2 | chr1:15287371 | chr1:14142921 | ENST00000311066 | 0 | 9 | 1162_1185 | 0 | 1683.0 | Zinc finger | C2H2-type 5 | |
| Tgene | PRDM2 | chr1:15287371 | chr1:14142921 | ENST00000311066 | 0 | 9 | 1191_1214 | 0 | 1683.0 | Zinc finger | C2H2-type 6 | |
| Tgene | PRDM2 | chr1:15287371 | chr1:14142921 | ENST00000311066 | 0 | 9 | 1333_1355 | 0 | 1683.0 | Zinc finger | C2H2-type 7%3B atypical | |
| Tgene | PRDM2 | chr1:15287371 | chr1:14142921 | ENST00000311066 | 0 | 9 | 1455_1478 | 0 | 1683.0 | Zinc finger | C2H2-type 8%3B atypical | |
| Tgene | PRDM2 | chr1:15287371 | chr1:14142921 | ENST00000311066 | 0 | 9 | 360_382 | 0 | 1683.0 | Zinc finger | C2H2-type 1 | |
| Tgene | PRDM2 | chr1:15287371 | chr1:14142921 | ENST00000311066 | 0 | 9 | 390_412 | 0 | 1683.0 | Zinc finger | C2H2-type 2 | |
| Tgene | PRDM2 | chr1:15287371 | chr1:14142921 | ENST00000311066 | 0 | 9 | 483_506 | 0 | 1683.0 | Zinc finger | C2H2-type 3 | |
| Tgene | PRDM2 | chr1:15287371 | chr1:14142921 | ENST00000376048 | 5 | 8 | 1134_1156 | 170 | 227.0 | Zinc finger | C2H2-type 4 | |
| Tgene | PRDM2 | chr1:15287371 | chr1:14142921 | ENST00000376048 | 5 | 8 | 1162_1185 | 170 | 227.0 | Zinc finger | C2H2-type 5 | |
| Tgene | PRDM2 | chr1:15287371 | chr1:14142921 | ENST00000376048 | 5 | 8 | 1191_1214 | 170 | 227.0 | Zinc finger | C2H2-type 6 | |
| Tgene | PRDM2 | chr1:15287371 | chr1:14142921 | ENST00000376048 | 5 | 8 | 1333_1355 | 170 | 227.0 | Zinc finger | C2H2-type 7%3B atypical | |
| Tgene | PRDM2 | chr1:15287371 | chr1:14142921 | ENST00000376048 | 5 | 8 | 1455_1478 | 170 | 227.0 | Zinc finger | C2H2-type 8%3B atypical | |
| Tgene | PRDM2 | chr1:15287371 | chr1:14142921 | ENST00000376048 | 5 | 8 | 360_382 | 170 | 227.0 | Zinc finger | C2H2-type 1 | |
| Tgene | PRDM2 | chr1:15287371 | chr1:14142921 | ENST00000376048 | 5 | 8 | 390_412 | 170 | 227.0 | Zinc finger | C2H2-type 2 | |
| Tgene | PRDM2 | chr1:15287371 | chr1:14142921 | ENST00000376048 | 5 | 8 | 483_506 | 170 | 227.0 | Zinc finger | C2H2-type 3 |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | KAZN | chr1:15287371 | chr1:14142921 | ENST00000361144 | + | 2 | 8 | 74_256 | 133 | 416.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | KAZN | chr1:15287371 | chr1:14142921 | ENST00000376030 | + | 2 | 15 | 74_256 | 139 | 776.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | KAZN | chr1:15287371 | chr1:14142921 | ENST00000400797 | + | 2 | 8 | 74_256 | 45 | 328.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | KAZN | chr1:15287371 | chr1:14142921 | ENST00000400798 | + | 2 | 8 | 74_256 | 45 | 328.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | KAZN | chr1:15287371 | chr1:14142921 | ENST00000422387 | + | 2 | 8 | 74_256 | 139 | 422.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | KAZN | chr1:15287371 | chr1:14142921 | ENST00000503743 | + | 3 | 9 | 74_256 | 139 | 422.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | KAZN | chr1:15287371 | chr1:14142921 | ENST00000361144 | + | 2 | 8 | 375_382 | 133 | 416.0 | Compositional bias | Note=Poly-Lys |
| Hgene | KAZN | chr1:15287371 | chr1:14142921 | ENST00000376030 | + | 2 | 15 | 375_382 | 139 | 776.0 | Compositional bias | Note=Poly-Lys |
| Hgene | KAZN | chr1:15287371 | chr1:14142921 | ENST00000400797 | + | 2 | 8 | 375_382 | 45 | 328.0 | Compositional bias | Note=Poly-Lys |
| Hgene | KAZN | chr1:15287371 | chr1:14142921 | ENST00000400798 | + | 2 | 8 | 375_382 | 45 | 328.0 | Compositional bias | Note=Poly-Lys |
| Hgene | KAZN | chr1:15287371 | chr1:14142921 | ENST00000422387 | + | 2 | 8 | 375_382 | 139 | 422.0 | Compositional bias | Note=Poly-Lys |
| Hgene | KAZN | chr1:15287371 | chr1:14142921 | ENST00000503743 | + | 3 | 9 | 375_382 | 139 | 422.0 | Compositional bias | Note=Poly-Lys |
| Hgene | KAZN | chr1:15287371 | chr1:14142921 | ENST00000361144 | + | 2 | 8 | 446_511 | 133 | 416.0 | Domain | SAM 1 |
| Hgene | KAZN | chr1:15287371 | chr1:14142921 | ENST00000361144 | + | 2 | 8 | 524_588 | 133 | 416.0 | Domain | SAM 2 |
| Hgene | KAZN | chr1:15287371 | chr1:14142921 | ENST00000361144 | + | 2 | 8 | 612_679 | 133 | 416.0 | Domain | SAM 3 |
| Hgene | KAZN | chr1:15287371 | chr1:14142921 | ENST00000376030 | + | 2 | 15 | 446_511 | 139 | 776.0 | Domain | SAM 1 |
| Hgene | KAZN | chr1:15287371 | chr1:14142921 | ENST00000376030 | + | 2 | 15 | 524_588 | 139 | 776.0 | Domain | SAM 2 |
| Hgene | KAZN | chr1:15287371 | chr1:14142921 | ENST00000376030 | + | 2 | 15 | 612_679 | 139 | 776.0 | Domain | SAM 3 |
| Hgene | KAZN | chr1:15287371 | chr1:14142921 | ENST00000400797 | + | 2 | 8 | 446_511 | 45 | 328.0 | Domain | SAM 1 |
| Hgene | KAZN | chr1:15287371 | chr1:14142921 | ENST00000400797 | + | 2 | 8 | 524_588 | 45 | 328.0 | Domain | SAM 2 |
| Hgene | KAZN | chr1:15287371 | chr1:14142921 | ENST00000400797 | + | 2 | 8 | 612_679 | 45 | 328.0 | Domain | SAM 3 |
| Hgene | KAZN | chr1:15287371 | chr1:14142921 | ENST00000400798 | + | 2 | 8 | 446_511 | 45 | 328.0 | Domain | SAM 1 |
| Hgene | KAZN | chr1:15287371 | chr1:14142921 | ENST00000400798 | + | 2 | 8 | 524_588 | 45 | 328.0 | Domain | SAM 2 |
| Hgene | KAZN | chr1:15287371 | chr1:14142921 | ENST00000400798 | + | 2 | 8 | 612_679 | 45 | 328.0 | Domain | SAM 3 |
| Hgene | KAZN | chr1:15287371 | chr1:14142921 | ENST00000422387 | + | 2 | 8 | 446_511 | 139 | 422.0 | Domain | SAM 1 |
| Hgene | KAZN | chr1:15287371 | chr1:14142921 | ENST00000422387 | + | 2 | 8 | 524_588 | 139 | 422.0 | Domain | SAM 2 |
| Hgene | KAZN | chr1:15287371 | chr1:14142921 | ENST00000422387 | + | 2 | 8 | 612_679 | 139 | 422.0 | Domain | SAM 3 |
| Hgene | KAZN | chr1:15287371 | chr1:14142921 | ENST00000503743 | + | 3 | 9 | 446_511 | 139 | 422.0 | Domain | SAM 1 |
| Hgene | KAZN | chr1:15287371 | chr1:14142921 | ENST00000503743 | + | 3 | 9 | 524_588 | 139 | 422.0 | Domain | SAM 2 |
| Hgene | KAZN | chr1:15287371 | chr1:14142921 | ENST00000503743 | + | 3 | 9 | 612_679 | 139 | 422.0 | Domain | SAM 3 |
| Tgene | PRDM2 | chr1:15287371 | chr1:14142921 | ENST00000235372 | 7 | 10 | 1052_1074 | 1678 | 2095.6666666666665 | Compositional bias | Note=Poly-Ser | |
| Tgene | PRDM2 | chr1:15287371 | chr1:14142921 | ENST00000235372 | 7 | 10 | 1361_1447 | 1678 | 2095.6666666666665 | Compositional bias | Note=Arg/Lys-rich (basic) | |
| Tgene | PRDM2 | chr1:15287371 | chr1:14142921 | ENST00000235372 | 7 | 10 | 268_296 | 1678 | 2095.6666666666665 | Compositional bias | Note=Asp/Glu-rich (acidic) | |
| Tgene | PRDM2 | chr1:15287371 | chr1:14142921 | ENST00000235372 | 7 | 10 | 933_1049 | 1678 | 2095.6666666666665 | Compositional bias | Note=Pro-rich | |
| Tgene | PRDM2 | chr1:15287371 | chr1:14142921 | ENST00000235372 | 7 | 10 | 28_141 | 1678 | 2095.6666666666665 | Domain | SET | |
| Tgene | PRDM2 | chr1:15287371 | chr1:14142921 | ENST00000376048 | 5 | 8 | 28_141 | 170 | 227.0 | Domain | SET | |
| Tgene | PRDM2 | chr1:15287371 | chr1:14142921 | ENST00000235372 | 7 | 10 | 1028_1052 | 1678 | 2095.6666666666665 | Motif | SH3-binding | |
| Tgene | PRDM2 | chr1:15287371 | chr1:14142921 | ENST00000235372 | 7 | 10 | 970_979 | 1678 | 2095.6666666666665 | Motif | SH3-binding | |
| Tgene | PRDM2 | chr1:15287371 | chr1:14142921 | ENST00000235372 | 7 | 10 | 985_998 | 1678 | 2095.6666666666665 | Motif | SH3-binding | |
| Tgene | PRDM2 | chr1:15287371 | chr1:14142921 | ENST00000235372 | 7 | 10 | 294_316 | 1678 | 2095.6666666666665 | Region | Note=Retinoblastoma protein binding | |
| Tgene | PRDM2 | chr1:15287371 | chr1:14142921 | ENST00000235372 | 7 | 10 | 1134_1156 | 1678 | 2095.6666666666665 | Zinc finger | C2H2-type 4 | |
| Tgene | PRDM2 | chr1:15287371 | chr1:14142921 | ENST00000235372 | 7 | 10 | 1162_1185 | 1678 | 2095.6666666666665 | Zinc finger | C2H2-type 5 | |
| Tgene | PRDM2 | chr1:15287371 | chr1:14142921 | ENST00000235372 | 7 | 10 | 1191_1214 | 1678 | 2095.6666666666665 | Zinc finger | C2H2-type 6 | |
| Tgene | PRDM2 | chr1:15287371 | chr1:14142921 | ENST00000235372 | 7 | 10 | 1333_1355 | 1678 | 2095.6666666666665 | Zinc finger | C2H2-type 7%3B atypical | |
| Tgene | PRDM2 | chr1:15287371 | chr1:14142921 | ENST00000235372 | 7 | 10 | 1455_1478 | 1678 | 2095.6666666666665 | Zinc finger | C2H2-type 8%3B atypical | |
| Tgene | PRDM2 | chr1:15287371 | chr1:14142921 | ENST00000235372 | 7 | 10 | 360_382 | 1678 | 2095.6666666666665 | Zinc finger | C2H2-type 1 | |
| Tgene | PRDM2 | chr1:15287371 | chr1:14142921 | ENST00000235372 | 7 | 10 | 390_412 | 1678 | 2095.6666666666665 | Zinc finger | C2H2-type 2 | |
| Tgene | PRDM2 | chr1:15287371 | chr1:14142921 | ENST00000235372 | 7 | 10 | 483_506 | 1678 | 2095.6666666666665 | Zinc finger | C2H2-type 3 |
Top |
Fusion Gene Sequence for KAZN-PRDM2 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >41288_41288_1_KAZN-PRDM2_KAZN_chr1_15287371_ENST00000361144_PRDM2_chr1_14142921_ENST00000376048_length(transcript)=2686nt_BP=635nt AGCCGTTCCCACGTGAGAGGCTCCGCGGCCGAATTCCTCGCGTGCAGCAGGCGCGGACCGCCCGGCGTCCGGCCGGACTGAGAGCCCTGG TCCGGCGCGCGCCGCCGGCCGGGCGGGAGGCGGGGGCGGGCTCGGCTGCGCCCCGATGCGGCGGCGACCTCCGGGTCTGTGAGCCCGGCG CGCGCCGTCGGAGCCCCTCGCGCAGCCGCTGGTAGCGTCCCCCCGGGCACCCGGCATGCGGGCGGCCGACTCGGGCTCGTGGGAGCGCGT CCGCCAGCTCGCGGCGCAGGGCGAGCCGGCGCCTTCCTGCGGGGCGGGGGCCGGGCCCGCGCGGCCCCCGGGACCCGCAGCCTGCGAGCA GTGCGTGGACGCGGCGGGGCCCGGCGATCGGCCCCGCGCCGGGGTTCCCCGGGTCCGAGCGGATGGCGACTGCAGCCAGCCCGTGCTCCT GCGGGAAGAAGTGTCGCGGCTCCAGGAGGAAGTTCACCTTCTCCGGCAGATGAAGGAGATGTTGGCGAAGGACCTGGAGGAGTCGCAGGG CGGCAAGTCCTCTGAGGTCCTCTCGGCCACCGAGCTCAGGGTCCAGCTGGCCCAGAAGGAGCAGGAGCTAGCCAGAGCCAAAGAAGCCTT GCAGGCTACAGCCTCCGCTTGGCGTCCCGATGCTCTCCACCAGCGGCCCCGTACATCACCAGGCAGTATAGGAAGGTCAAAGCTCCAGCT GCAGCCCAGTTCCAGGGACCATTCTTCAAAGAGTAGACACTCTGGCTGCTCCCTGACAGCACCTGAAGTGACCTGGAATCAGTGAAGCCA AAGGGACTGGCAGTCTGCCCTGCAGGGAGTACCGACCTATCCCAGTTGTGTGAGGCTGCGAGAGAAAGGGAGTGCATGTGCGCGCGTGCA TGTGTGCGTGCGTGTGTGTTCACGTGTTCTCGTGCGGGCGCGTGAGTGGTCTTCAAACGAGGGTCCCGATCCCCGGGGCGGCAGGAAGGG GGCCGACTCCACGCTGTCCTTTGGGATGATACTTGGATGCAGCTCTTGGGACCGTGTTCTGCAGCCCAGCCTTCCTGTTGGGGTGGGGCC TCTCCTACTATGCAATTTTTCAAGAGCTCCTTGACCCTGCTTTTTGCTTCTTGAGTTGTCTTTTGCCATTATGGGGACTTTGGTTTGACC CAGGGGTCAGCCTTAGGAAGGCCTTCAGGAGGAGGCCGAGTTCCCCTTCAGTACCACCCCTCTCTCCCCACCTTCCCTCTCCCGGCAACA TCTCTGGGAATCAACAGCATATTGACACGTTGGAGCCGAGCCTGAACATGCCCCTCGGCCCCAGCACATGGAAAACCCCCTTCCTTGCCT AAGGTGTCTGAGTTTCTGGCTCTTGAGGCATTTCCAGACTTGAAATTCTCATCAGTCCATTGCTCTTGAGTCTTTGCAGAGAACCTCAGA TCAGGTGCACCTGGGAGAAAGACTTTGTCCCCACTTACAGATCTATCTCCTCCCTTGGGAAGGGCAGGGAATGGGGACGGTGTATGGAGG GGAGGGATCTCCTGCGCCCTTCATTGCCACACTTGGTGGGACCATGAACATCTTTAGTGTCTGAGCTTCTCAAATTAGCTGCAATAGGAA AAAAACAAATTGGGAAATGAAAAAAAAATGGGAAGATTAAAAAGCACAGGGGGAAGAAGAAGAGATTTCGGAGGCCATCCTGCCAGGGGC GGACGGGGCTGACTCCTGCTCTCTGGAGGACGGTCAGTCCATGTCTCGGAGAAACGGGTGAGCTGAGCTTGGCGTTTGGACCCAGTTCAG TGAGGTTCTTGGGTTTTGTGCCTTTGGGGCAGACCCCAGGCAAGGATGTCTGAGACCACTTGGGCGCTGTTTTCTCAGCTCCAATTTCAA GAGTGAGCTATCAAACCCAGAGCGGAAGGAGGGAGCTCTGATGAGCACGGTTTGTCACACGATAAAGGGATTTTTTTTTTCAGGGCTACT ACGGTTGATCTTGCAACTCTGTAAATATGTATGTAGACACTTTTAAAAGCACGTATTTATGTCCCTGACTGTAAATGCTCCATTTTTAAA GTTTTATAACTTGTGTTATTTAATGAGTCAGTCAATCGGCTGCAGTATGGGATCTGATAAGGATCTAGGAGAAGGGTCTCATGCGGACCC TCACATGGGCAGAAAAATGGTGGTCATTGGCCGACATCACAGTTTTCCTGTTTCCCACCCAGCTAAAAACCGTTGTTTGCTTTAAATTTT CATAAACTGGAATCCTTTCACCCGCTCCTACAGCTAACCCTCACAAGCATGAAGTGCTGTGGCTGTTCCTTATCCTAATGATGCGCTTTT GTCCCGTAAATGTTAACACTCATGAAGCATACCCCGGCCTCTCAGTTCTTGAGGGCCTCCCCACCGCAGCAGCAAGGAAAGCTCACGAAC CCCAAACCTGGCAAGTCACCTGCAGCCCATGGTGAGCTCTGGGAAGTGTGGTTGAGGCCTTGGGGTCACTCCTTTTTTGCATGTGCAAAT GTGCTGGTCACCCTTCAACGCTCCCAGACGGTCAGGAAAACTGTTCCAATCATGAAAAGGGGGGATGATTTTGTAAAAGTGGCATTTCCT >41288_41288_1_KAZN-PRDM2_KAZN_chr1_15287371_ENST00000361144_PRDM2_chr1_14142921_ENST00000376048_length(amino acids)=201AA_BP= MLLLGQLDPELGGREDLRGLAALRLLQVLRQHLLHLPEKVNFLLEPRHFFPQEHGLAAVAIRSDPGNPGAGPIAGPRRVHALLAGCGSRG PRGPGPRPAGRRRLALRRELADALPRARVGRPHAGCPGGRYQRLREGLRRRAPGSQTRRSPPHRGAAEPAPASRPAGGARRTRALSPAGR -------------------------------------------------------------- >41288_41288_2_KAZN-PRDM2_KAZN_chr1_15287371_ENST00000376030_PRDM2_chr1_14142921_ENST00000376048_length(transcript)=2763nt_BP=712nt GCGCCCGCCCGCCGGGGTCTCGGCGATCGCTGCTCCTCCTCCTCCTTCTCCTCCTCTTTTTTCTCCTCCGCCTCCTCCCCCCGCCGCCTC GCCACCGCCGCGGCTAGGGCTGGAGGCGCCGCTGTCATTCCTGTGCCGGAGGAACCGGCGCTGCCGGTGCCTGGGGGTCGGGGCGCGGGC GAAGCCGGGCCGCGGAGGACACAACAGGTAGAGCCGGGGGTGCCCGGCCGCGCGCCCCCCGCGCATCATGCAGCTCTTTGTCACCTCTCT CGCCCCCAGGCCAAAATCCTGAGCATGATGGAAGACAATAAGCAGCTCGCGCTCCGCATCGATGGGGCGGTCCAGTCGGCCAGCCAGGAG GTGACCAACCTGCGAGCCGAACTCACGGCCACCAACCGGAGACTGGCGGAACTGAGCGGCGGCGGCGGCCCCGGCCCGGGCCCGGGAGCC GCGGCCAGCGCCTCGGCGGCGGGGGACTCGGCGGCGACGAACATGGAGAACCCCCAGCTTGGAGCGCAAGTGCTCCTGCGGGAAGAAGTG TCGCGGCTCCAGGAGGAAGTTCACCTTCTCCGGCAGATGAAGGAGATGTTGGCGAAGGACCTGGAGGAGTCGCAGGGCGGCAAGTCCTCT GAGGTCCTCTCGGCCACCGAGCTCAGGGTCCAGCTGGCCCAGAAGGAGCAGGAGCTAGCCAGAGCCAAAGAAGCCTTGCAGGCTACAGCC TCCGCTTGGCGTCCCGATGCTCTCCACCAGCGGCCCCGTACATCACCAGGCAGTATAGGAAGGTCAAAGCTCCAGCTGCAGCCCAGTTCC AGGGACCATTCTTCAAAGAGTAGACACTCTGGCTGCTCCCTGACAGCACCTGAAGTGACCTGGAATCAGTGAAGCCAAAGGGACTGGCAG TCTGCCCTGCAGGGAGTACCGACCTATCCCAGTTGTGTGAGGCTGCGAGAGAAAGGGAGTGCATGTGCGCGCGTGCATGTGTGCGTGCGT GTGTGTTCACGTGTTCTCGTGCGGGCGCGTGAGTGGTCTTCAAACGAGGGTCCCGATCCCCGGGGCGGCAGGAAGGGGGCCGACTCCACG CTGTCCTTTGGGATGATACTTGGATGCAGCTCTTGGGACCGTGTTCTGCAGCCCAGCCTTCCTGTTGGGGTGGGGCCTCTCCTACTATGC AATTTTTCAAGAGCTCCTTGACCCTGCTTTTTGCTTCTTGAGTTGTCTTTTGCCATTATGGGGACTTTGGTTTGACCCAGGGGTCAGCCT TAGGAAGGCCTTCAGGAGGAGGCCGAGTTCCCCTTCAGTACCACCCCTCTCTCCCCACCTTCCCTCTCCCGGCAACATCTCTGGGAATCA ACAGCATATTGACACGTTGGAGCCGAGCCTGAACATGCCCCTCGGCCCCAGCACATGGAAAACCCCCTTCCTTGCCTAAGGTGTCTGAGT TTCTGGCTCTTGAGGCATTTCCAGACTTGAAATTCTCATCAGTCCATTGCTCTTGAGTCTTTGCAGAGAACCTCAGATCAGGTGCACCTG GGAGAAAGACTTTGTCCCCACTTACAGATCTATCTCCTCCCTTGGGAAGGGCAGGGAATGGGGACGGTGTATGGAGGGGAGGGATCTCCT GCGCCCTTCATTGCCACACTTGGTGGGACCATGAACATCTTTAGTGTCTGAGCTTCTCAAATTAGCTGCAATAGGAAAAAAACAAATTGG GAAATGAAAAAAAAATGGGAAGATTAAAAAGCACAGGGGGAAGAAGAAGAGATTTCGGAGGCCATCCTGCCAGGGGCGGACGGGGCTGAC TCCTGCTCTCTGGAGGACGGTCAGTCCATGTCTCGGAGAAACGGGTGAGCTGAGCTTGGCGTTTGGACCCAGTTCAGTGAGGTTCTTGGG TTTTGTGCCTTTGGGGCAGACCCCAGGCAAGGATGTCTGAGACCACTTGGGCGCTGTTTTCTCAGCTCCAATTTCAAGAGTGAGCTATCA AACCCAGAGCGGAAGGAGGGAGCTCTGATGAGCACGGTTTGTCACACGATAAAGGGATTTTTTTTTTCAGGGCTACTACGGTTGATCTTG CAACTCTGTAAATATGTATGTAGACACTTTTAAAAGCACGTATTTATGTCCCTGACTGTAAATGCTCCATTTTTAAAGTTTTATAACTTG TGTTATTTAATGAGTCAGTCAATCGGCTGCAGTATGGGATCTGATAAGGATCTAGGAGAAGGGTCTCATGCGGACCCTCACATGGGCAGA AAAATGGTGGTCATTGGCCGACATCACAGTTTTCCTGTTTCCCACCCAGCTAAAAACCGTTGTTTGCTTTAAATTTTCATAAACTGGAAT CCTTTCACCCGCTCCTACAGCTAACCCTCACAAGCATGAAGTGCTGTGGCTGTTCCTTATCCTAATGATGCGCTTTTGTCCCGTAAATGT TAACACTCATGAAGCATACCCCGGCCTCTCAGTTCTTGAGGGCCTCCCCACCGCAGCAGCAAGGAAAGCTCACGAACCCCAAACCTGGCA AGTCACCTGCAGCCCATGGTGAGCTCTGGGAAGTGTGGTTGAGGCCTTGGGGTCACTCCTTTTTTGCATGTGCAAATGTGCTGGTCACCC TTCAACGCTCCCAGACGGTCAGGAAAACTGTTCCAATCATGAAAAGGGGGGATGATTTTGTAAAAGTGGCATTTCCTGGTCAGTGGTGGT >41288_41288_2_KAZN-PRDM2_KAZN_chr1_15287371_ENST00000376030_PRDM2_chr1_14142921_ENST00000376048_length(amino acids)=243AA_BP=187 MPVPGGRGAGEAGPRRTQQVEPGVPGRAPPAHHAALCHLSRPQAKILSMMEDNKQLALRIDGAVQSASQEVTNLRAELTATNRRLAELSG GGGPGPGPGAAASASAAGDSAATNMENPQLGAQVLLREEVSRLQEEVHLLRQMKEMLAKDLEESQGGKSSEVLSATELRVQLAQKEQELA -------------------------------------------------------------- >41288_41288_3_KAZN-PRDM2_KAZN_chr1_15287371_ENST00000400797_PRDM2_chr1_14142921_ENST00000376048_length(transcript)=2498nt_BP=447nt GCATCAGCAGCTGTCTGCCAGTGTTGACGGAGGGCTCATGGGTGCCAGGCCGCTTGCTGGGGCTTTTTATCTGTCCTCAGGCATTGGCAC AGCAGCCCTGGGAGATGAATACTATTGTGATCTTCATTCTCCACACAAGGACAGGGAAGTGACGTGCCAGGGTCAAACAACTACCCAAGG ACAGAGCCAGGATTTGAACCAGGCCATCCAGTTCCAGAACCACTGCTCTTCACTGTCATTGTGGCCATTTTACAGTGCTCCTGCGGGAAG AAGTGTCGCGGCTCCAGGAGGAAGTTCACCTTCTCCGGCAGATGAAGGAGATGTTGGCGAAGGACCTGGAGGAGTCGCAGGGCGGCAAGT CCTCTGAGGTCCTCTCGGCCACCGAGCTCAGGGTCCAGCTGGCCCAGAAGGAGCAGGAGCTAGCCAGAGCCAAAGAAGCCTTGCAGGCTA CAGCCTCCGCTTGGCGTCCCGATGCTCTCCACCAGCGGCCCCGTACATCACCAGGCAGTATAGGAAGGTCAAAGCTCCAGCTGCAGCCCA GTTCCAGGGACCATTCTTCAAAGAGTAGACACTCTGGCTGCTCCCTGACAGCACCTGAAGTGACCTGGAATCAGTGAAGCCAAAGGGACT GGCAGTCTGCCCTGCAGGGAGTACCGACCTATCCCAGTTGTGTGAGGCTGCGAGAGAAAGGGAGTGCATGTGCGCGCGTGCATGTGTGCG TGCGTGTGTGTTCACGTGTTCTCGTGCGGGCGCGTGAGTGGTCTTCAAACGAGGGTCCCGATCCCCGGGGCGGCAGGAAGGGGGCCGACT CCACGCTGTCCTTTGGGATGATACTTGGATGCAGCTCTTGGGACCGTGTTCTGCAGCCCAGCCTTCCTGTTGGGGTGGGGCCTCTCCTAC TATGCAATTTTTCAAGAGCTCCTTGACCCTGCTTTTTGCTTCTTGAGTTGTCTTTTGCCATTATGGGGACTTTGGTTTGACCCAGGGGTC AGCCTTAGGAAGGCCTTCAGGAGGAGGCCGAGTTCCCCTTCAGTACCACCCCTCTCTCCCCACCTTCCCTCTCCCGGCAACATCTCTGGG AATCAACAGCATATTGACACGTTGGAGCCGAGCCTGAACATGCCCCTCGGCCCCAGCACATGGAAAACCCCCTTCCTTGCCTAAGGTGTC TGAGTTTCTGGCTCTTGAGGCATTTCCAGACTTGAAATTCTCATCAGTCCATTGCTCTTGAGTCTTTGCAGAGAACCTCAGATCAGGTGC ACCTGGGAGAAAGACTTTGTCCCCACTTACAGATCTATCTCCTCCCTTGGGAAGGGCAGGGAATGGGGACGGTGTATGGAGGGGAGGGAT CTCCTGCGCCCTTCATTGCCACACTTGGTGGGACCATGAACATCTTTAGTGTCTGAGCTTCTCAAATTAGCTGCAATAGGAAAAAAACAA ATTGGGAAATGAAAAAAAAATGGGAAGATTAAAAAGCACAGGGGGAAGAAGAAGAGATTTCGGAGGCCATCCTGCCAGGGGCGGACGGGG CTGACTCCTGCTCTCTGGAGGACGGTCAGTCCATGTCTCGGAGAAACGGGTGAGCTGAGCTTGGCGTTTGGACCCAGTTCAGTGAGGTTC TTGGGTTTTGTGCCTTTGGGGCAGACCCCAGGCAAGGATGTCTGAGACCACTTGGGCGCTGTTTTCTCAGCTCCAATTTCAAGAGTGAGC TATCAAACCCAGAGCGGAAGGAGGGAGCTCTGATGAGCACGGTTTGTCACACGATAAAGGGATTTTTTTTTTCAGGGCTACTACGGTTGA TCTTGCAACTCTGTAAATATGTATGTAGACACTTTTAAAAGCACGTATTTATGTCCCTGACTGTAAATGCTCCATTTTTAAAGTTTTATA ACTTGTGTTATTTAATGAGTCAGTCAATCGGCTGCAGTATGGGATCTGATAAGGATCTAGGAGAAGGGTCTCATGCGGACCCTCACATGG GCAGAAAAATGGTGGTCATTGGCCGACATCACAGTTTTCCTGTTTCCCACCCAGCTAAAAACCGTTGTTTGCTTTAAATTTTCATAAACT GGAATCCTTTCACCCGCTCCTACAGCTAACCCTCACAAGCATGAAGTGCTGTGGCTGTTCCTTATCCTAATGATGCGCTTTTGTCCCGTA AATGTTAACACTCATGAAGCATACCCCGGCCTCTCAGTTCTTGAGGGCCTCCCCACCGCAGCAGCAAGGAAAGCTCACGAACCCCAAACC TGGCAAGTCACCTGCAGCCCATGGTGAGCTCTGGGAAGTGTGGTTGAGGCCTTGGGGTCACTCCTTTTTTGCATGTGCAAATGTGCTGGT CACCCTTCAACGCTCCCAGACGGTCAGGAAAACTGTTCCAATCATGAAAAGGGGGGATGATTTTGTAAAAGTGGCATTTCCTGGTCAGTG >41288_41288_3_KAZN-PRDM2_KAZN_chr1_15287371_ENST00000400797_PRDM2_chr1_14142921_ENST00000376048_length(amino acids)=122AA_BP= MPGVCPKGTKPKNLTELGPNAKLSSPVSPRHGLTVLQRAGVSPVRPWQDGLRNLFFFPLCFLIFPFFFHFPICFFPIAANLRSSDTKDVH -------------------------------------------------------------- >41288_41288_4_KAZN-PRDM2_KAZN_chr1_15287371_ENST00000400798_PRDM2_chr1_14142921_ENST00000376048_length(transcript)=2488nt_BP=437nt GGGAGAGGGAAGGTCAGGCTTGCCTCCTGTTCACAAGGACAACCTCTGTCTCCCTTGTGGCGGCTTCTATGAAGGTGGGGACTCCAGGCT TTCTCTCGTCTGCTTTTTGGATGCCTTCAAGAGAGGCCAGTTAGAGGATATCAATTGTGGCAATTCCCCAGAGCCCTCATCAGATGGCAT TAGCCCGTCATGTGCTGCAGTGGCACCCAGTTTGGAGGAAATTCGGAACCAGAAGAGTGTGACAGTGCTCCTGCGGGAAGAAGTGTCGCG GCTCCAGGAGGAAGTTCACCTTCTCCGGCAGATGAAGGAGATGTTGGCGAAGGACCTGGAGGAGTCGCAGGGCGGCAAGTCCTCTGAGGT CCTCTCGGCCACCGAGCTCAGGGTCCAGCTGGCCCAGAAGGAGCAGGAGCTAGCCAGAGCCAAAGAAGCCTTGCAGGCTACAGCCTCCGC TTGGCGTCCCGATGCTCTCCACCAGCGGCCCCGTACATCACCAGGCAGTATAGGAAGGTCAAAGCTCCAGCTGCAGCCCAGTTCCAGGGA CCATTCTTCAAAGAGTAGACACTCTGGCTGCTCCCTGACAGCACCTGAAGTGACCTGGAATCAGTGAAGCCAAAGGGACTGGCAGTCTGC CCTGCAGGGAGTACCGACCTATCCCAGTTGTGTGAGGCTGCGAGAGAAAGGGAGTGCATGTGCGCGCGTGCATGTGTGCGTGCGTGTGTG TTCACGTGTTCTCGTGCGGGCGCGTGAGTGGTCTTCAAACGAGGGTCCCGATCCCCGGGGCGGCAGGAAGGGGGCCGACTCCACGCTGTC CTTTGGGATGATACTTGGATGCAGCTCTTGGGACCGTGTTCTGCAGCCCAGCCTTCCTGTTGGGGTGGGGCCTCTCCTACTATGCAATTT TTCAAGAGCTCCTTGACCCTGCTTTTTGCTTCTTGAGTTGTCTTTTGCCATTATGGGGACTTTGGTTTGACCCAGGGGTCAGCCTTAGGA AGGCCTTCAGGAGGAGGCCGAGTTCCCCTTCAGTACCACCCCTCTCTCCCCACCTTCCCTCTCCCGGCAACATCTCTGGGAATCAACAGC ATATTGACACGTTGGAGCCGAGCCTGAACATGCCCCTCGGCCCCAGCACATGGAAAACCCCCTTCCTTGCCTAAGGTGTCTGAGTTTCTG GCTCTTGAGGCATTTCCAGACTTGAAATTCTCATCAGTCCATTGCTCTTGAGTCTTTGCAGAGAACCTCAGATCAGGTGCACCTGGGAGA AAGACTTTGTCCCCACTTACAGATCTATCTCCTCCCTTGGGAAGGGCAGGGAATGGGGACGGTGTATGGAGGGGAGGGATCTCCTGCGCC CTTCATTGCCACACTTGGTGGGACCATGAACATCTTTAGTGTCTGAGCTTCTCAAATTAGCTGCAATAGGAAAAAAACAAATTGGGAAAT GAAAAAAAAATGGGAAGATTAAAAAGCACAGGGGGAAGAAGAAGAGATTTCGGAGGCCATCCTGCCAGGGGCGGACGGGGCTGACTCCTG CTCTCTGGAGGACGGTCAGTCCATGTCTCGGAGAAACGGGTGAGCTGAGCTTGGCGTTTGGACCCAGTTCAGTGAGGTTCTTGGGTTTTG TGCCTTTGGGGCAGACCCCAGGCAAGGATGTCTGAGACCACTTGGGCGCTGTTTTCTCAGCTCCAATTTCAAGAGTGAGCTATCAAACCC AGAGCGGAAGGAGGGAGCTCTGATGAGCACGGTTTGTCACACGATAAAGGGATTTTTTTTTTCAGGGCTACTACGGTTGATCTTGCAACT CTGTAAATATGTATGTAGACACTTTTAAAAGCACGTATTTATGTCCCTGACTGTAAATGCTCCATTTTTAAAGTTTTATAACTTGTGTTA TTTAATGAGTCAGTCAATCGGCTGCAGTATGGGATCTGATAAGGATCTAGGAGAAGGGTCTCATGCGGACCCTCACATGGGCAGAAAAAT GGTGGTCATTGGCCGACATCACAGTTTTCCTGTTTCCCACCCAGCTAAAAACCGTTGTTTGCTTTAAATTTTCATAAACTGGAATCCTTT CACCCGCTCCTACAGCTAACCCTCACAAGCATGAAGTGCTGTGGCTGTTCCTTATCCTAATGATGCGCTTTTGTCCCGTAAATGTTAACA CTCATGAAGCATACCCCGGCCTCTCAGTTCTTGAGGGCCTCCCCACCGCAGCAGCAAGGAAAGCTCACGAACCCCAAACCTGGCAAGTCA CCTGCAGCCCATGGTGAGCTCTGGGAAGTGTGGTTGAGGCCTTGGGGTCACTCCTTTTTTGCATGTGCAAATGTGCTGGTCACCCTTCAA CGCTCCCAGACGGTCAGGAAAACTGTTCCAATCATGAAAAGGGGGGATGATTTTGTAAAAGTGGCATTTCCTGGTCAGTGGTGGTCTTCA >41288_41288_4_KAZN-PRDM2_KAZN_chr1_15287371_ENST00000400798_PRDM2_chr1_14142921_ENST00000376048_length(amino acids)=195AA_BP=139 MPPVHKDNLCLPCGGFYEGGDSRLSLVCFLDAFKRGQLEDINCGNSPEPSSDGISPSCAAVAPSLEEIRNQKSVTVLLREEVSRLQEEVH LLRQMKEMLAKDLEESQGGKSSEVLSATELRVQLAQKEQELARAKEALQATASAWRPDALHQRPRTSPGSIGRSKLQLQPSSRDHSSKSR -------------------------------------------------------------- >41288_41288_5_KAZN-PRDM2_KAZN_chr1_15287371_ENST00000422387_PRDM2_chr1_14142921_ENST00000376048_length(transcript)=2750nt_BP=699nt GGGGTCTCGGCGATCGCTGCTCCTCCTCCTCCTTCTCCTCCTCTTTTTTCTCCTCCGCCTCCTCCCCCCGCCGCCTCGCCACCGCCGCGG CTAGGGCTGGAGGCGCCGCTGTCATTCCTGTGCCGGAGGAACCGGCGCTGCCGGTGCCTGGGGGTCGGGGCGCGGGCGAAGCCGGGCCGC GGAGGACACAACAGGTAGAGCCGGGGGTGCCCGGCCGCGCGCCCCCCGCGCATCATGCAGCTCTTTGTCACCTCTCTCGCCCCCAGGCCA AAATCCTGAGCATGATGGAAGACAATAAGCAGCTCGCGCTCCGCATCGATGGGGCGGTCCAGTCGGCCAGCCAGGAGGTGACCAACCTGC GAGCCGAACTCACGGCCACCAACCGGAGACTGGCGGAACTGAGCGGCGGCGGCGGCCCCGGCCCGGGCCCGGGAGCCGCGGCCAGCGCCT CGGCGGCGGGGGACTCGGCGGCGACGAACATGGAGAACCCCCAGCTTGGAGCGCAAGTGCTCCTGCGGGAAGAAGTGTCGCGGCTCCAGG AGGAAGTTCACCTTCTCCGGCAGATGAAGGAGATGTTGGCGAAGGACCTGGAGGAGTCGCAGGGCGGCAAGTCCTCTGAGGTCCTCTCGG CCACCGAGCTCAGGGTCCAGCTGGCCCAGAAGGAGCAGGAGCTAGCCAGAGCCAAAGAAGCCTTGCAGGCTACAGCCTCCGCTTGGCGTC CCGATGCTCTCCACCAGCGGCCCCGTACATCACCAGGCAGTATAGGAAGGTCAAAGCTCCAGCTGCAGCCCAGTTCCAGGGACCATTCTT CAAAGAGTAGACACTCTGGCTGCTCCCTGACAGCACCTGAAGTGACCTGGAATCAGTGAAGCCAAAGGGACTGGCAGTCTGCCCTGCAGG GAGTACCGACCTATCCCAGTTGTGTGAGGCTGCGAGAGAAAGGGAGTGCATGTGCGCGCGTGCATGTGTGCGTGCGTGTGTGTTCACGTG TTCTCGTGCGGGCGCGTGAGTGGTCTTCAAACGAGGGTCCCGATCCCCGGGGCGGCAGGAAGGGGGCCGACTCCACGCTGTCCTTTGGGA TGATACTTGGATGCAGCTCTTGGGACCGTGTTCTGCAGCCCAGCCTTCCTGTTGGGGTGGGGCCTCTCCTACTATGCAATTTTTCAAGAG CTCCTTGACCCTGCTTTTTGCTTCTTGAGTTGTCTTTTGCCATTATGGGGACTTTGGTTTGACCCAGGGGTCAGCCTTAGGAAGGCCTTC AGGAGGAGGCCGAGTTCCCCTTCAGTACCACCCCTCTCTCCCCACCTTCCCTCTCCCGGCAACATCTCTGGGAATCAACAGCATATTGAC ACGTTGGAGCCGAGCCTGAACATGCCCCTCGGCCCCAGCACATGGAAAACCCCCTTCCTTGCCTAAGGTGTCTGAGTTTCTGGCTCTTGA GGCATTTCCAGACTTGAAATTCTCATCAGTCCATTGCTCTTGAGTCTTTGCAGAGAACCTCAGATCAGGTGCACCTGGGAGAAAGACTTT GTCCCCACTTACAGATCTATCTCCTCCCTTGGGAAGGGCAGGGAATGGGGACGGTGTATGGAGGGGAGGGATCTCCTGCGCCCTTCATTG CCACACTTGGTGGGACCATGAACATCTTTAGTGTCTGAGCTTCTCAAATTAGCTGCAATAGGAAAAAAACAAATTGGGAAATGAAAAAAA AATGGGAAGATTAAAAAGCACAGGGGGAAGAAGAAGAGATTTCGGAGGCCATCCTGCCAGGGGCGGACGGGGCTGACTCCTGCTCTCTGG AGGACGGTCAGTCCATGTCTCGGAGAAACGGGTGAGCTGAGCTTGGCGTTTGGACCCAGTTCAGTGAGGTTCTTGGGTTTTGTGCCTTTG GGGCAGACCCCAGGCAAGGATGTCTGAGACCACTTGGGCGCTGTTTTCTCAGCTCCAATTTCAAGAGTGAGCTATCAAACCCAGAGCGGA AGGAGGGAGCTCTGATGAGCACGGTTTGTCACACGATAAAGGGATTTTTTTTTTCAGGGCTACTACGGTTGATCTTGCAACTCTGTAAAT ATGTATGTAGACACTTTTAAAAGCACGTATTTATGTCCCTGACTGTAAATGCTCCATTTTTAAAGTTTTATAACTTGTGTTATTTAATGA GTCAGTCAATCGGCTGCAGTATGGGATCTGATAAGGATCTAGGAGAAGGGTCTCATGCGGACCCTCACATGGGCAGAAAAATGGTGGTCA TTGGCCGACATCACAGTTTTCCTGTTTCCCACCCAGCTAAAAACCGTTGTTTGCTTTAAATTTTCATAAACTGGAATCCTTTCACCCGCT CCTACAGCTAACCCTCACAAGCATGAAGTGCTGTGGCTGTTCCTTATCCTAATGATGCGCTTTTGTCCCGTAAATGTTAACACTCATGAA GCATACCCCGGCCTCTCAGTTCTTGAGGGCCTCCCCACCGCAGCAGCAAGGAAAGCTCACGAACCCCAAACCTGGCAAGTCACCTGCAGC CCATGGTGAGCTCTGGGAAGTGTGGTTGAGGCCTTGGGGTCACTCCTTTTTTGCATGTGCAAATGTGCTGGTCACCCTTCAACGCTCCCA GACGGTCAGGAAAACTGTTCCAATCATGAAAAGGGGGGATGATTTTGTAAAAGTGGCATTTCCTGGTCAGTGGTGGTCTTCAAGACGACA >41288_41288_5_KAZN-PRDM2_KAZN_chr1_15287371_ENST00000422387_PRDM2_chr1_14142921_ENST00000376048_length(amino acids)=243AA_BP=187 MPVPGGRGAGEAGPRRTQQVEPGVPGRAPPAHHAALCHLSRPQAKILSMMEDNKQLALRIDGAVQSASQEVTNLRAELTATNRRLAELSG GGGPGPGPGAAASASAAGDSAATNMENPQLGAQVLLREEVSRLQEEVHLLRQMKEMLAKDLEESQGGKSSEVLSATELRVQLAQKEQELA -------------------------------------------------------------- >41288_41288_6_KAZN-PRDM2_KAZN_chr1_15287371_ENST00000503743_PRDM2_chr1_14142921_ENST00000376048_length(transcript)=2678nt_BP=627nt GGGGTCTCGGCGATCGCTGCTCCTCCTCCTCCTTCTCCTCCTCTTTTTTCTCCTCCGCCTCCTCCCCCCGCCGCCTCGCCACCGCCGCGG CTAGGGCTGGAGGCGCCGCTGTCATTCCTGTGCCGGAGGAACCGGCGCTGCCGGTGCCTGGGGGTCGGGGCGCGGGCGAAGCCGGGCCGC GGAGGACACAACAGGCCAAAATCCTGAGCATGATGGAAGACAATAAGCAGCTCGCGCTCCGCATCGATGGGGCGGTCCAGTCGGCCAGCC AGGAGGTGACCAACCTGCGAGCCGAACTCACGGCCACCAACCGGAGACTGGCGGAACTGAGCGGCGGCGGCGGCCCCGGCCCGGGCCCGG GAGCCGCGGCCAGCGCCTCGGCGGCGGGGGACTCGGCGGCGACGAACATGGAGAACCCCCAGCTTGGAGCGCAAGTGCTCCTGCGGGAAG AAGTGTCGCGGCTCCAGGAGGAAGTTCACCTTCTCCGGCAGATGAAGGAGATGTTGGCGAAGGACCTGGAGGAGTCGCAGGGCGGCAAGT CCTCTGAGGTCCTCTCGGCCACCGAGCTCAGGGTCCAGCTGGCCCAGAAGGAGCAGGAGCTAGCCAGAGCCAAAGAAGCCTTGCAGGCTA CAGCCTCCGCTTGGCGTCCCGATGCTCTCCACCAGCGGCCCCGTACATCACCAGGCAGTATAGGAAGGTCAAAGCTCCAGCTGCAGCCCA GTTCCAGGGACCATTCTTCAAAGAGTAGACACTCTGGCTGCTCCCTGACAGCACCTGAAGTGACCTGGAATCAGTGAAGCCAAAGGGACT GGCAGTCTGCCCTGCAGGGAGTACCGACCTATCCCAGTTGTGTGAGGCTGCGAGAGAAAGGGAGTGCATGTGCGCGCGTGCATGTGTGCG TGCGTGTGTGTTCACGTGTTCTCGTGCGGGCGCGTGAGTGGTCTTCAAACGAGGGTCCCGATCCCCGGGGCGGCAGGAAGGGGGCCGACT CCACGCTGTCCTTTGGGATGATACTTGGATGCAGCTCTTGGGACCGTGTTCTGCAGCCCAGCCTTCCTGTTGGGGTGGGGCCTCTCCTAC TATGCAATTTTTCAAGAGCTCCTTGACCCTGCTTTTTGCTTCTTGAGTTGTCTTTTGCCATTATGGGGACTTTGGTTTGACCCAGGGGTC AGCCTTAGGAAGGCCTTCAGGAGGAGGCCGAGTTCCCCTTCAGTACCACCCCTCTCTCCCCACCTTCCCTCTCCCGGCAACATCTCTGGG AATCAACAGCATATTGACACGTTGGAGCCGAGCCTGAACATGCCCCTCGGCCCCAGCACATGGAAAACCCCCTTCCTTGCCTAAGGTGTC TGAGTTTCTGGCTCTTGAGGCATTTCCAGACTTGAAATTCTCATCAGTCCATTGCTCTTGAGTCTTTGCAGAGAACCTCAGATCAGGTGC ACCTGGGAGAAAGACTTTGTCCCCACTTACAGATCTATCTCCTCCCTTGGGAAGGGCAGGGAATGGGGACGGTGTATGGAGGGGAGGGAT CTCCTGCGCCCTTCATTGCCACACTTGGTGGGACCATGAACATCTTTAGTGTCTGAGCTTCTCAAATTAGCTGCAATAGGAAAAAAACAA ATTGGGAAATGAAAAAAAAATGGGAAGATTAAAAAGCACAGGGGGAAGAAGAAGAGATTTCGGAGGCCATCCTGCCAGGGGCGGACGGGG CTGACTCCTGCTCTCTGGAGGACGGTCAGTCCATGTCTCGGAGAAACGGGTGAGCTGAGCTTGGCGTTTGGACCCAGTTCAGTGAGGTTC TTGGGTTTTGTGCCTTTGGGGCAGACCCCAGGCAAGGATGTCTGAGACCACTTGGGCGCTGTTTTCTCAGCTCCAATTTCAAGAGTGAGC TATCAAACCCAGAGCGGAAGGAGGGAGCTCTGATGAGCACGGTTTGTCACACGATAAAGGGATTTTTTTTTTCAGGGCTACTACGGTTGA TCTTGCAACTCTGTAAATATGTATGTAGACACTTTTAAAAGCACGTATTTATGTCCCTGACTGTAAATGCTCCATTTTTAAAGTTTTATA ACTTGTGTTATTTAATGAGTCAGTCAATCGGCTGCAGTATGGGATCTGATAAGGATCTAGGAGAAGGGTCTCATGCGGACCCTCACATGG GCAGAAAAATGGTGGTCATTGGCCGACATCACAGTTTTCCTGTTTCCCACCCAGCTAAAAACCGTTGTTTGCTTTAAATTTTCATAAACT GGAATCCTTTCACCCGCTCCTACAGCTAACCCTCACAAGCATGAAGTGCTGTGGCTGTTCCTTATCCTAATGATGCGCTTTTGTCCCGTA AATGTTAACACTCATGAAGCATACCCCGGCCTCTCAGTTCTTGAGGGCCTCCCCACCGCAGCAGCAAGGAAAGCTCACGAACCCCAAACC TGGCAAGTCACCTGCAGCCCATGGTGAGCTCTGGGAAGTGTGGTTGAGGCCTTGGGGTCACTCCTTTTTTGCATGTGCAAATGTGCTGGT CACCCTTCAACGCTCCCAGACGGTCAGGAAAACTGTTCCAATCATGAAAAGGGGGGATGATTTTGTAAAAGTGGCATTTCCTGGTCAGTG >41288_41288_6_KAZN-PRDM2_KAZN_chr1_15287371_ENST00000503743_PRDM2_chr1_14142921_ENST00000376048_length(amino acids)=219AA_BP=163 MPVPGGRGAGEAGPRRTQQAKILSMMEDNKQLALRIDGAVQSASQEVTNLRAELTATNRRLAELSGGGGPGPGPGAAASASAAGDSAATN MENPQLGAQVLLREEVSRLQEEVHLLRQMKEMLAKDLEESQGGKSSEVLSATELRVQLAQKEQELARAKEALQATASAWRPDALHQRPRT -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for KAZN-PRDM2 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
| Hgene | KAZN | chr1:15287371 | chr1:14142921 | ENST00000361144 | + | 2 | 8 | 174_333 | 133.33333333333334 | 416.0 | PPL |
| Hgene | KAZN | chr1:15287371 | chr1:14142921 | ENST00000376030 | + | 2 | 15 | 174_333 | 139.33333333333334 | 776.0 | PPL |
| Hgene | KAZN | chr1:15287371 | chr1:14142921 | ENST00000400797 | + | 2 | 8 | 174_333 | 45.333333333333336 | 328.0 | PPL |
| Hgene | KAZN | chr1:15287371 | chr1:14142921 | ENST00000400798 | + | 2 | 8 | 174_333 | 45.333333333333336 | 328.0 | PPL |
| Hgene | KAZN | chr1:15287371 | chr1:14142921 | ENST00000422387 | + | 2 | 8 | 174_333 | 139.33333333333334 | 422.0 | PPL |
| Hgene | KAZN | chr1:15287371 | chr1:14142921 | ENST00000503743 | + | 3 | 9 | 174_333 | 139.33333333333334 | 422.0 | PPL |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for KAZN-PRDM2 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for KAZN-PRDM2 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
