
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:KCNAB1-KHDRBS1 (FusionGDB2 ID:41342) |
Fusion Gene Summary for KCNAB1-KHDRBS1 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: KCNAB1-KHDRBS1 | Fusion gene ID: 41342 | Hgene | Tgene | Gene symbol | KCNAB1 | KHDRBS1 | Gene ID | 7881 | 10657 |
| Gene name | potassium voltage-gated channel subfamily A member regulatory beta subunit 1 | KH RNA binding domain containing, signal transduction associated 1 | |
| Synonyms | AKR6A3|KCNA1B|KV-BETA-1|Kvb1.3|hKvBeta3|hKvb3 | Sam68|p62|p68 | |
| Cytomap | 3q25.31 | 1p35.2 | |
| Type of gene | protein-coding | protein-coding | |
| Description | voltage-gated potassium channel subunit beta-1K(+) channel subunit beta-1K+ channel Beta1a chainpotassium channel beta 3 chainpotassium channel beta3 subunitpotassium channel shaker chain beta 1apotassium channel, voltage gated subfamily A regulator | KH domain-containing, RNA-binding, signal transduction-associated protein 1GAP-associated tyrosine phosphoprotein p62 (Sam68)KH domain containing, RNA binding, signal transduction associated 1p21 Ras GTPase-activating protein-associated p62src-associa | |
| Modification date | 20200313 | 20200327 | |
| UniProtAcc | Q14722 | Q07666 | |
| Ensembl transtripts involved in fusion gene | ENST00000497291, ENST00000302490, ENST00000389634, ENST00000389636, ENST00000471742, ENST00000490337, | ENST00000327300, ENST00000307714, ENST00000492989, | |
| Fusion gene scores | * DoF score | 14 X 14 X 3=588 | 11 X 8 X 6=528 |
| # samples | 14 | 12 | |
| ** MAII score | log2(14/588*10)=-2.0703893278914 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(12/528*10)=-2.13750352374993 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: KCNAB1 [Title/Abstract] AND KHDRBS1 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | KCNAB1(156249286)-KHDRBS1(32502511), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | KCNAB1 | GO:1901379 | regulation of potassium ion transmembrane transport | 7890032 |
| Tgene | KHDRBS1 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome | 24514149 |
| Tgene | KHDRBS1 | GO:0007166 | cell surface receptor signaling pathway | 9045636 |
| Tgene | KHDRBS1 | GO:0031647 | regulation of protein stability | 21613532 |
| Tgene | KHDRBS1 | GO:0045948 | positive regulation of translational initiation | 21613532 |
| Tgene | KHDRBS1 | GO:0046833 | positive regulation of RNA export from nucleus | 21613532 |
 Fusion gene breakpoints across KCNAB1 (5'-gene) Fusion gene breakpoints across KCNAB1 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
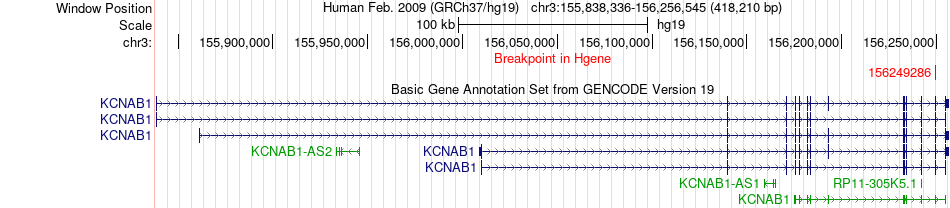 |
 Fusion gene breakpoints across KHDRBS1 (3'-gene) Fusion gene breakpoints across KHDRBS1 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
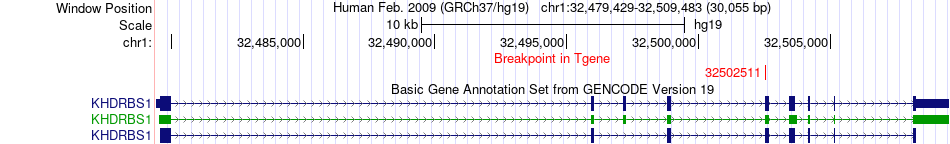 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | SARC | TCGA-IW-A3M6-01A | KCNAB1 | chr3 | 156249286 | - | KHDRBS1 | chr1 | 32502511 | + |
Top |
Fusion Gene ORF analysis for KCNAB1-KHDRBS1 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 3UTR-3CDS | ENST00000497291 | ENST00000327300 | KCNAB1 | chr3 | 156249286 | - | KHDRBS1 | chr1 | 32502511 | + |
| 3UTR-3UTR | ENST00000497291 | ENST00000307714 | KCNAB1 | chr3 | 156249286 | - | KHDRBS1 | chr1 | 32502511 | + |
| 3UTR-3UTR | ENST00000497291 | ENST00000492989 | KCNAB1 | chr3 | 156249286 | - | KHDRBS1 | chr1 | 32502511 | + |
| 5CDS-3UTR | ENST00000302490 | ENST00000307714 | KCNAB1 | chr3 | 156249286 | - | KHDRBS1 | chr1 | 32502511 | + |
| 5CDS-3UTR | ENST00000302490 | ENST00000492989 | KCNAB1 | chr3 | 156249286 | - | KHDRBS1 | chr1 | 32502511 | + |
| 5CDS-3UTR | ENST00000389634 | ENST00000307714 | KCNAB1 | chr3 | 156249286 | - | KHDRBS1 | chr1 | 32502511 | + |
| 5CDS-3UTR | ENST00000389634 | ENST00000492989 | KCNAB1 | chr3 | 156249286 | - | KHDRBS1 | chr1 | 32502511 | + |
| 5CDS-3UTR | ENST00000389636 | ENST00000307714 | KCNAB1 | chr3 | 156249286 | - | KHDRBS1 | chr1 | 32502511 | + |
| 5CDS-3UTR | ENST00000389636 | ENST00000492989 | KCNAB1 | chr3 | 156249286 | - | KHDRBS1 | chr1 | 32502511 | + |
| 5CDS-3UTR | ENST00000471742 | ENST00000307714 | KCNAB1 | chr3 | 156249286 | - | KHDRBS1 | chr1 | 32502511 | + |
| 5CDS-3UTR | ENST00000471742 | ENST00000492989 | KCNAB1 | chr3 | 156249286 | - | KHDRBS1 | chr1 | 32502511 | + |
| 5CDS-3UTR | ENST00000490337 | ENST00000307714 | KCNAB1 | chr3 | 156249286 | - | KHDRBS1 | chr1 | 32502511 | + |
| 5CDS-3UTR | ENST00000490337 | ENST00000492989 | KCNAB1 | chr3 | 156249286 | - | KHDRBS1 | chr1 | 32502511 | + |
| In-frame | ENST00000302490 | ENST00000327300 | KCNAB1 | chr3 | 156249286 | - | KHDRBS1 | chr1 | 32502511 | + |
| In-frame | ENST00000389634 | ENST00000327300 | KCNAB1 | chr3 | 156249286 | - | KHDRBS1 | chr1 | 32502511 | + |
| In-frame | ENST00000389636 | ENST00000327300 | KCNAB1 | chr3 | 156249286 | - | KHDRBS1 | chr1 | 32502511 | + |
| In-frame | ENST00000471742 | ENST00000327300 | KCNAB1 | chr3 | 156249286 | - | KHDRBS1 | chr1 | 32502511 | + |
| In-frame | ENST00000490337 | ENST00000327300 | KCNAB1 | chr3 | 156249286 | - | KHDRBS1 | chr1 | 32502511 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000490337 | KCNAB1 | chr3 | 156249286 | - | ENST00000327300 | KHDRBS1 | chr1 | 32502511 | + | 3053 | 1234 | 64 | 1794 | 576 |
| ENST00000389636 | KCNAB1 | chr3 | 156249286 | - | ENST00000327300 | KHDRBS1 | chr1 | 32502511 | + | 2936 | 1117 | 34 | 1677 | 547 |
| ENST00000471742 | KCNAB1 | chr3 | 156249286 | - | ENST00000327300 | KHDRBS1 | chr1 | 32502511 | + | 3173 | 1354 | 217 | 1914 | 565 |
| ENST00000302490 | KCNAB1 | chr3 | 156249286 | - | ENST00000327300 | KHDRBS1 | chr1 | 32502511 | + | 3806 | 1987 | 820 | 2547 | 575 |
| ENST00000389634 | KCNAB1 | chr3 | 156249286 | - | ENST00000327300 | KHDRBS1 | chr1 | 32502511 | + | 2848 | 1029 | 0 | 1589 | 529 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000490337 | ENST00000327300 | KCNAB1 | chr3 | 156249286 | - | KHDRBS1 | chr1 | 32502511 | + | 0.000994336 | 0.99900573 |
| ENST00000389636 | ENST00000327300 | KCNAB1 | chr3 | 156249286 | - | KHDRBS1 | chr1 | 32502511 | + | 0.001435159 | 0.9985649 |
| ENST00000471742 | ENST00000327300 | KCNAB1 | chr3 | 156249286 | - | KHDRBS1 | chr1 | 32502511 | + | 0.000745632 | 0.9992544 |
| ENST00000302490 | ENST00000327300 | KCNAB1 | chr3 | 156249286 | - | KHDRBS1 | chr1 | 32502511 | + | 0.001432549 | 0.99856746 |
| ENST00000389634 | ENST00000327300 | KCNAB1 | chr3 | 156249286 | - | KHDRBS1 | chr1 | 32502511 | + | 0.000737198 | 0.99926275 |
Top |
Fusion Genomic Features for KCNAB1-KHDRBS1 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
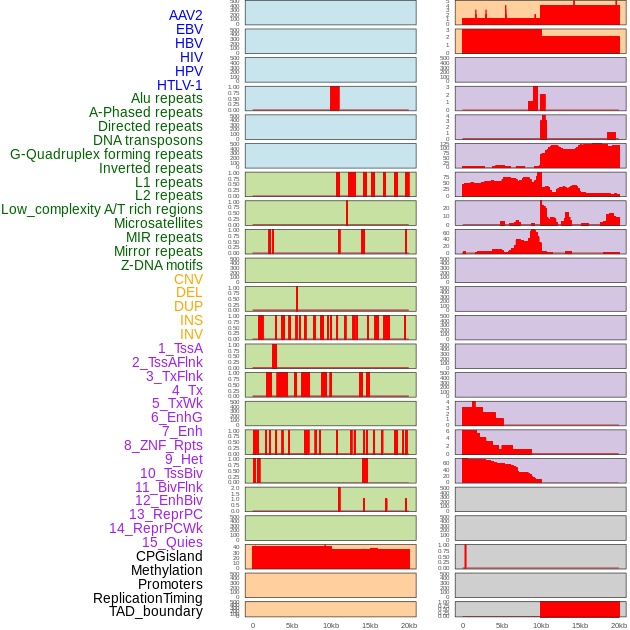 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for KCNAB1-KHDRBS1 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr3:156249286/chr1:32502511) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| KCNAB1 | KHDRBS1 |
| FUNCTION: Cytoplasmic potassium channel subunit that modulates the characteristics of the channel-forming alpha-subunits (PubMed:7499366, PubMed:7603988, PubMed:17156368,PubMed:17540341, PubMed:19713757). Modulates action potentials via its effect on the pore-forming alpha subunits (By similarity). Promotes expression of the pore-forming alpha subunits at the cell membrane, and thereby increases channel activity (By similarity). Mediates closure of delayed rectifier potassium channels by physically obstructing the pore via its N-terminal domain and increases the speed of channel closure for other family members (PubMed:9763623). Promotes the closure of KCNA1, KCNA2 and KCNA5 channels (PubMed:7499366, PubMed:7890032, PubMed:7603988, PubMed:7649300, PubMed:8938711, PubMed:12077175, PubMed:12130714, PubMed:15361858, PubMed:17540341, PubMed:19713757). Accelerates KCNA4 channel closure (PubMed:7890032, PubMed:7649300, PubMed:7890764, PubMed:9763623). Accelerates the closure of heteromeric channels formed by KCNA1 and KCNA4 (PubMed:17156368). Accelerates the closure of heteromeric channels formed by KCNA2, KCNA5 and KCNA6 (By similarity). Isoform KvB1.2 has no effect on KCNA1, KCNA2 or KCNB1 (PubMed:7890032, PubMed:7890764). Enhances KCNB1 and KCNB2 channel activity (By similarity). Binds NADPH; this is required for efficient down-regulation of potassium channel activity (PubMed:17540341). Has NADPH-dependent aldoketoreductase activity (By similarity). Oxidation of the bound NADPH strongly decreases N-type inactivation of potassium channel activity (By similarity). {ECO:0000250|UniProtKB:P63143, ECO:0000250|UniProtKB:P63144, ECO:0000269|PubMed:12077175, ECO:0000269|PubMed:12130714, ECO:0000269|PubMed:15361858, ECO:0000269|PubMed:17156368, ECO:0000269|PubMed:17540341, ECO:0000269|PubMed:19713757, ECO:0000269|PubMed:7499366, ECO:0000269|PubMed:7603988, ECO:0000269|PubMed:7649300, ECO:0000269|PubMed:7890032, ECO:0000269|PubMed:7890764, ECO:0000269|PubMed:8938711, ECO:0000269|PubMed:9763623, ECO:0000305}. | FUNCTION: Recruited and tyrosine phosphorylated by several receptor systems, for example the T-cell, leptin and insulin receptors. Once phosphorylated, functions as an adapter protein in signal transduction cascades by binding to SH2 and SH3 domain-containing proteins. Role in G2-M progression in the cell cycle. Represses CBP-dependent transcriptional activation apparently by competing with other nuclear factors for binding to CBP. Also acts as a putative regulator of mRNA stability and/or translation rates and mediates mRNA nuclear export. Positively regulates the association of constitutive transport element (CTE)-containing mRNA with large polyribosomes and translation initiation. According to some authors, is not involved in the nucleocytoplasmic export of unspliced (CTE)-containing RNA species according to (PubMed:22253824). RNA-binding protein that plays a role in the regulation of alternative splicing and influences mRNA splice site selection and exon inclusion. Binds to RNA containing 5'-[AU]UAA-3' as a bipartite motif spaced by more than 15 nucleotides. Binds poly(A). Can regulate CD44 alternative splicing in a Ras pathway-dependent manner (By similarity). In cooperation with HNRNPA1 modulates alternative splicing of BCL2L1 by promoting splicing toward isoform Bcl-X(S), and of SMN1 (PubMed:17371836, PubMed:20186123). Can regulate alternative splicing of NRXN1 and NRXN3 in the laminin G-like domain 6 containing the evolutionary conserved neurexin alternative spliced segment 4 (AS4) involved in neurexin selective targeting to postsynaptic partners. In a neuronal activity-dependent manner cooperates synergistically with KHDRBS2/SLIM-1 in regulation of NRXN1 exon skipping at AS4. The cooperation with KHDRBS2/SLIM-1 is antagonistic for regulation of NXRN3 alternative splicing at AS4 (By similarity). {ECO:0000250|UniProtKB:Q60749, ECO:0000269|PubMed:15021911, ECO:0000269|PubMed:17371836, ECO:0000269|PubMed:20186123, ECO:0000269|PubMed:20610388, ECO:0000269|PubMed:22253824, ECO:0000269|PubMed:26758068}.; FUNCTION: Isoform 3, which is expressed in growth-arrested cells only, inhibits S phase. {ECO:0000269|PubMed:9013542}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | KCNAB1 | chr3:156249286 | chr1:32502511 | ENST00000302490 | - | 13 | 14 | 108_109 | 372 | 402.0 | Nucleotide binding | NADP |
| Hgene | KCNAB1 | chr3:156249286 | chr1:32502511 | ENST00000302490 | - | 13 | 14 | 240_241 | 372 | 402.0 | Nucleotide binding | NADP |
| Hgene | KCNAB1 | chr3:156249286 | chr1:32502511 | ENST00000302490 | - | 13 | 14 | 295_300 | 372 | 402.0 | Nucleotide binding | NADP |
| Hgene | KCNAB1 | chr3:156249286 | chr1:32502511 | ENST00000471742 | - | 13 | 14 | 108_109 | 379 | 409.0 | Nucleotide binding | NADP |
| Hgene | KCNAB1 | chr3:156249286 | chr1:32502511 | ENST00000471742 | - | 13 | 14 | 240_241 | 379 | 409.0 | Nucleotide binding | NADP |
| Hgene | KCNAB1 | chr3:156249286 | chr1:32502511 | ENST00000471742 | - | 13 | 14 | 295_300 | 379 | 409.0 | Nucleotide binding | NADP |
| Hgene | KCNAB1 | chr3:156249286 | chr1:32502511 | ENST00000471742 | - | 13 | 14 | 375_381 | 379 | 409.0 | Nucleotide binding | NADP |
| Hgene | KCNAB1 | chr3:156249286 | chr1:32502511 | ENST00000490337 | - | 13 | 14 | 108_109 | 390 | 420.0 | Nucleotide binding | NADP |
| Hgene | KCNAB1 | chr3:156249286 | chr1:32502511 | ENST00000490337 | - | 13 | 14 | 240_241 | 390 | 420.0 | Nucleotide binding | NADP |
| Hgene | KCNAB1 | chr3:156249286 | chr1:32502511 | ENST00000490337 | - | 13 | 14 | 295_300 | 390 | 420.0 | Nucleotide binding | NADP |
| Hgene | KCNAB1 | chr3:156249286 | chr1:32502511 | ENST00000490337 | - | 13 | 14 | 375_381 | 390 | 420.0 | Nucleotide binding | NADP |
| Tgene | KHDRBS1 | chr3:156249286 | chr1:32502511 | ENST00000327300 | 3 | 9 | 282_292 | 257 | 444.0 | Compositional bias | DMA/Gly-rich | |
| Tgene | KHDRBS1 | chr3:156249286 | chr1:32502511 | ENST00000327300 | 3 | 9 | 295_301 | 257 | 444.0 | Compositional bias | Pro-rich | |
| Tgene | KHDRBS1 | chr3:156249286 | chr1:32502511 | ENST00000327300 | 3 | 9 | 302_332 | 257 | 444.0 | Compositional bias | Arg/Gly-rich | |
| Tgene | KHDRBS1 | chr3:156249286 | chr1:32502511 | ENST00000327300 | 3 | 9 | 334_363 | 257 | 444.0 | Compositional bias | Pro-rich | |
| Tgene | KHDRBS1 | chr3:156249286 | chr1:32502511 | ENST00000492989 | 2 | 8 | 282_292 | 218 | 405.0 | Compositional bias | DMA/Gly-rich | |
| Tgene | KHDRBS1 | chr3:156249286 | chr1:32502511 | ENST00000492989 | 2 | 8 | 295_301 | 218 | 405.0 | Compositional bias | Pro-rich | |
| Tgene | KHDRBS1 | chr3:156249286 | chr1:32502511 | ENST00000492989 | 2 | 8 | 302_332 | 218 | 405.0 | Compositional bias | Arg/Gly-rich | |
| Tgene | KHDRBS1 | chr3:156249286 | chr1:32502511 | ENST00000492989 | 2 | 8 | 334_363 | 218 | 405.0 | Compositional bias | Pro-rich |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | KCNAB1 | chr3:156249286 | chr1:32502511 | ENST00000302490 | - | 13 | 14 | 375_381 | 372 | 402.0 | Nucleotide binding | NADP |
| Tgene | KHDRBS1 | chr3:156249286 | chr1:32502511 | ENST00000327300 | 3 | 9 | 34_41 | 257 | 444.0 | Compositional bias | Pro-rich | |
| Tgene | KHDRBS1 | chr3:156249286 | chr1:32502511 | ENST00000327300 | 3 | 9 | 44_55 | 257 | 444.0 | Compositional bias | DMA/Gly-rich | |
| Tgene | KHDRBS1 | chr3:156249286 | chr1:32502511 | ENST00000327300 | 3 | 9 | 59_89 | 257 | 444.0 | Compositional bias | Pro-rich | |
| Tgene | KHDRBS1 | chr3:156249286 | chr1:32502511 | ENST00000492989 | 2 | 8 | 34_41 | 218 | 405.0 | Compositional bias | Pro-rich | |
| Tgene | KHDRBS1 | chr3:156249286 | chr1:32502511 | ENST00000492989 | 2 | 8 | 44_55 | 218 | 405.0 | Compositional bias | DMA/Gly-rich | |
| Tgene | KHDRBS1 | chr3:156249286 | chr1:32502511 | ENST00000492989 | 2 | 8 | 59_89 | 218 | 405.0 | Compositional bias | Pro-rich | |
| Tgene | KHDRBS1 | chr3:156249286 | chr1:32502511 | ENST00000327300 | 3 | 9 | 171_197 | 257 | 444.0 | Domain | KH | |
| Tgene | KHDRBS1 | chr3:156249286 | chr1:32502511 | ENST00000492989 | 2 | 8 | 171_197 | 218 | 405.0 | Domain | KH | |
| Tgene | KHDRBS1 | chr3:156249286 | chr1:32502511 | ENST00000327300 | 3 | 9 | 100_260 | 257 | 444.0 | Region | Involved in homodimerization | |
| Tgene | KHDRBS1 | chr3:156249286 | chr1:32502511 | ENST00000492989 | 2 | 8 | 100_260 | 218 | 405.0 | Region | Involved in homodimerization |
Top |
Fusion Gene Sequence for KCNAB1-KHDRBS1 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >41342_41342_1_KCNAB1-KHDRBS1_KCNAB1_chr3_156249286_ENST00000302490_KHDRBS1_chr1_32502511_ENST00000327300_length(transcript)=3806nt_BP=1987nt AGTTCCAACTGTTGTGCTGCAGCAGATTTGGTCTGAGTCTGGGCAGAGCCCCTTTTTATGGAGCAACCCTCCCGATTTGTTACAGCAAGA GCCTGGTGCAAGTGATCTTTGCCTTCCCATCGCACACAAAGGCAAAGAGCTCCTTAAAGGGACCTGCCTGCTCCTCCGTGCAGCTGCAAA CCCGTGTCGGTGCAGGCTCAGCAACCCAGGCTTTCGCTCGAGAATGAGTGGGGAGGGGTCTTGAGGAGCCCCCACGTCCTCCCGGAGCGG AGGCGGGAGGGAGCTGACAGCCGGAGTGGATGGAGACCACCAGTCCGAGGGGACCCGCTTGCGATTAGCTGCTTCTTGGGATGCTCCGCG CGCCAGTTGAGCATCCTGCCTGTGGTCCAGGGGATGCTGGCGGCGCATCTTGCGGTAAGCCTGCGCTGATTTGAGCCATCGCCCAGAAGC GAGCCCGCATTCAGACACCGCGGACCGGCTCCGCGAAGGGCTGCGTGCAGTCACCATGAATGCTCAGCTCTAGGCTTCTGTAAAAACCTC CCCCACTGCATGGATTTGGCTGTTTAATCAGCCGAGATTACAGCCCGGGAAGATTTGCAAACCTGATTCTCCCTCCAGCTGCTGTGACAA CTTCGGGGCCGTCTGTTTACTTCTCTGGGTCCCCGGGGACTCTTTCCTGCATCGCAGCCCCTTGTGCCACTTTGTGGGCTTGGCTCGAAA GTCAGCCGCCAGGACTCCTCTGACGGCATCCCCAGGAAGGGGGAGCAAGGAGGGCTTAAAAGAAAAGCAGAAATCCCCGCAACTGCTCAA CCTCTCGTGACTGCCTGTGTTCTGGGGTTCTGAGAGGGACCGTGCGCTGCCTGGGGAAGCAATGCAAGTCTCCATAGCCTGCACAGAGCA CAATTTGAAGAGTCGGAATGGTGAGGACCGACTTCTGAGCAAGCAGAGCTCCACCGCCCCCAATGTGGTGAACGCAGCCCGGGCCAAATT CCGCACGGTCGCTATCATCGCGCGCAGCCTGGGGACGTTCACGCCTCAGCATCACATTTCTCTCAAAGAGTCCACCGCAAAGCAGACTGG CATGAAATATAGGAATCTTGGAAAATCAGGACTCAGAGTTTCTTGCTTGGGTCTTGGAACATGGGTGACATTTGGAGGTCAAATTTCAGA TGAGGTTGCTGAACGGCTGATGACCATCGCCTATGAAAGTGGTGTTAACCTCTTTGATACTGCCGAAGTCTATGCTGCTGGAAAGGCTGA AGTGATTCTGGGGAGCATCATCAAGAAGAAAGGCTGGAGGAGGTCCAGTCTGGTCATAACAACCAAACTCTACTGGGGTGGAAAAGCTGA AACAGAAAGAGGGCTGTCAAGAAAGCATATTATTGAAGGATTGAAGGGCTCCCTCCAGAGGCTGCAGCTCGAGTATGTGGATGTGGTCTT TGCAAATCGACCGGACAGTAACACTCCCATGGAAGAAATTGTCCGAGCCATGACACATGTGATAAACCAAGGCATGGCGATGTACTGGGG CACCTCGAGATGGAGTGCTATGGAGATCATGGAAGCCTATTCTGTAGCAAGACAGTTCAATATGATCCCACCGGTCTGTGAACAAGCTGA GTACCATCTTTTCCAGAGAGAGAAAGTGGAGGTCCAGCTGCCAGAGCTCTACCACAAAATAGGTGTTGGCGCAATGACATGGTCTCCACT TGCCTGTGGAATCATCTCAGGAAAATACGGAAACGGGGTGCCTGAAAGTTCCAGGGCTTCACTGAAGTGCTACCAGTGGTTGAAAGAAAG AATTGTAAGTGAAGAAGGGAGAAAACAGCAAAACAAGCTAAAAGACCTTTCCCCAATTGCGGAGCGTCTGGGATGCACACTACCTCAGCT AGCTGTTGCGTGGTGCCTGAGAAATGAAGGTGTGAGTTCTGTGCTCCTGGGATCATCCACTCCTGAACAACTCATTGAAAACCTTGGTGC CATTCAGGATATGATGGATGATATCTGTCAGGAGCAATTTCTAGAGCTGTCCTACTTGAATGGAGTACCTGAACCCTCTCGTGGACGTGG GGTGCCAGTGAGAGGCCGGGGAGCTGCACCTCCTCCACCACCTGTTCCCAGGGGCCGTGGTGTTGGACCACCTCGGGGGGCTTTGGTACG TGGTACACCAGTAAGGGGAGCCATCACCAGAGGTGCCACTGTGACTCGAGGCGTGCCACCCCCACCTACTGTGAGGGGTGCTCCAGCACC AAGAGCACGGACAGCGGGCATCCAGAGGATACCTTTGCCTCCACCTCCTGCACCAGAAACATATGAAGAATATGGATATGATGATACATA CGCAGAACAAAGTTACGAAGGCTACGAAGGCTATTACAGCCAGAGTCAAGGGGACTCAGAATATTATGACTATGGACATGGGGAGGTTCA AGATTCTTATGAAGCTTATGGCCAGGACGACTGGAATGGGACCAGGCCGTCGCTGAAGGCCCCTCCTGCTAGGCCAGTGAAGGGAGCATA CAGAGAGCACCCATATGGACGTTATTAAAAACAAACATGAGGGGAAAATATCAGTTATGAGCAAAGTTGTTACTGATTTCTTGTATCTCC CAGGATTCCTGTTGCTTTACCCACAACAGACAAGTAATTGTCTAAGTGTTTTTCTTCGTGGTCCCCTTCTTCTCCCCACCTTATTCCATT CTTAACTCTGCATTCTGGCTTCTGTATGTAGTATTTTAAAATGAGTTAAAATAGATTTAGGAATATTGAATTAATTTTTTAAGTGTGTAG ATGCTTTTTTCTTTGTTGTTTAAATATAAACAGAAGTGTACCTTTTATAATAAAAAAAAGAAGTTGAGTAAAAAAAAAAAACACACAAAC CTGTTAGTTTCAAAAATGACATTGCTTGCTTAAAGGTTCTGAAGTAAAGGCTTGTTAAGTTTCTCTTAGTTTTGATTTGAGGCATCCCGT AAAGTTGTAGTTGCAGAATCCCAAACTAGGCTACATTTCAAAATTCAGGGCTGTTTAAGATTTAAAATCACAAACATTAACGGCAGTAGG CACCACCATGTAAAAGTGAGCTCAGACGTCTCTAAAAAATGTTTCCTTTATAAAAGCACATGGCGGTTGAATCTTAAGGTTAAATTTTAA TATGAAAGATCCTCATGAATTAAATAGTTGATGCAATTTTTAACGTTAATTGATATAAAAAAAAAAACAACAAAATTAGGCTTGTAAAAC TGACTTTTTCATTACGTGGGTTTTGAAATCTAGCCCCAGACATACTGTGTTGAGAGATACTTAGAGGGAGGGAGTAGGTTTTGAAGAGGT TGATGGTGGTGGGGAGGGAAGGCCTCCTGAATTGAGTTTGATGCAGAGCTTTTTAGCCATGAAGAATCTTTCAGTCATAGTACTAATAAT TAAATTTTCAGTATTTAAAAAGACAAAGTATTTTGTCCATTTGAGATTCTGCACTCCATGAAAAGTTCACTTGGACGCTGGGGCCAAAAG CTGTTGATTTTCTTAAGTTGACGGTTGTCAATATATCGAACTGTTCCCAAGTTAGTCAAGTATGTCTCAACACTAGCATGATATAAAAAG GGACACTGCAGCTGAATGAAAAAGGAATCAAAATCCACTTTGTACATAAGTTAAAGTCCTAATTGGATTTGTACCGTCCTCCCATTTTGT TCTCGGAAGATTAAATGCTACATGTGTAAGTCTGCCTAAATAGGTAGCTTAAACTTATGTCAAAATGTCTGCAGCAGTTTGTCAATAAAG >41342_41342_1_KCNAB1-KHDRBS1_KCNAB1_chr3_156249286_ENST00000302490_KHDRBS1_chr1_32502511_ENST00000327300_length(amino acids)=575AA_BP=388 MPVFWGSERDRALPGEAMQVSIACTEHNLKSRNGEDRLLSKQSSTAPNVVNAARAKFRTVAIIARSLGTFTPQHHISLKESTAKQTGMKY RNLGKSGLRVSCLGLGTWVTFGGQISDEVAERLMTIAYESGVNLFDTAEVYAAGKAEVILGSIIKKKGWRRSSLVITTKLYWGGKAETER GLSRKHIIEGLKGSLQRLQLEYVDVVFANRPDSNTPMEEIVRAMTHVINQGMAMYWGTSRWSAMEIMEAYSVARQFNMIPPVCEQAEYHL FQREKVEVQLPELYHKIGVGAMTWSPLACGIISGKYGNGVPESSRASLKCYQWLKERIVSEEGRKQQNKLKDLSPIAERLGCTLPQLAVA WCLRNEGVSSVLLGSSTPEQLIENLGAIQDMMDDICQEQFLELSYLNGVPEPSRGRGVPVRGRGAAPPPPPVPRGRGVGPPRGALVRGTP VRGAITRGATVTRGVPPPPTVRGAPAPRARTAGIQRIPLPPPPAPETYEEYGYDDTYAEQSYEGYEGYYSQSQGDSEYYDYGHGEVQDSY -------------------------------------------------------------- >41342_41342_2_KCNAB1-KHDRBS1_KCNAB1_chr3_156249286_ENST00000389634_KHDRBS1_chr1_32502511_ENST00000327300_length(transcript)=2848nt_BP=1029nt ATGCAAGTCTCCATAGCCTGCACAGAGCACAATTTGAAGAGTCGGAATGGTGAGGACCGACTTCTGAGCAAGCAGAGCTCCACCGCCCCC AATGTGGTGAACGCAGCCCGGGCCAAATTCCGCACGGTCGCTATCATCGCGCGCAGCCTGGGGACGTTCACGCCTCAGCATCACATTTCT CTCAAAGAGTCCACCGCAAAGCAGACTGGCATGAAATATAGGAATCTTGGAAAATCAGGACTCAGAGTTTCTTGCTTGGGTCTTGGAACA TGGGTGACATTTGGAGGTCAAATTTCAGATGAGGTTGCTGAACGGCTGATGACCATCGCCTATGAAAGTGGTGTTAACCTCTTTGATACT GCCGAAGTCTATGCTGCTGGAAAGGCTGAAGTGATTCTGGGGAGCATCATCAAGAAGAAAGGCTGGAGGAGGTCCAGTCTGGTCATAACA ACCAAACTCTACTGGGGTGGAAAAGCTGAAACAGAAAGAGGGCTGTCAAGAAAGCATATTATTGAAGAAATTGTCCGAGCCATGACACAT GTGATAAACCAAGGCATGGCGATGTACTGGGGCACCTCGAGATGGAGTGCTATGGAGATCATGGAAGCCTATTCTGTAGCAAGACAGTTC AATATGATCCCACCGGTCTGTGAACAAGCTGAGTACCATCTTTTCCAGAGAGAGAAAGTGGAGGTCCAGCTGCCAGAGCTCTACCACAAA ATAGGTGTTGGCGCAATGACATGGTCTCCACTTGCCTGTGGAATCATCTCAGGAAAATACGGAAACGGGGTGCCTGAAAGTTCCAGGGCT TCACTGAAGTGCTACCAGTGGTTGAAAGAAAGAATTGTAAGTGAAGAAGGGAGAAAACAGCAAAACAAGCTAAAAGACCTTTCCCCAATT GCGGAGCGTCTGGGATGCACACTACCTCAGCTAGCTGTTGCGTGGTGCCTGAGAAATGAAGGTGTGAGTTCTGTGCTCCTGGGATCATCC ACTCCTGAACAACTCATTGAAAACCTTGGTGCCATTCAGGATATGATGGATGATATCTGTCAGGAGCAATTTCTAGAGCTGTCCTACTTG AATGGAGTACCTGAACCCTCTCGTGGACGTGGGGTGCCAGTGAGAGGCCGGGGAGCTGCACCTCCTCCACCACCTGTTCCCAGGGGCCGT GGTGTTGGACCACCTCGGGGGGCTTTGGTACGTGGTACACCAGTAAGGGGAGCCATCACCAGAGGTGCCACTGTGACTCGAGGCGTGCCA CCCCCACCTACTGTGAGGGGTGCTCCAGCACCAAGAGCACGGACAGCGGGCATCCAGAGGATACCTTTGCCTCCACCTCCTGCACCAGAA ACATATGAAGAATATGGATATGATGATACATACGCAGAACAAAGTTACGAAGGCTACGAAGGCTATTACAGCCAGAGTCAAGGGGACTCA GAATATTATGACTATGGACATGGGGAGGTTCAAGATTCTTATGAAGCTTATGGCCAGGACGACTGGAATGGGACCAGGCCGTCGCTGAAG GCCCCTCCTGCTAGGCCAGTGAAGGGAGCATACAGAGAGCACCCATATGGACGTTATTAAAAACAAACATGAGGGGAAAATATCAGTTAT GAGCAAAGTTGTTACTGATTTCTTGTATCTCCCAGGATTCCTGTTGCTTTACCCACAACAGACAAGTAATTGTCTAAGTGTTTTTCTTCG TGGTCCCCTTCTTCTCCCCACCTTATTCCATTCTTAACTCTGCATTCTGGCTTCTGTATGTAGTATTTTAAAATGAGTTAAAATAGATTT AGGAATATTGAATTAATTTTTTAAGTGTGTAGATGCTTTTTTCTTTGTTGTTTAAATATAAACAGAAGTGTACCTTTTATAATAAAAAAA AGAAGTTGAGTAAAAAAAAAAAACACACAAACCTGTTAGTTTCAAAAATGACATTGCTTGCTTAAAGGTTCTGAAGTAAAGGCTTGTTAA GTTTCTCTTAGTTTTGATTTGAGGCATCCCGTAAAGTTGTAGTTGCAGAATCCCAAACTAGGCTACATTTCAAAATTCAGGGCTGTTTAA GATTTAAAATCACAAACATTAACGGCAGTAGGCACCACCATGTAAAAGTGAGCTCAGACGTCTCTAAAAAATGTTTCCTTTATAAAAGCA CATGGCGGTTGAATCTTAAGGTTAAATTTTAATATGAAAGATCCTCATGAATTAAATAGTTGATGCAATTTTTAACGTTAATTGATATAA AAAAAAAAACAACAAAATTAGGCTTGTAAAACTGACTTTTTCATTACGTGGGTTTTGAAATCTAGCCCCAGACATACTGTGTTGAGAGAT ACTTAGAGGGAGGGAGTAGGTTTTGAAGAGGTTGATGGTGGTGGGGAGGGAAGGCCTCCTGAATTGAGTTTGATGCAGAGCTTTTTAGCC ATGAAGAATCTTTCAGTCATAGTACTAATAATTAAATTTTCAGTATTTAAAAAGACAAAGTATTTTGTCCATTTGAGATTCTGCACTCCA TGAAAAGTTCACTTGGACGCTGGGGCCAAAAGCTGTTGATTTTCTTAAGTTGACGGTTGTCAATATATCGAACTGTTCCCAAGTTAGTCA AGTATGTCTCAACACTAGCATGATATAAAAAGGGACACTGCAGCTGAATGAAAAAGGAATCAAAATCCACTTTGTACATAAGTTAAAGTC CTAATTGGATTTGTACCGTCCTCCCATTTTGTTCTCGGAAGATTAAATGCTACATGTGTAAGTCTGCCTAAATAGGTAGCTTAAACTTAT >41342_41342_2_KCNAB1-KHDRBS1_KCNAB1_chr3_156249286_ENST00000389634_KHDRBS1_chr1_32502511_ENST00000327300_length(amino acids)=529AA_BP=342 MQVSIACTEHNLKSRNGEDRLLSKQSSTAPNVVNAARAKFRTVAIIARSLGTFTPQHHISLKESTAKQTGMKYRNLGKSGLRVSCLGLGT WVTFGGQISDEVAERLMTIAYESGVNLFDTAEVYAAGKAEVILGSIIKKKGWRRSSLVITTKLYWGGKAETERGLSRKHIIEEIVRAMTH VINQGMAMYWGTSRWSAMEIMEAYSVARQFNMIPPVCEQAEYHLFQREKVEVQLPELYHKIGVGAMTWSPLACGIISGKYGNGVPESSRA SLKCYQWLKERIVSEEGRKQQNKLKDLSPIAERLGCTLPQLAVAWCLRNEGVSSVLLGSSTPEQLIENLGAIQDMMDDICQEQFLELSYL NGVPEPSRGRGVPVRGRGAAPPPPPVPRGRGVGPPRGALVRGTPVRGAITRGATVTRGVPPPPTVRGAPAPRARTAGIQRIPLPPPPAPE -------------------------------------------------------------- >41342_41342_3_KCNAB1-KHDRBS1_KCNAB1_chr3_156249286_ENST00000389636_KHDRBS1_chr1_32502511_ENST00000327300_length(transcript)=2936nt_BP=1117nt AGTGACTTCCAGTCTTCTCTGAAAGATCTCCACGATGCTGGCAGCCCGGACAGGGGCAGCGGGGAGTCAGATCTCAGAGGAGAACACCAA GTTAAGGAGACAGTCTGGGTTTTCTGTAGCAGGGAAAGACAAATCTCCCAAGAAAGCCTCAGAAAACGCTAAAGACAGCAGCCTTAGTCC CTCAGGGGAAAGCCAGCTCAGGGCGCGTCAACTGGCTCTGCTGCGCGAAGTGGAGATGAACTGGTACCTAAAGCTCTGCGACCTGTCCAG CGAGCACACCACCGTCTGCACCACAGGCATGCCGCACAGGAATCTTGGAAAATCAGGACTCAGAGTTTCTTGCTTGGGTCTTGGAACATG GGTGACATTTGGAGGTCAAATTTCAGATGAGGTTGCTGAACGGCTGATGACCATCGCCTATGAAAGTGGTGTTAACCTCTTTGATACTGC CGAAGTCTATGCTGCTGGAAAGGCTGAAGTGATTCTGGGGAGCATCATCAAGAAGAAAGGCTGGAGGAGGTCCAGTCTGGTCATAACAAC CAAACTCTACTGGGGTGGAAAAGCTGAAACAGAAAGAGGGCTGTCAAGAAAGCATATTATTGAAGAAATTGTCCGAGCCATGACACATGT GATAAACCAAGGCATGGCGATGTACTGGGGCACCTCGAGATGGAGTGCTATGGAGATCATGGAAGCCTATTCTGTAGCAAGACAGTTCAA TATGATCCCACCGGTCTGTGAACAAGCTGAGTACCATCTTTTCCAGAGAGAGAAAGTGGAGGTCCAGCTGCCAGAGCTCTACCACAAAAT AGGTGTTGGCGCAATGACATGGTCTCCACTTGCCTGTGGAATCATCTCAGGAAAATACGGAAACGGGGTGCCTGAAAGTTCCAGGGCTTC ACTGAAGTGCTACCAGTGGTTGAAAGAAAGAATTGTAAGTGAAGAAGGGAGAAAACAGCAAAACAAGCTAAAAGACCTTTCCCCAATTGC GGAGCGTCTGGGATGCACACTACCTCAGCTAGCTGTTGCGTGGTGCCTGAGAAATGAAGGTGTGAGTTCTGTGCTCCTGGGATCATCCAC TCCTGAACAACTCATTGAAAACCTTGGTGCCATTCAGGATATGATGGATGATATCTGTCAGGAGCAATTTCTAGAGCTGTCCTACTTGAA TGGAGTACCTGAACCCTCTCGTGGACGTGGGGTGCCAGTGAGAGGCCGGGGAGCTGCACCTCCTCCACCACCTGTTCCCAGGGGCCGTGG TGTTGGACCACCTCGGGGGGCTTTGGTACGTGGTACACCAGTAAGGGGAGCCATCACCAGAGGTGCCACTGTGACTCGAGGCGTGCCACC CCCACCTACTGTGAGGGGTGCTCCAGCACCAAGAGCACGGACAGCGGGCATCCAGAGGATACCTTTGCCTCCACCTCCTGCACCAGAAAC ATATGAAGAATATGGATATGATGATACATACGCAGAACAAAGTTACGAAGGCTACGAAGGCTATTACAGCCAGAGTCAAGGGGACTCAGA ATATTATGACTATGGACATGGGGAGGTTCAAGATTCTTATGAAGCTTATGGCCAGGACGACTGGAATGGGACCAGGCCGTCGCTGAAGGC CCCTCCTGCTAGGCCAGTGAAGGGAGCATACAGAGAGCACCCATATGGACGTTATTAAAAACAAACATGAGGGGAAAATATCAGTTATGA GCAAAGTTGTTACTGATTTCTTGTATCTCCCAGGATTCCTGTTGCTTTACCCACAACAGACAAGTAATTGTCTAAGTGTTTTTCTTCGTG GTCCCCTTCTTCTCCCCACCTTATTCCATTCTTAACTCTGCATTCTGGCTTCTGTATGTAGTATTTTAAAATGAGTTAAAATAGATTTAG GAATATTGAATTAATTTTTTAAGTGTGTAGATGCTTTTTTCTTTGTTGTTTAAATATAAACAGAAGTGTACCTTTTATAATAAAAAAAAG AAGTTGAGTAAAAAAAAAAAACACACAAACCTGTTAGTTTCAAAAATGACATTGCTTGCTTAAAGGTTCTGAAGTAAAGGCTTGTTAAGT TTCTCTTAGTTTTGATTTGAGGCATCCCGTAAAGTTGTAGTTGCAGAATCCCAAACTAGGCTACATTTCAAAATTCAGGGCTGTTTAAGA TTTAAAATCACAAACATTAACGGCAGTAGGCACCACCATGTAAAAGTGAGCTCAGACGTCTCTAAAAAATGTTTCCTTTATAAAAGCACA TGGCGGTTGAATCTTAAGGTTAAATTTTAATATGAAAGATCCTCATGAATTAAATAGTTGATGCAATTTTTAACGTTAATTGATATAAAA AAAAAAACAACAAAATTAGGCTTGTAAAACTGACTTTTTCATTACGTGGGTTTTGAAATCTAGCCCCAGACATACTGTGTTGAGAGATAC TTAGAGGGAGGGAGTAGGTTTTGAAGAGGTTGATGGTGGTGGGGAGGGAAGGCCTCCTGAATTGAGTTTGATGCAGAGCTTTTTAGCCAT GAAGAATCTTTCAGTCATAGTACTAATAATTAAATTTTCAGTATTTAAAAAGACAAAGTATTTTGTCCATTTGAGATTCTGCACTCCATG AAAAGTTCACTTGGACGCTGGGGCCAAAAGCTGTTGATTTTCTTAAGTTGACGGTTGTCAATATATCGAACTGTTCCCAAGTTAGTCAAG TATGTCTCAACACTAGCATGATATAAAAAGGGACACTGCAGCTGAATGAAAAAGGAATCAAAATCCACTTTGTACATAAGTTAAAGTCCT AATTGGATTTGTACCGTCCTCCCATTTTGTTCTCGGAAGATTAAATGCTACATGTGTAAGTCTGCCTAAATAGGTAGCTTAAACTTATGT >41342_41342_3_KCNAB1-KHDRBS1_KCNAB1_chr3_156249286_ENST00000389636_KHDRBS1_chr1_32502511_ENST00000327300_length(amino acids)=547AA_BP=360 MLAARTGAAGSQISEENTKLRRQSGFSVAGKDKSPKKASENAKDSSLSPSGESQLRARQLALLREVEMNWYLKLCDLSSEHTTVCTTGMP HRNLGKSGLRVSCLGLGTWVTFGGQISDEVAERLMTIAYESGVNLFDTAEVYAAGKAEVILGSIIKKKGWRRSSLVITTKLYWGGKAETE RGLSRKHIIEEIVRAMTHVINQGMAMYWGTSRWSAMEIMEAYSVARQFNMIPPVCEQAEYHLFQREKVEVQLPELYHKIGVGAMTWSPLA CGIISGKYGNGVPESSRASLKCYQWLKERIVSEEGRKQQNKLKDLSPIAERLGCTLPQLAVAWCLRNEGVSSVLLGSSTPEQLIENLGAI QDMMDDICQEQFLELSYLNGVPEPSRGRGVPVRGRGAAPPPPPVPRGRGVGPPRGALVRGTPVRGAITRGATVTRGVPPPPTVRGAPAPR ARTAGIQRIPLPPPPAPETYEEYGYDDTYAEQSYEGYEGYYSQSQGDSEYYDYGHGEVQDSYEAYGQDDWNGTRPSLKAPPARPVKGAYR -------------------------------------------------------------- >41342_41342_4_KCNAB1-KHDRBS1_KCNAB1_chr3_156249286_ENST00000471742_KHDRBS1_chr1_32502511_ENST00000327300_length(transcript)=3173nt_BP=1354nt ATGCACAGGCCTGGGCAGGGACACACAACCAACAGCTCTGTCGGGAACTTAGGAGCCTGCTCGCCTACAAAAATGATAAGAGAGATCTAA AGAAAACAAGCTAAACCTTTTGAGGGACATACTGAAGCCAGATAACCCAAGGTATTCACAGCAAGATACAGTGAGTCTTAAAGTTAAGCA CCGTGCAATTAGCTTTGCTTCCTTGGGTTTTTGAAACATGCATCTGTATAAACCTGCCTGTGCAGACATCCCGAGCCCCAAGCTGGGTCT GCCAAAATCCAGTGAATCGGCTCTAAAATGTAGATGGCACCTAGCAGTGACCAAGACTCAGCCTCAGGCGGCCTGCAAACCTGTGAGGCC CAGTGGAGCAGCCGAACAGAAATATGTGGAAAAGTTTCTACGTGTTCATGGAATTTCGTTGCAGGAAACCACCAGAGCAGAGACGGGCAT GGCATACAGGAATCTTGGAAAATCAGGACTCAGAGTTTCTTGCTTGGGTCTTGGAACATGGGTGACATTTGGAGGTCAAATTTCAGATGA GGTTGCTGAACGGCTGATGACCATCGCCTATGAAAGTGGTGTTAACCTCTTTGATACTGCCGAAGTCTATGCTGCTGGAAAGGCTGAAGT GATTCTGGGGAGCATCATCAAGAAGAAAGGCTGGAGGAGGTCCAGTCTGGTCATAACAACCAAACTCTACTGGGGTGGAAAAGCTGAAAC AGAAAGAGGGCTGTCAAGAAAGCATATTATTGAAGGATTGAAGGGCTCCCTCCAGAGGCTGCAGCTCGAGTATGTGGATGTGGTCTTTGC AAATCGACCGGACAGTAACACTCCCATGGAAGAAATTGTCCGAGCCATGACACATGTGATAAACCAAGGCATGGCGATGTACTGGGGCAC CTCGAGATGGAGTGCTATGGAGATCATGGAAGCCTATTCTGTAGCAAGACAGTTCAATATGATCCCACCGGTCTGTGAACAAGCTGAGTA CCATCTTTTCCAGAGAGAGAAAGTGGAGGTCCAGCTGCCAGAGCTCTACCACAAAATAGGTGTTGGCGCAATGACATGGTCTCCACTTGC CTGTGGAATCATCTCAGGAAAATACGGAAACGGGGTGCCTGAAAGTTCCAGGGCTTCACTGAAGTGCTACCAGTGGTTGAAAGAAAGAAT TGTAAGTGAAGAAGGGAGAAAACAGCAAAACAAGCTAAAAGACCTTTCCCCAATTGCGGAGCGTCTGGGATGCACACTACCTCAGCTAGC TGTTGCGTGGTGCCTGAGAAATGAAGGTGTGAGTTCTGTGCTCCTGGGATCATCCACTCCTGAACAACTCATTGAAAACCTTGGTGCCAT TCAGGATATGATGGATGATATCTGTCAGGAGCAATTTCTAGAGCTGTCCTACTTGAATGGAGTACCTGAACCCTCTCGTGGACGTGGGGT GCCAGTGAGAGGCCGGGGAGCTGCACCTCCTCCACCACCTGTTCCCAGGGGCCGTGGTGTTGGACCACCTCGGGGGGCTTTGGTACGTGG TACACCAGTAAGGGGAGCCATCACCAGAGGTGCCACTGTGACTCGAGGCGTGCCACCCCCACCTACTGTGAGGGGTGCTCCAGCACCAAG AGCACGGACAGCGGGCATCCAGAGGATACCTTTGCCTCCACCTCCTGCACCAGAAACATATGAAGAATATGGATATGATGATACATACGC AGAACAAAGTTACGAAGGCTACGAAGGCTATTACAGCCAGAGTCAAGGGGACTCAGAATATTATGACTATGGACATGGGGAGGTTCAAGA TTCTTATGAAGCTTATGGCCAGGACGACTGGAATGGGACCAGGCCGTCGCTGAAGGCCCCTCCTGCTAGGCCAGTGAAGGGAGCATACAG AGAGCACCCATATGGACGTTATTAAAAACAAACATGAGGGGAAAATATCAGTTATGAGCAAAGTTGTTACTGATTTCTTGTATCTCCCAG GATTCCTGTTGCTTTACCCACAACAGACAAGTAATTGTCTAAGTGTTTTTCTTCGTGGTCCCCTTCTTCTCCCCACCTTATTCCATTCTT AACTCTGCATTCTGGCTTCTGTATGTAGTATTTTAAAATGAGTTAAAATAGATTTAGGAATATTGAATTAATTTTTTAAGTGTGTAGATG CTTTTTTCTTTGTTGTTTAAATATAAACAGAAGTGTACCTTTTATAATAAAAAAAAGAAGTTGAGTAAAAAAAAAAAACACACAAACCTG TTAGTTTCAAAAATGACATTGCTTGCTTAAAGGTTCTGAAGTAAAGGCTTGTTAAGTTTCTCTTAGTTTTGATTTGAGGCATCCCGTAAA GTTGTAGTTGCAGAATCCCAAACTAGGCTACATTTCAAAATTCAGGGCTGTTTAAGATTTAAAATCACAAACATTAACGGCAGTAGGCAC CACCATGTAAAAGTGAGCTCAGACGTCTCTAAAAAATGTTTCCTTTATAAAAGCACATGGCGGTTGAATCTTAAGGTTAAATTTTAATAT GAAAGATCCTCATGAATTAAATAGTTGATGCAATTTTTAACGTTAATTGATATAAAAAAAAAAACAACAAAATTAGGCTTGTAAAACTGA CTTTTTCATTACGTGGGTTTTGAAATCTAGCCCCAGACATACTGTGTTGAGAGATACTTAGAGGGAGGGAGTAGGTTTTGAAGAGGTTGA TGGTGGTGGGGAGGGAAGGCCTCCTGAATTGAGTTTGATGCAGAGCTTTTTAGCCATGAAGAATCTTTCAGTCATAGTACTAATAATTAA ATTTTCAGTATTTAAAAAGACAAAGTATTTTGTCCATTTGAGATTCTGCACTCCATGAAAAGTTCACTTGGACGCTGGGGCCAAAAGCTG TTGATTTTCTTAAGTTGACGGTTGTCAATATATCGAACTGTTCCCAAGTTAGTCAAGTATGTCTCAACACTAGCATGATATAAAAAGGGA CACTGCAGCTGAATGAAAAAGGAATCAAAATCCACTTTGTACATAAGTTAAAGTCCTAATTGGATTTGTACCGTCCTCCCATTTTGTTCT CGGAAGATTAAATGCTACATGTGTAAGTCTGCCTAAATAGGTAGCTTAAACTTATGTCAAAATGTCTGCAGCAGTTTGTCAATAAAGTTT >41342_41342_4_KCNAB1-KHDRBS1_KCNAB1_chr3_156249286_ENST00000471742_KHDRBS1_chr1_32502511_ENST00000327300_length(amino acids)=565AA_BP=378 MHLYKPACADIPSPKLGLPKSSESALKCRWHLAVTKTQPQAACKPVRPSGAAEQKYVEKFLRVHGISLQETTRAETGMAYRNLGKSGLRV SCLGLGTWVTFGGQISDEVAERLMTIAYESGVNLFDTAEVYAAGKAEVILGSIIKKKGWRRSSLVITTKLYWGGKAETERGLSRKHIIEG LKGSLQRLQLEYVDVVFANRPDSNTPMEEIVRAMTHVINQGMAMYWGTSRWSAMEIMEAYSVARQFNMIPPVCEQAEYHLFQREKVEVQL PELYHKIGVGAMTWSPLACGIISGKYGNGVPESSRASLKCYQWLKERIVSEEGRKQQNKLKDLSPIAERLGCTLPQLAVAWCLRNEGVSS VLLGSSTPEQLIENLGAIQDMMDDICQEQFLELSYLNGVPEPSRGRGVPVRGRGAAPPPPPVPRGRGVGPPRGALVRGTPVRGAITRGAT VTRGVPPPPTVRGAPAPRARTAGIQRIPLPPPPAPETYEEYGYDDTYAEQSYEGYEGYYSQSQGDSEYYDYGHGEVQDSYEAYGQDDWNG -------------------------------------------------------------- >41342_41342_5_KCNAB1-KHDRBS1_KCNAB1_chr3_156249286_ENST00000490337_KHDRBS1_chr1_32502511_ENST00000327300_length(transcript)=3053nt_BP=1234nt AGAAAAATCAATTCAGATTACTTTGATGACAGTGACTTCCAGTCTTCTCTGAAAGATCTCCACGATGCTGGCAGCCCGGACAGGGGCAGC GGGGAGTCAGATCTCAGAGGAGAACACCAAGTTAAGGAGACAGTCTGGGTTTTCTGTAGCAGGGAAAGACAAATCTCCCAAGAAAGCCTC AGAAAACGCTAAAGACAGCAGCCTTAGTCCCTCAGGGGAAAGCCAGCTCAGGGCGCGTCAACTGGCTCTGCTGCGCGAAGTGGAGATGAA CTGGTACCTAAAGCTCTGCGACCTGTCCAGCGAGCACACCACCGTCTGCACCACAGGCATGCCGCACAGGAATCTTGGAAAATCAGGACT CAGAGTTTCTTGCTTGGGTCTTGGAACATGGGTGACATTTGGAGGTCAAATTTCAGATGAGGTTGCTGAACGGCTGATGACCATCGCCTA TGAAAGTGGTGTTAACCTCTTTGATACTGCCGAAGTCTATGCTGCTGGAAAGGCTGAAGTGATTCTGGGGAGCATCATCAAGAAGAAAGG CTGGAGGAGGTCCAGTCTGGTCATAACAACCAAACTCTACTGGGGTGGAAAAGCTGAAACAGAAAGAGGGCTGTCAAGAAAGCATATTAT TGAAGGATTGAAGGGCTCCCTCCAGAGGCTGCAGCTCGAGTATGTGGATGTGGTCTTTGCAAATCGACCGGACAGTAACACTCCCATGGA AGAAATTGTCCGAGCCATGACACATGTGATAAACCAAGGCATGGCGATGTACTGGGGCACCTCGAGATGGAGTGCTATGGAGATCATGGA AGCCTATTCTGTAGCAAGACAGTTCAATATGATCCCACCGGTCTGTGAACAAGCTGAGTACCATCTTTTCCAGAGAGAGAAAGTGGAGGT CCAGCTGCCAGAGCTCTACCACAAAATAGGTGTTGGCGCAATGACATGGTCTCCACTTGCCTGTGGAATCATCTCAGGAAAATACGGAAA CGGGGTGCCTGAAAGTTCCAGGGCTTCACTGAAGTGCTACCAGTGGTTGAAAGAAAGAATTGTAAGTGAAGAAGGGAGAAAACAGCAAAA CAAGCTAAAAGACCTTTCCCCAATTGCGGAGCGTCTGGGATGCACACTACCTCAGCTAGCTGTTGCGTGGTGCCTGAGAAATGAAGGTGT GAGTTCTGTGCTCCTGGGATCATCCACTCCTGAACAACTCATTGAAAACCTTGGTGCCATTCAGGATATGATGGATGATATCTGTCAGGA GCAATTTCTAGAGCTGTCCTACTTGAATGGAGTACCTGAACCCTCTCGTGGACGTGGGGTGCCAGTGAGAGGCCGGGGAGCTGCACCTCC TCCACCACCTGTTCCCAGGGGCCGTGGTGTTGGACCACCTCGGGGGGCTTTGGTACGTGGTACACCAGTAAGGGGAGCCATCACCAGAGG TGCCACTGTGACTCGAGGCGTGCCACCCCCACCTACTGTGAGGGGTGCTCCAGCACCAAGAGCACGGACAGCGGGCATCCAGAGGATACC TTTGCCTCCACCTCCTGCACCAGAAACATATGAAGAATATGGATATGATGATACATACGCAGAACAAAGTTACGAAGGCTACGAAGGCTA TTACAGCCAGAGTCAAGGGGACTCAGAATATTATGACTATGGACATGGGGAGGTTCAAGATTCTTATGAAGCTTATGGCCAGGACGACTG GAATGGGACCAGGCCGTCGCTGAAGGCCCCTCCTGCTAGGCCAGTGAAGGGAGCATACAGAGAGCACCCATATGGACGTTATTAAAAACA AACATGAGGGGAAAATATCAGTTATGAGCAAAGTTGTTACTGATTTCTTGTATCTCCCAGGATTCCTGTTGCTTTACCCACAACAGACAA GTAATTGTCTAAGTGTTTTTCTTCGTGGTCCCCTTCTTCTCCCCACCTTATTCCATTCTTAACTCTGCATTCTGGCTTCTGTATGTAGTA TTTTAAAATGAGTTAAAATAGATTTAGGAATATTGAATTAATTTTTTAAGTGTGTAGATGCTTTTTTCTTTGTTGTTTAAATATAAACAG AAGTGTACCTTTTATAATAAAAAAAAGAAGTTGAGTAAAAAAAAAAAACACACAAACCTGTTAGTTTCAAAAATGACATTGCTTGCTTAA AGGTTCTGAAGTAAAGGCTTGTTAAGTTTCTCTTAGTTTTGATTTGAGGCATCCCGTAAAGTTGTAGTTGCAGAATCCCAAACTAGGCTA CATTTCAAAATTCAGGGCTGTTTAAGATTTAAAATCACAAACATTAACGGCAGTAGGCACCACCATGTAAAAGTGAGCTCAGACGTCTCT AAAAAATGTTTCCTTTATAAAAGCACATGGCGGTTGAATCTTAAGGTTAAATTTTAATATGAAAGATCCTCATGAATTAAATAGTTGATG CAATTTTTAACGTTAATTGATATAAAAAAAAAAACAACAAAATTAGGCTTGTAAAACTGACTTTTTCATTACGTGGGTTTTGAAATCTAG CCCCAGACATACTGTGTTGAGAGATACTTAGAGGGAGGGAGTAGGTTTTGAAGAGGTTGATGGTGGTGGGGAGGGAAGGCCTCCTGAATT GAGTTTGATGCAGAGCTTTTTAGCCATGAAGAATCTTTCAGTCATAGTACTAATAATTAAATTTTCAGTATTTAAAAAGACAAAGTATTT TGTCCATTTGAGATTCTGCACTCCATGAAAAGTTCACTTGGACGCTGGGGCCAAAAGCTGTTGATTTTCTTAAGTTGACGGTTGTCAATA TATCGAACTGTTCCCAAGTTAGTCAAGTATGTCTCAACACTAGCATGATATAAAAAGGGACACTGCAGCTGAATGAAAAAGGAATCAAAA TCCACTTTGTACATAAGTTAAAGTCCTAATTGGATTTGTACCGTCCTCCCATTTTGTTCTCGGAAGATTAAATGCTACATGTGTAAGTCT >41342_41342_5_KCNAB1-KHDRBS1_KCNAB1_chr3_156249286_ENST00000490337_KHDRBS1_chr1_32502511_ENST00000327300_length(amino acids)=576AA_BP=389 MLAARTGAAGSQISEENTKLRRQSGFSVAGKDKSPKKASENAKDSSLSPSGESQLRARQLALLREVEMNWYLKLCDLSSEHTTVCTTGMP HRNLGKSGLRVSCLGLGTWVTFGGQISDEVAERLMTIAYESGVNLFDTAEVYAAGKAEVILGSIIKKKGWRRSSLVITTKLYWGGKAETE RGLSRKHIIEGLKGSLQRLQLEYVDVVFANRPDSNTPMEEIVRAMTHVINQGMAMYWGTSRWSAMEIMEAYSVARQFNMIPPVCEQAEYH LFQREKVEVQLPELYHKIGVGAMTWSPLACGIISGKYGNGVPESSRASLKCYQWLKERIVSEEGRKQQNKLKDLSPIAERLGCTLPQLAV AWCLRNEGVSSVLLGSSTPEQLIENLGAIQDMMDDICQEQFLELSYLNGVPEPSRGRGVPVRGRGAAPPPPPVPRGRGVGPPRGALVRGT PVRGAITRGATVTRGVPPPPTVRGAPAPRARTAGIQRIPLPPPPAPETYEEYGYDDTYAEQSYEGYEGYYSQSQGDSEYYDYGHGEVQDS -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for KCNAB1-KHDRBS1 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
| Tgene | KHDRBS1 | chr3:156249286 | chr1:32502511 | ENST00000327300 | 3 | 9 | 351_443 | 257.0 | 444.0 | HNRNPA1 | |
| Tgene | KHDRBS1 | chr3:156249286 | chr1:32502511 | ENST00000492989 | 2 | 8 | 351_443 | 218.0 | 405.0 | HNRNPA1 | |
| Tgene | KHDRBS1 | chr3:156249286 | chr1:32502511 | ENST00000327300 | 3 | 9 | 400_420 | 257.0 | 444.0 | ZBTB7A | |
| Tgene | KHDRBS1 | chr3:156249286 | chr1:32502511 | ENST00000492989 | 2 | 8 | 400_420 | 218.0 | 405.0 | ZBTB7A |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for KCNAB1-KHDRBS1 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for KCNAB1-KHDRBS1 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
